“Paralyze resistance with persistence”, Woody Hayes
The discovery in 2002 of frequent mutation of BRAF in cancer was a milestone event in our understanding of the genetics of melanoma (1). This observation, amongst others, led to the current understanding that melanoma is a collection of genetically divergent neoplasms united by common histopathologic criteria (2). Moreover, the implicit promise of this discovery was that inhibition of oncogenic BRAF signaling would improve the treatment of patients with metastatic disease, which had largely defied the best efforts of medical oncology. However, the clinical failure of initial RAF or MEK inhibitor trials, combined with evidence that BRAF was mutated in benign nevi, led to skepticism that BRAF→MEK→ERK signaling was important for melanoma maintenance (3-6). In 2010, this early promise was finally realized when Plexxicon and Roche scientists in collaboration with a world-wide consortium of medical oncologists, described the properties of PLX-4032, a pan-RAF inhibitor that elicited striking tumor regressions in Phase I clinical trials (7, 8). Indeed, the remarkable (~80%) response rate of patients to PLX-4302 garnered considerable attention from the press and excitement in the melanoma research community (9). One of the remarkable, but unsung, aspects of PLX-4032′s success was the critical role that drug formulation played in obtaining sufficiently sustained inhibition of BRAF→MEK→ERK signaling in patients (7).
Although the response rate to PLX-4032 in Phase I was striking, it rapidly became apparent that prospects for curing patients with BRAF mutant melanomas would be limited by the twin problems of primary and acquired drug resistance (10-12). Indeed, analysis of Gleevec resistant chronic myelogenous leukemia (CML) or Tarceva resistant non-small cell lung cancer (NSCLC) suggested that the most likely resistance mechanism would be secondary mutations in oncogenic BRAF that substitute another amino acid for the “gatekeeper” threonine at position 529 (T529). Such substitutions in drug resistant CML or NSCLC replace the analogous threonine in BCR-ABL or the EGF receptor respectively with another amino acid compatible with ATP binding, hydrolysis and phosphotransferase activity but which prevents stable binding of the drug to the protein’s ATP binding site (e.g. T315I in BCR-ABL, T790M in EGFR) (13, 14). Moreover, experimental second-site substitution of threonine 529 for methionine into BRAFV600E, the most common mutationally activated form of the protein, gave rise to BRAFT529M,V600E that was highly oncogenic and resistant to multiple RAF inhibitors (15). Hence, the recent publication of a raft of papers describing mechanisms of acquired RAF inhibitor resistance are very surprising since none of these papers report the strongly predicted mechanism of resistance (10-12). Indeed, these reports indicate that there are multiple mechanisms of RAF inhibitor resistance, some of which render RAF→MEK→ERK signaling drug resistant and some of which appear to bypass a requirement for this pathway altogether. Importantly, some of these latter mechanisms may themselves be amenable to pharmacological targeting, holding out hope for new strategies to target RAF inhibitor resistant melanoma.
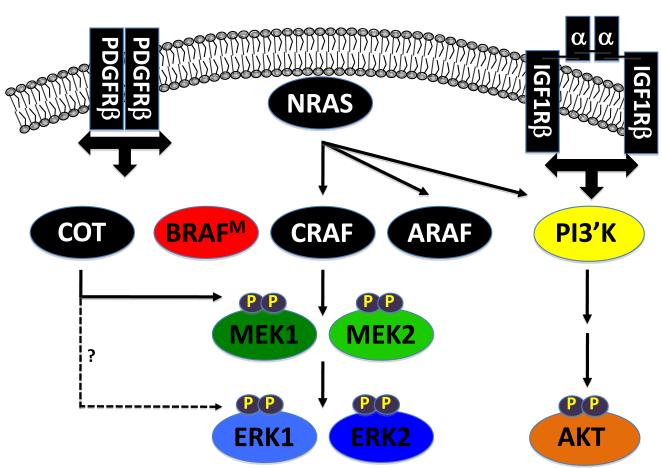
In many, but not all, cases of acquired RAF inhibitor resistance, melanoma cells display reactivation of the ERK1/2 MAP kinase pathway. Hence, some resistant melanoma cells require this pathway for proliferation such that activation of parallel signaling pathways is insufficient to compensate for inhibition of BRAF→MEK→ERK signaling and some do not. One strategy employed to identify mechanisms of drug resistance was to select cultured cells in gradually increasing concentrations of RAF inhibitor. By this strategy, Nazarian et al. using PLX-4032 (10) and Villanueva et al. using SB590885 (11) identified three general mechanisms of RAF inhibitor resistance: 1. Deregulated receptor tyrosine kinase (RTK) activity, e.g. Platelet-derived growth factor receptor β (PDGRβ) or Insulin-like growth factor 1 receptor (IGF1R); 2. Mutational activation of NRAS or; 3. Switching amongst RAF isoforms (Fig. 1)
Figure 1. Mechanisms of RAF inhibitor resistance.
Three recently published papers identified multiple mechanisms of RAF inhibitor resistance using cultured cells and patient biopsy specimens (10-12). Nazarian et al., and Villaneuva et al., identified enhanced receptor tyrosine kinase (RTK) signaling, particularly PDGFRβ and IGF1 receptor, as mechanisms of resistance. Nazarian et al., also documented mutational activation of NRAS as an additional mechanism. By ectopic over-expression of cDNAs in sensitive cells, Johanessen et al., identified nine protein kinases as having the ability to confer RAF inhibitor resistance, of which one was a control (MEK1[DD]). Most prominent were the serine kinases COT/TPL2 and CRAF, the former appearing to be responsible for examples of both primary and acquired RAF inhibitor resistance. Johanessen et al., propose that, despite its designation as MAP3K8, COT may be able to directly phosphorylate ERK1/2 in the manner of a MAP2K. Johanessen et al’s observation that over-expressed or mutationally activated CRAF can promote RAF inhibitor resistance echoed observations by Villaneuva et al. that RAF inhibitor resistant cells employ can use either CRAF or ARAF for re-activation of the MEK1/2→ERK1/2 MAP kinase signaling pathway. Proteins implicated in RAF inhibitor resistance are indicated in black.
RAF inhibitor resistance due to enhanced PDGFRβ signaling was not due to either mutation or amplification of the PDGFRB gene. Furthermore, these RAF inhibitor resistant cells displayed an mRNA expression profile characteristic of PDGF signaling and distinct from the parental sensitive cells. Using paired biopsy specimens from patients on clinical trials, Nazarian et al claim that 4/12 patients with RAF inhibitor resistant disease displayed evidence of elevated PDFGRβ expression.
In other RAF inhibitor resistant cells a separate resistance mechanism was detected, namely elevated expression of mutationally activated NRAS. Indeed, in a single patient, one PLX-4032 resistant tumor expressed NRASQ61K and another expressed NRASQ61R, underlining the extent of micro-heterogeneity displayed by melanoma. Importantly, mutational activation of NRAS and alterations in PDGFRβ signaling were not detected in the same RAF inhibitor resistant cells or tumors. Moreover, unlike the situation with PDGFRβ signaling, RAF inhibitor resistant cells expressing mutant NRAS remained sensitive to MEK1/2 inhibition and therefore dependent on RAF→MEK→ERK signaling for proliferation.
To unequivocally rule out second-site mutation of oncogenic BRAF, Nazarian et al. sequenced BRAF from nine RAF inhibitor resistant primary melanomas by “ultra-deep” (5 tumors from 5 patients, median ~127x coverage) and “deep” (5 tumors from 2 patients, median ~10x coverage) DNA sequencing. Although T1799A mutations in exon 15 encoding mutationally activated BRAFV600E were readily detected, none of these specimens displayed second-site BRAF mutations. Indeed, the authors made special efforts to independently sequence BRAF exon 13 to rule out second-site mutations affecting T529. Hence, although second site BRAF mutations might yet be identified when larger numbers of tumors or resistance to additional RAF inhibitors are analyzed, Nazarian et al rule out this simple mechanism of PLX-4032 resistance for now.
That activated PDGFRβ or mutated NRAS was important for RAF inhibitor resistance was demonstrated by over-expression of the relevant proteins, which rendered sensitive cells more resistant to PLX-4032. Furthermore, RNAi-mediated knockdown of PDGFRβ or mutated NRAS in appropriate cell lines led to growth inhibition. Interestingly, PDGFRβ-mediated RAF inhibitor resistant cells were also highly resistant to pharmacological MEK inhibition, implying that these cells have bypassed their original requirement for RAF→MEK→ERK signaling for proliferation. This appears superficially similar to the ability of elevated c-MET activation to confer Tarceva resistance in NSCLC cells expressing mutated EGFR (16). However, in that situation, elevated c-MET signaling is due to gene amplification, which is not the case with PDGFRB in RAF inhibitor resistant melanomas. By contrast, mutational activation of NRAS served to re-activate RAF→MEK→ERK signaling in the face of potent BRAF inhibition. Perhaps most surprisingly, cells with PDGFRβ-mediated RAF inhibitor resistance were also resistant to the well-characterized PDGFR inhibitor Gleevec, even when combined with the RAF inhibitor. If clinically authenticated, this result does not bode well for pharmacological targeting of PDGFR signaling in the relevant subset of RAF inhibitor resistant melanomas.
Although the manuscript from Villanueva et al., identified alternate mechanisms of RAF inhibitor resistance, a common theme of deregulated RTK signaling emerged. In this case, RAF inhibitor resistant cells displayed activation of the IGF1R signaling axis. As with PDGFRβ, enhanced IGF1R signaling was not due to mutation or amplification of the IGF1R gene. Moreover, whereas the parental melanoma cells relied exclusively on BRAF for MEK→ERK activation, resistant cells displayed elevated expression of both A- and CRAF and an ability to use either of these RAF isoforms to sustain ERK activity in the face of SB590885 treatment. Despite the importance of IGF1R, ectopic expression of IGF1 was insufficient to render cells resistant to the inhibitory effects of SB590885 on pERK. Overall, these data suggest that the resistant cells are co-dependent on both RAF→MEK→ERK and IGF1R mediated signaling events for RAF inhibitor resistance. Indeed, treatment of RAF inhibitor resistant cells with a combination of a MEK1/2 and an IGF1R inhibitor led to more potent cell killing than either agent alone.
The potential clinical significance of the work of Villanueva et al came from analysis of paired sets of pre- and post-resistance patient specimens from five patients whose BRAFV600E expressing melanomas relapsed 4-15 months after PLX-4032 treatment. One patient showed elevated IGF1R and pAKT in the resistant tumor. Moreover, a second patient’s resistant tumor displayed homozygous loss of PTEN combined with elevated pAKT that was not detected in the pre-treatment sample. These data suggest, but do not unequivocally prove, that activation of the PI3′-kinase→PDK→AKT signaling axis may diminish the response of BRAFV600E expressing melanomas to PLX-4032. Although this observation is consistent with the ability of PTEN silencing to diminish responses to pathway-targeted breast cancer and glioblastoma multiforme therapy, it remains unclear whether simple elevation of pAKT is sufficient to confer RAF inhibitor resistance in melanoma (17, 18). The results of the ongoing PLX-4032 Phase III clinical trial may ultimately shed light on whether PTEN status or pAKT is predictive of primary or acquired RAF inhibitor resistance. Regardless, given the perceived importance of PTEN silencing in the progression of BRAF mutant melanomas and the rapid development of PI3′- kinase pathway targeted therapeutics, it is likely that some combination of RAF plus PI3′- kinase pathway inhibitors will be tested clinically in the near future (19-21).
Using a complementary approach, Johanessen et al., employed cDNA expression screening to identify kinases with the ability to confer resistance to PLX-4720, a compound closely related to PLX-4032 (12, (22). By this means, ectopic expression of 9 out of 597 protein, lipid, carbohydrate or nucleotide kinases tested conferred RAF inhibitor resistance to cultured melanoma cells, of which one was a control (gain of function MEK1[DD]). In an echo of the other two papers, three of the cDNAs encoded protein tyrosine kinases: AXL, ERBB2 and FGR and the remaining five encoded serine/threonine kinases: COT/TPL2 (MAP3K8), CRAF, PAK3 and Protein kinase C epsilon (PRKCE) and eta (PRKCH). The identification of COT/TPL2 was of particular interest since, like all three RAFs, it is a MAP3K that links upstream signals such as Toll receptor activation to ERK1/2 and JNK activation leading to elevated NF-κB activity. Interestingly, COT is localized on chromosome 10 and the authors identified three RAF inhibitor naïve cell lines that contained DNA copy number gains for COT, all of which displayed de novo RAF inhibitor resistance. Unlike the other papers that focused solely on acquired resistance, these data suggest that elevated COT expression may be a mechanism for primary RAF inhibitor resistance. Whether selection for increased COT copy number in melanoma is driven by COT itself or by closely related genes such as ZEB1 remains to be determined. But, the fact that inhibition of COT in a BRAFV600E expressing melanoma cell line led to inhibition of pERK and reduced cell viability suggests the cells are co-dependent on both protein kinases for signaling and survival. Validation of the importance of COT came from identification of patient-derived primary tumors displaying acquired RAF inhibitor resistance in which COT mRNA was increased comparable to that observed in cell lines with de novo resistance. Since COT is a MAP3K that activates ERK1/2 through its action on MEK1/2, one might reasonably expect that cells with COT-mediated RAF inhibitor resistance should remain sensitive to MEK inhibition. Surprisingly, that appeared not to be the case. To explain this result, the authors propose that COT may in fact be a MAP2K and thereby able to directly activate ERK1 by dual threonine 202 and tyrosine 204 phosphorylation in a MEK1/2 independent manner. Resolution of the biochemical activity of COT as a serine-specific MAP3K or a dual-specificity threonine/tyrosine MAP2K will require additional experimentation.
Implications for future therapy of BRAF mutated cancers
The fact that no single predominant mechanism of RAF inhibitor resistance emerged from the various studies is potentially problematic but not insurmountable for clinical translation. First and foremost, a much larger number of pre-treatment and post-resistance primary patient specimens must be analyzed to determine if a predominant mechanism of RAF inhibitor resistance might emerge. Indeed, RAF inhibitor resistance mechanisms have only been identified in small numbers of PLX-4032 treated melanoma patients and it remains possible that mechanisms of primary or acquired resistance will be drug and/or disease selective. Indeed, the data beg the question as to why second-site T529 alterations were not detected in PLX-4032 resistant tumors given the strong likelihood that such second-site BRAF mutations exist in tumors. The data hint at the possibility that the action of PLX-4032 in patients may be more complex than previously thought. Although originally identified as a BRAFV600E inhibitor, more detailed characterization of PLX-4032 revealed that it is a pan-RAF inhibitor that also potently inhibits the unrelated protein tyrosine kinases SRMS (Src-related kinase lacking C-terminal regulatory tyrosine and N-terminal myristylation sites) and ACK1 (Activated CDC42 kinase 1) (7). Consequently, analysis of clinical resistance mechanisms to this and other RAF inhibitors in melanoma and other cancers expressing mutated BRAF will be important and informative.
Another factor driving resistance mechanisms may be the “mutational stew” that evolves as cancer progresses and, to that end, melanoma may be particularly heterogeneous. Stratton and colleagues reported full genome sequencing of COLO829 melanoma cells and identified ~33,000 genetic alterations, of which ~30,000 carried a signature of ultraviolet (UV) B light-induced mutation (23). Moreover, many melanoma patients are treated with systemic Dacarbazine (DTIC) or Temozolomide chemotherapy, DNA alkylating agents known to induce large numbers of mutations (24). Hence, potential RAF inhibitor resistance mechanisms may be influenced by UVB exposure during melanoma progression, prior chemotherapy or both. Hence, assuming that PLX-4032 may ultimately be approved for use in the adjuvant or neo-adjuvant setting, it will be interesting to determine mechanisms of resistance in otherwise chemo-naïve patients. Moreover, such considerations may have important implications in the choice, dose and schedule for the combined use of RAF inhibitors and other types of cancer therapy.
In many, but not all circumstances, RAF inhibitor resistant cells remain sensitive to inhibition of the MEK→ERK pathway. Hence, the use of MEK or ERK inhibitors, either alone in combination with RAF inhibition, may prove to be fruitful. Clearly, if RTK activation is a common mechanism of RAF inhibitor resistance, it may be possible to employ the growing armamentarium of RTK inhibitors such as Gleevec (inhibits ABL, PDGFR, KIT), Herceptin (inhibits ERBB2) and the various antibody and small molecule inhibitors of IGF1R and PI3′-kinase→PDK→AKT signaling that are currently in development. Moreover, validation of COT/TPL2 as a bona fide target in primary or RAF inhibitor resistant melanoma will likely spur further development of inhibitors targeting this protein kinase (25). But the apparent heterogeneity of RAF inhibitor resistance mechanisms suggests that accurate diagnostic tools will be required to determine which mechanism is involved in any given patient. Furthermore, the fact that some patients might relapse with multiple tumors with separate resistance mechanisms is a daunting prospect. It is clear, although the use of BRAF inhibitors is finally showing promise in the clinic, it remains unclear how the anti-tumor activity of such agents can be best optimized to provide durable remissions to melanoma patients. However, fulfillment of the promise of RAF pathway directed cancer therapy will doutbtless require continued persistence in both the lab and the clinic.
LITERATURE CITED
- 1.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Broekaert SM, Roy R, Okamoto I, et al. Genetic and morphologic features for melanoma classification. Pigment Cell Melanoma Res. 23:763–70. doi: 10.1111/j.1755-148X.2010.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 4.Flaherty KT, Schiller J, Schuchter LM, et al. A phase I trial of the oral, multikinase inhibitor sorafenib in combination with carboplatin and paclitaxel. Clin Cancer Res. 2008;14:4836–42. doi: 10.1158/1078-0432.CCR-07-4123. [DOI] [PubMed] [Google Scholar]
- 5.Amaravadi RK, Schuchter LM, McDermott DF, et al. Phase II Trial of Temozolomide and Sorafenib in Advanced Melanoma Patients with or without Brain Metastases. Clin Cancer Res. 2009;15:7711–8. doi: 10.1158/1078-0432.CCR-09-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adjei AA, Cohen RB, Franklin W, et al. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J Clin Oncol. 2008;26:2139–46. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bollag G, Hirth P, Tsai J, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaherty KT, Puzanov I, Kim KB, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 363:809–19. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmon A. A Drug Trial Cycle: Recovery, Relapse, Reinvention. New York Times. 2010 February 23-25th; [Google Scholar]
- 10.Nazarian R, Shi H, Wang Q, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010 doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villanueva J, Vultur A, Lee JT, et al. Acquired Resistance to BRAF Inhibitors Mediated by a RAF Kinase Switch in Melanoma Can Be Overcome by Cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johannessen CM, Boehm JS, Kim SY, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010 doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whittaker S, Kirk R, Hayward R, et al. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 16.Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 17.Nagata Y, Lan KH, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6:117–27. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Mellinghoff IK, Wang MY, Vivanco I, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–24. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 19.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–41. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–52. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dancey JE. Molecular targeting: PI3 kinase pathway. Ann Oncol. 2004;15(Suppl 4):iv233–9. doi: 10.1093/annonc/mdh932. [DOI] [PubMed] [Google Scholar]
- 22.Tsai J, Lee JT, Wang W, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.The Cancer Genome Analysis Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cusack K, Allen H, Bischoff A, et al. Identification of a selective thieno[2,3-c]pyridine inhibitor of COT kinase and TNF-alpha production. Bioorg Med Chem Lett. 2009;19:1722–5. doi: 10.1016/j.bmcl.2009.01.088. [DOI] [PubMed] [Google Scholar]