Abstract
T cells bearing αβ receptors recognize antigenic peptides bound to class I and class II glycoproteins encoded in the major histocompatibility complex (MHC). Cytotoxic and helper T cells respond respectively to peptide antigens derived from endogenous sources presented by MHC class I, and exogenous sources presented by MHC II, on antigen presenting cells. Differences in the MHC class I and class II structures and their maturation pathways have evolved to optimize antigen presentation to their respective T cells. A main focus of our laboratory is on efforts to understand molecular events in processing of antigen for presentation by MHC class II. The different stages of MHC class II—interactions with molecular chaperons involved in folding and traffic from the ER through the antigen-loading compartments, peptide exchange, and transport to the cell surface have been investigated. Through intense research on biophysical and biochemical properties of MHC class II molecules, we have learned that the conformational heterogeneity of MHC class II induced upon binding to different peptides is a key regulator in antigen presentation and epitope selection, and a determinant of the ability of MHC class II to participate in peptide association or dissociation and interaction with the peptide editor HLA-DM.
Keywords: MHC class II, Kinetics, Conformational heterogeneity, Immunodominance, H-bonds, Peptide-receptive conformation
Kinetic and conformational heterogeneity of MHC class II heterodimers
The crystallographic structure of the MHC class I and class II proteins have led many immunologists to view MHC molecules as possessing fully formed binding sites into which suitable peptides fit. In reality, however, MHC molecules are structurally unstable under physiological conditions without the bound peptides [1]. Our laboratory has long been working on the kinetics and structural variability of MHC class II proteins interacting with peptides and/or accessory proteins and chaperons in antigen processing. The overall conclusion resulting from nearly two decades of work by us and many other laboratories could be said to be that the conformation of the MHC class II molecule is an important factor in determining its ability to participate in peptide association or dissociation [2–10]. Binding of peptide to MHC class II involves several conformational changes; among these is a short-lived “peptide-receptive” conformation [11–13] to which peptide can bind with rapid and monophasic kinetics. We suggest that the conformational instability of the peptide-binding groove of MHC class II is mother nature's design of choice for serving the optimal goal of exogenous antigen capture and its presentation to CD4 T cells.
Our first observations indicating that binding of peptides to MHC class II molecules was a complex process and involved multiple kinetic steps was published in 1989 [4]. Using the pigeon cytochrome-c peptide pCytc(88–104) and I-Ek reconstituted in planar lipid bilayers on glass slides, as it was established in Harden McConnell's laboratory [14], we investigated the kinetics of peptide-MHC reactions. We reported formation of two types of IEk-pCytc peptide complexes; one complex had slow apparent association and dissociation kinetics, very similar to those reported for the chicken ovalbumin peptide and I-Ad [15], and the second complex formed and dissociated about a hundred times more rapidly. The short-lived complex was reported as a kinetic intermediate in the formation of the long-lived complex [4]. The idea of complex kinetic behavior instigated conformational complexity or heterogeneity in binding and dissociation of peptide/MHC complexes.
Work from McConnell's laboratory demonstrated that dissociation of peptides from MHC class II induced a “floppy” conformation that was different from the “compact” conformation of most purified MHC class II molecules and was detected by a simple SDS–PAGE gel assay when the samples were not boiled (Gentle SDS–PAGE, as we like to call it) [8, 16]. A follow-up of those experiments demonstrated that at low pH of endosomal compartments (5.00–6.00) a brief exposure to the appropriate peptide promoted formation of compact, SDS-resistant conformation in proportion to stably bound peptide, indicating that peptide was important in determining class II MHC structure [2, 3]. Our results also indicated that efficient generation of long-lived peptide/class II complexes involves two stages: initial conditioning of MHC class II in an acidic environment, forming a floppy MHC that enhanced the ability of class II to enter a compact conformation, upon binding to specific peptides [3]. Those data demonstrated that MHC class II molecules required peptides for proper folding and that the binding of peptide induced conformational changes in MHC II molecules and made them resistant to denaturation in the presence of SDS. The terms SDS-stable or compact MHC versus SDS-sensitive, or floppy MHC heterodimers, became standard terminologies in immunology since.
Gentle SDS–PAGE assay provided a convenient tool for exploring the MHC class II cellular trafficking and antigen processing. Different conformations of MHC class II molecules from synthesis to maturation within APCs were pioneered by Germain's and Watts’ laboratories [17, 18]. The results demonstrated striking parallels between the findings using purified molecules, and the events taking place within antigen presenting cells, as newly synthesized MHC molecules migrated as peptide-free SDS-sensitive chains, while the mature peptide loaded MHC II appeared as compact heterodimers [17]. Extension of these findings to other antigens and MHC alleles generated a wealth of knowledge and enable mapping of different steps in trafficking of MHC class II, dissociation of newly synthesized MHC II from invariant chain Ii, exchange of Ii peptide (CLIP) with antigenic peptides and the localization of all the components of antigen processing to certain high density vesicular compartment called MIIC are some of the application of this simple SDS–PAGE assay.
Further investigation toward understanding the correlations between the kinetics and structural forms of MHC class II suggested that the early kinetic form was a necessary step in preserving MHC class II viability [5]. Using soluble HLA-DR1 molecules as a model, we showed that the two kinetic and structural forms of MHC class II were linked. Insect cell-derived HLA-DR1 class II molecules showed fast occupancy with rapidly dissociating peptide while remaining sensitive to SDS-induced chain dissociation. The same DR1 molecules slowly and quantitatively formed long-lived complexes resistant to SDS-induced denaturation. Surprisingly, low-affinity interaction with peptide protects class II from denaturation at physiological temperature, a finding that helped with understanding the role of invariant chain in the intracellular behavior of class II molecules [5, 19, 20].
The biochemical mechanism of SDS stability
Despite the benefits of using the SDS stability assay, an understanding of its biochemical mechanism was lacking. To assess the mechanism of complex stability to SDS-induced MHC class II chain dissociation, we used the well-characterized HLA-DR1 molecule. Pocket 1(P1) of DR1 plays the most important role in the peptide interactions, as shown by binding studies [21] and x-ray crystal structures of DR1/HA306–318 [22] and DR1/A2 [23]. P1 is a deep pocket lined with a series of hydrophobic residues that constitute about 85% of the solvent accessible area. The P1 anchor residues, Tyr308 in HA306–318 peptide and Trp307 in A2 peptide, are almost completely buried in pocket 1. A strong preference of pocket 1 for the aromatic side chain residues (Tyr, Trp, Phe) has been reported [21, 24]. Long aliphatic side chains (Met, Leu, Ile, Val) also bind, although less efficiently [24]. In SDS–PAGE experiments with HLA-DR1, only the peptides that had aromatic or long aliphatic side chains as the P1 anchor were able to form SDS-stable complexes [25–27]. Our experiments with several HA306–318-derived peptides indicated that hydrophobic interactions between the P1 residues and the bulky P1 anchor and the resulting burial of these residues were primarily responsible for SDS stability [28]. To further explore whether the SDS sensitivity was primarily due to the exposed hydrophobic regions, we mutated residue DR1β86Gly at the bottom of P1 to Tyr, presumably reducing the depth of the pocket and the exposure of hydrophobic residues and increasing the contacts between the MHC II alpha and beta chains that form P1. In direct contrast to wild-type DR1, the peptide-free mutant DR1 existed as a stable α/β heterodimer in SDS. Moreover, the presence of a smaller hydrophobic residue, such as Ala, as P1 anchor with no contribution from any other anchor was sufficient to enhance the SDS stability of the mutant complexes, demonstrating that the basis of SDS resistance may be localized to P1 interactions [6]. Knowing that SDS primarily binds to hydrophobic regions of proteins, and that incompletely folded proteins, as intermediates in the folding pathway, are often characterized by the presence of exposed non-polar patches, it is reasonable to predict that the incompletely folded nascent MHC class II molecules should be SDS-sensitive, as shown to be the case [28].
A peptide-receptive conformation generated by dissociation of low-affinity MHC/peptide complex
The findings described above laid the foundation for further investigation of the biological significance of different conformations for MHC class II [6, 28]. Having learned that substituting P1 anchor residue of HA306–318 to alanine would convert it to a poor binding peptide with dissociation rates orders of magnitude faster, and whose complexes with HLADR1 resemble a kinetic intermediate, we took on a study to address the impacts of short-lived complexes on formation of long-lived complexes. We first used low-affinity variants of HA306–318, which lacked the principal anchor, to generate nascent DR1 molecules in correct conformations. A new fluorescence assay that enabled simultaneous detection of two different peptide complexes indicated that this nascent molecule formed a stable complex with the high affinity HA peptide at the same rate at which it was generated. Rigorous kinetic analyses indicated that the stable peptide-binding reaction had to be extremely rapid, almost spontaneous with peptide dissociation, to result in single exponential kinetic rates similar to the dissociation rate of the short-lived complex. The rate of complex formation was the same whether HA was in stoichiometric concentration or 30-fold excess relative to DR1. This apparent lack of concentration dependence could result from an extremely fast intrinsic peptide-binding rate, further evidence for the near spontaneity of this reaction. These data suggest that the MHC molecules exist in at least two different conformations with respect to their peptide-binding ability: one very receptive to binding, and the other not receptive to binding [11].
From what we discussed above, it became clear that the rate-limiting step in the formation of stable peptide/MHC complexes occurred in the generation of a receptive class II conformation [11, 12]. A receptive conformation could be created when a resident peptide dissociated. At this time, a second peptide could bind class II nearly spontaneously and stoichiometrically. However, the receptive molecule was flexible with a very short half-life, and in the absence of any free peptide, rapidly reverted to a “closed”, slow peptide-binding conformation [11]. From this understanding of events in peptide binding and the MHC class II groove flexibility a clear role for the class II invariant chain peptide, CLIP, as a surrogate short-lived peptide for shaping of MHC class II emerged. In peptide-loading compartments, CLIP dissociates, leaving a receptive groove “open” for efficient peptide binding. This is in agreement with previous findings that empty soluble DR1 molecules aggregate in the absence of peptides [19, 29], or invariant chains [30], and with the speculations on the role for the invariant chain [31].
Relevance of different MHC class II conformations to HLA-DM function
HLA-DM had been shown to have a role in enhancing of the peptide loading and or exchange of peptides. However, despite its discovery for near a decade, the mechanism of its action had remained a mystery. It was not clear how HLA-DM could dissociate some peptide/MHC complexes but not the others. Our previously described DR1βG86Y, which remained permanently in receptive or “open” form partially resembled wild-type DR1 in complex with a peptide that had tyrosine as the main anchor. The molecule was rigid and open, and thus could bind and dissociate peptides efficiently. As DM seems to be involved in class II peptide-loading processes, we considered different conformations of MHC as potential ligands for DM. By probing kinetic and conformational intermediates of the wild-type, and DR1βG86Y/peptide complexes, and examining their reactivity with DM, we proposed that DM recognizes and targets the flexible hydrophobic pocket 1 of DR1 and converts the molecule into a peptide-receptive conformation. However, a more rigid conformation, generated upon filling of pocket 1, was less susceptible to DM effects [32]. We found out that the key to this discrimination was in the recognition of differences in conformation of MHC molecules bound to different peptides. When peptides that bound to MHC molecules and induced a conformation that was rather flexible, such as CLIP, or peptides with small P1 anchor residues, the complex was recognized by HLA-DM, which catalyzed their dissociation. In contrast, when a peptide formed complexes that had compact conformation, DM did not recognize them as targets and did not interact with and hence did not dissociate them. Independent reports from other investigators in the field were in agreement with those conclusions [13, 33–38].
HLA-DM effector function
The observations described above supported the notion that DM had a recognition mechanism for its targets based on the conformation of the target [32]. However, the effector mechanism remained to be discovered. In an effort to address the effector mechanism, we generated a second mutant DR1 [39]. Hydrogen bonds (H-bonds) are known to be formed between conserved amino acids of the MHC II molecules and the backbone of the bound peptide [22, 23, 40, 41]. Wilson et al. [42] have shown that by mutating two acidic residues to amides in the core of the H-bond network in I-Ek, the kinetics of peptide exchange was enhanced at low pH. McFarland et al. reported that by mutating βHis 81 to Asn, the stability of peptide/I-Ad was reduced [43]. Saito et al. [44] demonstrated that the formation of H-bonds between βHis81 of I-Ek and peptides significantly contributed to the thermal stability of the complex. However, the possibility that H-bonds could play a role in DM effector function had not been demonstrated. We hypothesized that upon recognition of a suitable MHC II P1 pocket structure, DM causes conformational changes that result in the breaking of one or more H-bonds formed between the peptide backbone and the MHC II groove, destabilizing the bound peptide.
The mutant DR1βH81N molecule appeared functional in all respects tested, e.g. complexes of DR1βH81N-HA306–318 and DR1βH81N-CLIP were SDS stable and migrated similar to DR1wt complexes on a gel; also, DR1βH81N bound peptides with association curves similar to DR1wt. However, these complexes dissociated very rapidly and independent of peptide sequence. The rates of dissociation of these complexes were similar to the accelerated dissociation rates seen with DR1wt/peptide complexes in the presence of DM, and DM completely failed to accelerate the dissociation rate of DR1βH81N/peptide complexes. This was not due to a lack of recognition of complexes by DM, because we observed that the physical interaction of DM with DR1βH81N in solution was normal and DM could still mediate conversion DR1βH81N into a peptide-receptive conformation, allowing for quicker binding to peptide. Finally, we generated a compensatory mutant DR1βH81N/βV85H that potentially reintroduced an appropriate His-mediated H-bond with peptide (i.e. a possible target for DM), and we observed that with DR1βH81N/βV85H/peptide complexes, DM-mediated peptide dissociation from the MHC II molecule was reconstituted [39].
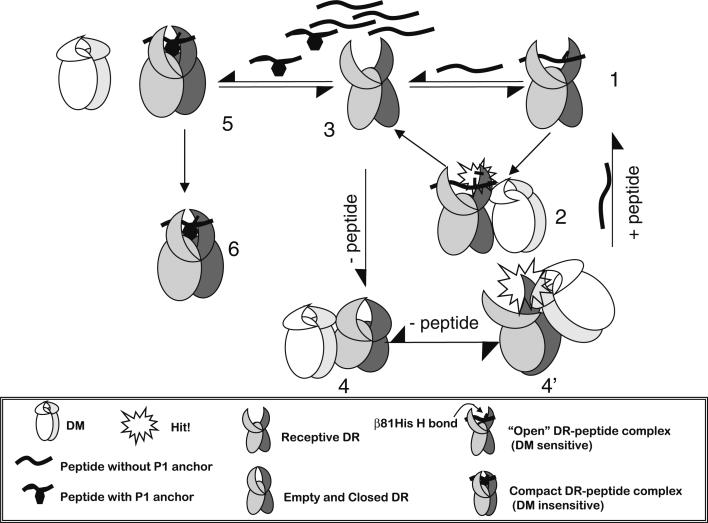
We propose that the DR1βH81N molecule with the perturbed H-bond represents a “post DM effected” transitional state; this may explain why peptides dissociated from this mutant with DM-mediated kinetics even in the absence of DM, and why the addition of DM had no further effect. We suggested that DM effects peptide/MHC II complex dissociation by a “hit-and-run” mechanism, where a transient interaction between DM and DR1 causes a conformational change in DR1 leading to the perturbation of the β81His H-bond, and this results in destabilization of bound peptide [39, 45]. It is noteworthy that DM generates but does not stabilize and maintain the peptide-receptive conformation of MHC class II in the absence of peptides (Fig. 1) [46].
Fig. 1.
A model for heterogeneity of MHC class II conformations as the basis for DM recognition its effector function. A newly synthesized MHC class II, or DR1, molecule, occupied here by a peptide that does not fill Pocket 1, is in a ‘floppy’ or ‘open,’ DM-sensitive conformation (1). DM can interact transiently with the molecule by using the proposed ‘hit-and-run’ mechanism [39, 46] and can induce local conformational changes that lead to break in the b81His hydrogen bond between the peptide and DR1, resulting in the release of peptide (2). This generates a peptide-receptive DR1 (3), upon which several events might follow. The molecule can bind another peptide that is similar to the one described above and can then go through another round of DM-mediated dissociation (steps 1 and 2). Alternatively, in the absence of peptide, the DR1 molecule might close and become inactive over time under physiological conditions (4). The empty DR1 might now be rescued by a DM ‘hit’ to generate the peptide-receptive form again (4’). Finally, if DR1 binds a peptide that fills P1, the molecule then changes to a tight, DM-insensitive conformation (5). DM cannot interact productively with this complex, and the DR1 bound to peptide is exported to the cell surface (6)
We suggest that these two mutants (DR1βH81N and DRβG86Y) reveal the basis of the recognition and the effector functions of DM, and that this two-step functionality of DM may explain some of the earlier data. Depending on sequence, various peptide side chains will fit differently into the peptide-binding groove of MHC class II molecules, resulting in slightly different peptide-MHC class II conformations. While this may result in variable kinetic stabilities of the complexes, we propose that the actual criterion and a true predictor for DM recognition are the conformation of the complex, which is independent of its intrinsic kinetic stability. If DM recognizes and interacts with the peptide/MHC complex, it mediates its effector function, which is to generate an open peptide-receptive conformation. This opening up of the groove weakens the H-bonds between MHC class II and peptide, allowing for rapid dissociation. If there is a peptide in the milieu, it can now bind rapidly, otherwise the groove of MHC class II will close, as the lifetime of this intermediate is quite short [11, 46]. The molecule stays as such until another transient interaction with DM occurs. We suspect that empty MHC class II is conformationally similar to a “DM susceptible” complex [7] (Fig. 1). This may explain why DM can also interact with empty MHC class II molecules and convert them into a receptive conformation, allowing for rapid peptide binding. Once a peptide binds that converts MHC class II into a compact DM-insensitive conformation, DM can no longer recognize and interact with the molecule and is ineffective in mediating peptide dissociation.
Similarities with Tapasin function
Intriguingly, Tapasin, a chaperon for peptide loading to MHC class I, has been shown to follow a similar mechanism for its function in selecting peptides for presentation by MHC class I [45, 47]. Both TPN and DM stabilize peptide-deficient MHC molecules. The stabilizing effect ensures that the antigen groove is maintained in a conformation receptive to capturing candidate peptides. TPN and DM act by opening the antigen-binding groove of MHC molecules, allowing for MHC to scan for a good fitting peptide at a faster rate. Equally striking, TPN and DM catalyze the exchange of peptides by disrupting conserved hydrogen bonds between peptides and MHC residues. Finally, once MHC class I and class II molecules have captured optimal peptides, both molecules undergo conformational changes that render them insensitive to further productive interactions with TPN and DM, respectively. Thus, despite differences in structure, molecular chaperones and maturation pathways, MHC class I and class II molecules seem to be subjected to similar and stringent peptide-selection processes that ultimately influence the repertoire presented at the cell surface. As such, an MHC molecule would have to bind to a large number of peptide sequences of a given protein antigen, but only a small subset of these peptides can impart conformational changes in MHC/peptide complexes that render the newly formed complexes insensitive to TPN or DM. Thus, the observation that both MHC class I and class II antigen processing systems are regulated by very similar molecular mechanisms presents a powerful argument for its validity.
Acknowledgments
SS-N wishes to specially thank Dr. Harden McConnell for his remarkable mentorship and for rightfully pointing out the importance of kinetic experiments. Funding for the research described here was from Cancer Research Institute, NIAID and NIGM. This work was supported by R01GM53549 and R01AI063764 grants to SS-N.
Footnotes
Present Address: S. Natarajan Dendreon, Seattle, WA, USA
Present Address: C.-L. Chou Program in Neuroscience, The University of Arizona, College of Pharmacy, Tucson, AZ 85721, USA
Present Address: I. Z. Hartman Department of Molecular Genetics, University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd., Dallas, TX 75390-9046, USA
Present Address: K. Narayan Laboratory of Cell Biology, NCI, NIH, Bethesda, MD 20892-4256, USA
References
- 1.Germain RN, Margulies DH. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–50. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- 2.Sadegh-Nasseri S, Germain RN. How MHC class II molecules work: peptide-dependent completion of protein folding. Immunol Today. 1992;13(2):43–6. doi: 10.1016/0167-5699(92)90131-P. [DOI] [PubMed] [Google Scholar]
- 3.Sadegh-Nasseri S, Germain RN. A role for peptide in determining MHC class II structure. Nature. 1991;353(6340):167–70. doi: 10.1038/353167a0. [DOI] [PubMed] [Google Scholar]
- 4.Sadegh-Nasseri S, McConnell HM. A kinetic intermediate in the reaction of an antigenic peptide and I-Ek. Nature. 1989;337(6204):274–6. doi: 10.1038/337274a0. [DOI] [PubMed] [Google Scholar]
- 5.Sadegh-Nasseri S, Stern LJ, Wiley DC, Germain RN. MHC class II function preserved by low-affinity peptide interactions preceding stable binding. Nature. 1994;370(6491):647–50. doi: 10.1038/370647a0. [DOI] [PubMed] [Google Scholar]
- 6.Sato AK, Zarutskie JA, Rushe MM, Lomakin A, Natarajan SK, Sadegh-Nasseri S, et al. Determinants of the peptide-induced conformational change in the human class II major histocompatibility complex protein HLA-DR1. J Biol Chem. 2000;275(3):2165–73. doi: 10.1074/jbc.275.3.2165. [DOI] [PubMed] [Google Scholar]
- 7.Carven GJ, Stern LJ. Probing the ligand-induced conformational change in HLA-DR1 by selective chemical modification and mass spectrometric mapping. Biochemistry. 2005;44(42):13625–37. doi: 10.1021/bi050972p. [DOI] [PubMed] [Google Scholar]
- 8.Dornmair K, Rothenhausler B, McConnell HM. Structural intermediates in the reactions of antigenic peptides with MHC molecules. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):409–16. doi: 10.1101/sqb.1989.054.01.050. [DOI] [PubMed] [Google Scholar]
- 9.Witt SN, McConnell HM. Formation and dissociation of short-lived class II MHC-peptide complexes. Biochemistry. 1994;33(7):1861–8. doi: 10.1021/bi00173a032. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt L, Boniface JJ, Davis MM, McConnell HM. Conformational isomers of a class II MHC-peptide complex in solution. J Mol Biol. 1999;286(1):207–18. doi: 10.1006/jmbi.1998.2463. [DOI] [PubMed] [Google Scholar]
- 11.Natarajan SK, Assadi M, Sadegh-Nasseri S. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J Immunol. 1999;162(7):4030–6. [PubMed] [Google Scholar]
- 12.Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, et al. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9(5):699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 13.Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The kinetic basis of peptide exchange catalysis by HLA-dm. Proc Natl Acad Sci USA. 2001;98(22):12450–5. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watts TH, Brian AA, Kappler JW, Marrack P, McConnell HM. Antigen presentation by supported planar membranes containing affinity-purified I-Ad. Proc Natl Acad Sci USA. 1984;81(23):7564–8. doi: 10.1073/pnas.81.23.7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buus S, Sette A, Colon SM, Jenis DM, Grey HM. Isolation and characterization of antigen-IA complexes involved in T cell recognition. Cell. 1986;47(6):1071–7. doi: 10.1016/0092-8674(86)90822-6. [DOI] [PubMed] [Google Scholar]
- 16.Dornmair K, McConnell HM. Refolding and reassembly of separate alpha and beta chains of class II molecules of the major histocompatibility complex leads to increased peptide-binding capacity. Proc Natl Acad Sci USA. 1990;87(11):4134–8. doi: 10.1073/pnas.87.11.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Germain RN, Hendrix LR. MHC class II structure, occupancy and surface expression determined by post-endoplasmic reticulum antigen binding. Nature. 1991;353(6340):134–9. doi: 10.1038/353134a0. [DOI] [PubMed] [Google Scholar]
- 18.Davidson HW, Reid PA, Lanzavecchia A, Watts C. Processed antigen binds to newly synthesized MHC class II molecules in antigen-specific b lymphocytes. Cell. 1991;67(1):105–16. doi: 10.1016/0092-8674(91)90575-j. [DOI] [PubMed] [Google Scholar]
- 19.Sadegh-Nasseri S. Peptide, invariant chain, or molecular aggregation preserves class II from functional inactivation. In: Humphreys RE, Pierce SK, editors. Antigen processing and presentation. Vol. 1. Academic Press; San Diego: 1994. pp. 170–87. [Google Scholar]
- 20.Park SJ, Sadegh-Nasseri S, Wiley DC. Invariant chain made in Escherichia coli has an exposed n-terminal segment that blocks antigen binding to HLA-DR1 and a trimeric c-terminal segment that binds empty HLA-DR1. Proc Natl Acad Sci USA. 1995;92(24):11289–93. doi: 10.1073/pnas.92.24.11289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jardetzky TS, Gorga JC, Busch R, Rothbard J, Strominger JL, Wiley DC. Peptide binding to HLA-DR1: a peptide with most residues substituted to alanine retains MHC binding. EMBO J. 1990;9(6):1797–803. doi: 10.1002/j.1460-2075.1990.tb08304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368(6468):215–21. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 23.Murthy VL, Stern LJ. The class II MHC protein HLA-DR1 in complex with an endogenous peptide: implications for the structural basis of the specificity of peptide binding. Structure. 1997;5(10):1385–96. doi: 10.1016/s0969-2126(97)00288-8. [DOI] [PubMed] [Google Scholar]
- 24.Hammer J, Valsasnini P, Tolba K, Bolin D, Higelin J, Takacs B, et al. Promiscuous and allele-specific anchors in HLA-DR-binding peptides. Cell. 1993;74(1):197–203. doi: 10.1016/0092-8674(93)90306-b. [DOI] [PubMed] [Google Scholar]
- 25.Wu S, Gorski J, Eckels DD, Newton-Nash DK. T cell recognition of MHC class II-associated peptides is independent of peptide affinity for MHC and sodium dodecyl sulfate stability of the peptide/MHC complex. Effects of conservative amino acid substitutions at anchor position 1 of influenza matrix protein19–31. J Immunol. 1996;156(10):3815–20. [PubMed] [Google Scholar]
- 26.Verreck FA, Vermeulen C, Poel AV, Jorritsma P, Amons R, Coligan JE, et al. The generation of SDS-stable HLA DR dimers is independent of efficient peptide binding. Int Immunol. 1996;8(3):397–404. doi: 10.1093/intimm/8.3.397. [DOI] [PubMed] [Google Scholar]
- 27.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, et al. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358(6389):764–8. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 28.Natarajan SK, Stern LJ, Sadegh-Nasseri S. Sodium dodecyl sulfate stability of HLA-DR1 complexes correlates with burial of hydrophobic residues in pocket 1. J Immunol. 1999;162(6):3463–70. [PubMed] [Google Scholar]
- 29.Stern LJ, Wiley DC. The human class II MHC protein HLA-DR1 assembles as empty alpha beta heterodimers in the absence of antigenic peptide. Cell. 1992;68(3):465–77. doi: 10.1016/0092-8674(92)90184-e. [DOI] [PubMed] [Google Scholar]
- 30.Germain RN, Rinker AG., Jr Peptide binding inhibits protein aggregation of invariant-chain free class II dimers and promotes surface expression of occupied molecules. Nature. 1993;363(6431):725–8. doi: 10.1038/363725a0. [DOI] [PubMed] [Google Scholar]
- 31.Romagnoli P, Germain RN. The clip region of invariant chain plays a critical role in regulating major histocompatibility complex class II folding, transport, and peptide occupancy. J Exp Med. 1994;180(3):1107–13. doi: 10.1084/jem.180.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chou CL, Sadegh-Nasseri S. HLA-dm recognizes the flexible conformation of major histocompatibility complex class II. J Exp Med. 2000;192(12):1697–706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pu Z, Lovitch SB, Bikoff EK, Unanue ER. T cells distinguish MHC-peptide complexes formed in separate vesicles and edited by H2-DM. Immunity. 2004;20(4):467–76. doi: 10.1016/s1074-7613(04)00073-1. [DOI] [PubMed] [Google Scholar]
- 34.Marin-Esteban V, Falk K, Rotzschke O. “Chemical analogues” Of HLA-DM can induce a peptide-receptive state in HLA-DR molecules. J Biol Chem. 2004;279(49):50684–90. doi: 10.1074/jbc.M407598200. [DOI] [PubMed] [Google Scholar]
- 35.Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19(2):183–92. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 36.Belmares MP, Busch R, Mellins ED, McConnell HM. Formation of two peptide/MHC II isomers is catalyzed differentially by HLA-DM. Biochemistry. 2003;42(3):838–47. doi: 10.1021/bi020466p. [DOI] [PubMed] [Google Scholar]
- 37.Stratikos E, Mosyak L, Zaller DM, Wiley DC. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-dm catalysis. J Exp Med. 2002;196(2):173–83. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doebele RC, Busch R, Scott HM, Pashine A, Mellins ED. Determination of the HLA-DM interaction site on HLA-DR molecules. Immunity. 2000;13(4):517–27. doi: 10.1016/s1074-7613(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 39.Narayan K, Chou CL, Kim A, Hartman IZ, Dalai S, Khoruzhenko S, et al. HLA-DM targets the hydrogen bond between the histidine at position beta81 and peptide to dissociate HLA-DR-peptide complexes. Nat Immunol. 2007;8(1):92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fremont DH, HenDRickson WA, Marrack P, Kappler J. Structures of an MHC class II molecule with covalently bound single peptides. Science. 1996;272(5264):1001–4. doi: 10.1126/science.272.5264.1001. [DOI] [PubMed] [Google Scholar]
- 41.Fremont DH, Monnaie D, Nelson CA, HenDRickson WA, Unanue ER. Crystal structure of I-Ak in complex with a dominant epitope of lysozyme. Immunity. 1998;8(3):305–17. doi: 10.1016/s1074-7613(00)80536-1. [DOI] [PubMed] [Google Scholar]
- 42.Wilson N, Fremont D, Marrack P, Kappler J. Mutations changing the kinetics of class II MHC peptide exchange. Immunity. 2001;14(5):513–22. doi: 10.1016/s1074-7613(01)00140-6. [DOI] [PubMed] [Google Scholar]
- 43.McFarland BJ, Beeson C, Sant AJ. Cutting edge: a single, essential hydrogen bond controls the stability of peptide-MHC class II complexes. J Immunol. 1999;163(7):3567–71. [PubMed] [Google Scholar]
- 44.Saito K, Oda M, Sarai A, Azuma T, Kozono H. Contribution of a single hydrogen bond between betahis81 of MHC class II I-E(k) and the bound peptide to the PH-dependent thermal stability. Microbiol Immunol. 2004;48(1):53–7. doi: 10.1111/j.1348-0421.2004.tb03487.x. [DOI] [PubMed] [Google Scholar]
- 45.Sadegh-Nasseri S, Chen M, Narayan K, Bouvier M. The convergent roles of tapasin and HLA-DM in antigen presentation. Trends Immunol. 2008;29(3):141–7. doi: 10.1016/j.it.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. HLA-DM mediates peptide exchange by interacting transiently and repeatedly with HLA-DR1. Mol Immunol. 2009;46(15):3157–62. doi: 10.1016/j.molimm.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26(6):1681–90. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]