Abstract
Cytotoxic and helper T cells respond to peptides derived from endogenous and exogenous sources that bind to major histocompatibility complex (MHC) class I and class II molecules and are presented on antigen-presenting cells. MHC class I and class II structures and maturation pathways have evolved to optimize antigen presentation to their respective T cells. The accessory proteins tapasin and HLA-DM (DM) crucially influence the selection of peptides that bind to the MHC molecules. We discuss here the dynamic interactions of tapasin and DM with their corresponding MHC molecules that indicate striking parallels. Utilization of a common mode of peptide selection by two different, but related, biological systems argue for its mechanistic validity.
The major histocompatibility complex class I assembly complex
The cell-surface presentation of antigenic peptides by class I major histocompatibility complex (MHC) molecules is the culmination of a complex process that takes place in the endoplasmic reticulum (ER) and involves a multiprotein complex that includes tapasin (TPN), the thiol-disulfide oxidoreductase ERp57, the chaperone calreticulin (CRT), and the transporter associated with antigen processing (TAP). This so-called ‘class I assembly complex’ influences the peptide repertoire presented by MHC class I molecules to CD8+ cytotoxic T cells (CTLs). This is an important process that affects viral immunity, tumorigenicity, autoimmunity, and transplantation.
A crucial role for tapasin in the MHC class I assembly complex: evidence from cell-based studies
A large number of cell-based studies have examined the function of TPN, and numerous roles have been attributed to it. It has been suggested that TPN recruits and stabilizes peptide-deficient MHC class I molecules [1–6]. Evidence that TPN bridges TAP to peptide-deficient MHC class I molecules [7–9] and enhances the ability of TAP to transport peptides into the ER has also been provided [10,11]. Other studies have emphasized a role for TPN in influencing the selection of peptides that load onto MHC class I molecules. More specifically, it was suggested that TPN edits peptides in favor of high-affinity binders [12–16], shapes the cell-surface presentation of peptides according to their intrinsic half-lives [17], and broadens the initial pool of bound peptides [5]. Although not explicitly demonstrated, it was also suggested that TPN might be involved in catalyzing the dissociation of peptides from MHC class I molecules [13,18].
The crucial role of TPN within the MHC class I assembly complex was further highlighted by the finding in cell-based studies that TPN associates covalently with ERp57 through an intermolecular disulfide bond [16]. It was initially thought that this association catalyzes the isomerization of the conserved disulfide bond in the α2 domain of MHC class I heavy chains [16]. More recently, however, it was shown that the covalent association of TPN to ERp57 protects the disulfide bond in the α2 domain against reduction by ERp57 and therefore helps maintain MHC class I molecule structure [19]. From a different perspective, it was also suggested that ERp57, covalently linked to TPN, acts as a structural component within the MHC class I assembly complex [6,18,19]. Thus, although the non-redox and structural roles of ERp57 within the assembly complex have yet to be clearly understood, its effects are optimally manifested through a covalent association with TPN.
In conclusion, cell-based studies have identified multiple functions for TPN that all point to a crucial role in altering the peptide repertoire presented by MHC class I molecules. These studies, however, could not provide a mechanistic understanding of how TPN-mediated effects on the peptide repertoire are manifested through these proposed functions.
Studies of tapasin in cell-free systems
The transient nature of the MHC class I assembly complex in the ER implies that interactions within this complex are intrinsically weak. To circumvent the problem of weak intrinsic affinities, we developed a cell-free system consisting of soluble TPN and human leukocyte antigen (HLA)-B*0801 (a human MHC class I haplotype) molecules that incorporate Jun and Fos leucine peptides, respectively [20]. Kinetics studies with the zippered TPNjun-HLA-B*0801fos complex clearly showed that TPNjun acts as a catalyst by increasing both the association and dissociation rates of peptides. Results were also consistent with a chaperone role for TPNjun owing to its stabilizing effects on immature HLA-B*0801fos molecules. Overall, these studies provided convincing evidence that TPN can act on MHC class I molecules in the absence of CRT, ERp57, or TAP, emphasizing that TPN alone possesses all of the intrinsic properties for influencing peptide selection. Finally, using a competitive peptide binding assay, we showed that both the catalytic and chaperone functions of TPNjun enable the protein to alter the selection of peptides presented by HLA-B*0801fos molecules [20]. More recently, using a reconstituted MHC class I assembly complex, it was shown that the chaperone, catalytic, and editing activities of TPN were compromised unless ERp57 was added to the system [21]. In the context of our results with the TPNjun-HLA-B*0801fos complex [20], these observations suggested that ERp57 was crucial in maintaining the structural integrity of the MHC class I assembly complex in solution. This reinforces the view that ERp57 acts as a key structural component of the MHC class I assembly complex, as discussed above.
Studies with the TPNjun-HLA-B*0801fos complex showed that although all peptides were sensitive to TPNjun during the association reactions (i.e. class I loading), only a subset of the peptides was sensitive to TPNjun during the dissociation reactions [20]. Perhaps most important, the intrinsic off rates of the peptide only modestly correlated with the magnitude of their sensitivities to TPNjun. Furthermore, the use of modified synthetic peptides demonstrated that disruption of conserved hydrogen bonds at the C-terminal end, but not at the N-terminal end, of the antigen groove is a crucial event in a TPNjun-mediated peptide dissociation mechanism [20]. A possible role for hydrogen bonds at the C-terminal end of the groove in the function of TPN was hypothesized previously [18]. Peptide-sequence-dependent MHC class I interactions along the entire length of the groove were also shown to contribute to TPNjun-mediated effects [20]. Overall, an important implication of these results is that MHC class I-peptide interactions at the C terminus of the groove are especially important in determining the fate of bound peptides under the action of TPN. This is consistent with mutagenesis studies of MHC class I heavy chains that have highlighted the importance of certain residues at the C-terminal end of the groove for maintaining the stability of the MHC class I assembly complex [22]. We therefore suggest that this localized action of TPN on the antigen groove is a plausible explanation for why intrinsic peptide off rates, which are global measures of intrinsic kinetics stabilities, cannot be strictly correlated with the magnitude of their sensitivities to TPN.
In conclusion, studies of TPN in cell-based and cell-free systems demonstrated that stabilization of interactions between TPN and peptide-deficient MHC class I molecules, whether through the Jun and Fos leucine peptides or through the covalent association of ERp57 with TPN together with CRT, is crucial for enabling TPN to exert its catalytic and chaperone functions toward MHC class I molecules.
A mechanistic model for peptide selection under the action of tapasin
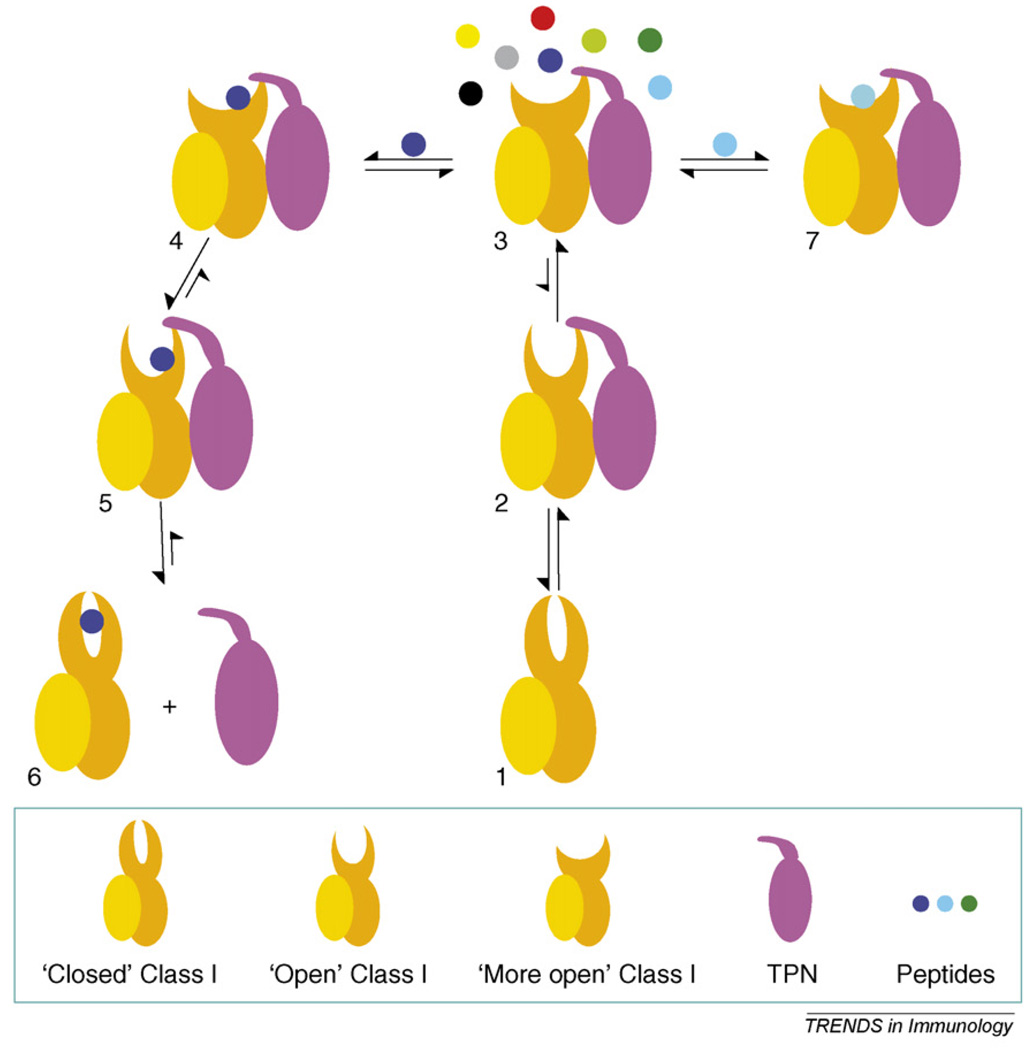
We recently proposed a mechanistic model describing peptide selection under the action of TPN (see Figure 1, steps 1–7) [20]. In this model, TPN perturbs a pre-existing dynamic equilibrium between the closed (1) and open (2) forms of peptide-deficient MHC class I molecules by preferentially associating with the open form. This association stabilizes open MHC class I molecules and prevents their inactivation into closed molecules. In this model, TPN widens the antigen groove of open MHC class I molecules (3); a more widely open binding groove permits peptides to associate and dissociate faster. From a structural point of view, a more open binding groove perturbs the shapes of the binding pockets and decreases somewhat their specificities toward peptides. Consequently, it is probable that different peptides (in terms of amino acid sequences, lengths, number, etc.) have an opportunity to be captured in the presence of TPN. Consistent with this, the effect of TPN on broadening the spectrum of peptides presented by HLA-B8 in TPN-positive and -deficient cells has been demonstrated [5]. If a bound candidate peptide establishes sufficiently stabilizing interactions with MHC residues along the entire length of the groove (4), but especially with those at the C-terminal region of the cleft, it will induce conformational changes in the binding cleft (5) that cause the release of TPN (6). The newly formed peptide-filled molecule (6), which is no longer sensitive to TPN, will probably be transported to the cell surface. However, if a bound candidate peptide is unable to promote conformational changes in the antigen groove (7), it will not as easily disengage TPN. Such bound peptides will eventually dissociate from the MHC class I molecule, and rounds of peptide selection under the action of TPN (3) will continue until a peptide capable of releasing TPN is captured into the groove. Although several aspects of this model remain to be elucidated (e.g. just how are the MHC class I assembly proteins released from the newly formed MHC class I-peptide molecules?), the capture of a peptide that can ‘compete away’ TPN is the trigger point for completing MHC class I maturation (Figure 1).
Figure 1.
A model of tapasin-mediated peptide selection by class I molecules. Tapasin (TPN) perturbs an equilibrium between the closed (1) and open (2) forms of empty class I molecules by binding to and stabilizing the open form. TPN widens the antigen-binding groove of open class I molecules (3), causing peptides to associate (or dissociate) faster. A more open groove probably allows different peptides to be captured in the initial binding step. If a bound candidate peptide can establish sufficiently stabilizing interactions with MHC residues along the groove (4), it will induce conformational changes in the class I molecule (5) that disengage TPN (6). The newly formed peptide-filled class I molecule (6), which is no longer sensitive to the action of TPN, will be transported to the cell surface. However, if a bound candidate peptide is unable to induce conformational changes in the class I molecule (7), owing to its inability to establish energetically stabilizing interactions with MHC residues, it will not as easily disengage TPN. Such bound candidate peptides will eventually dissociate, and another round of peptide selection under the action of TPN (3) will take place.
Thus, in this model, by widening the antigen groove, TPN first permits MHC class I molecules to rapidly capture a different pool of peptides. Then, by stabilizing the antigen groove, TPN provides an ‘energy barrier’ that restricts this initial selection only to those peptides that can provide at least the same level of stabilization that TPN imparts onto the groove. A TPN-mediated peptide selection mechanism thus serves to produce ‘edited’ MHC class I-peptide molecules that are characterized by a certain minimal kinetic stability. The stability threshold, imposed by the presence of TPN, might have been adapted through evolution to ensure that MHC class I-peptide molecules formed in the ER display sufficient kinetic stabilities to persist over time at the cell surface. Although a thorough analysis of TPN-mediated effects on peptide selection by using a large set of carefully selected synthetic epitopes remains to be carried out, the ultimate role of TPN is to influence the spectrum of peptides presented by MHC class I molecules.
MHC class II synthesis and assembly with chaperones
The MHC class II antigen-processing pathway has evolved to optimize the processing of exogenous antigens taken up by antigen presenting cells (APCs) and their presentation to CD4+ helper T cells. An important early step in this process is the assembly of newly synthesized MHC class II αβ heterodimers and the Invariant Chain (Ii). Ii acts as a chaperone by ensuring the proper folding of nascent MHC class II molecules in the ER and by sorting through the Trans-Golgi Network to the low pH endosomal compartments in the APC [23]. As MHC class II molecules move toward vesicular compartments enriched for exogenous antigens, Ii is gradually cleaved away, leaving behind a portion called Class II-associated Invariant chain peptide (CLIP), which remains bound to the peptide-binding groove. The main function of CLIP was originally regarded as a means to prevent premature binding of self-peptides to the groove of MHC class II molecules [24]. However, better understanding of the conformational flexibility of the MHC class II-peptide-binding groove underscored a less appreciated role for CLIP as a ‘place-keeper’ of the groove, whose removal would generate a conformation of MHC class II molecules that is peptide receptive. A peptide-receptive MHC class II molecule binds peptides extremely rapidly and stably [25,26]. In the endosomal compartments specialized for the capture of exogenous antigenic peptides, HLA-DM (DM) plays a vital role in exchanging CLIP for an exogenous antigen-derived peptide. This section of the review will discuss recent advances in understanding various properties of DM and the intriguing parallels revealed between peptide exchange and editing in MHC class I and MHC class II antigen-processing pathways.
Biochemical and structural characterization of DM
The role of DM in antigen processing has been explored for years and is still actively researched. In solution as well as in cells, DM is now known to be active at moderately acidic pH [27] and capable of accelerating the dissociation of CLIP and other peptides from MHC class II molecules [27,28]. In addition, DM can accelerate the rate of association of peptides to MHC class II molecules [28,29], validating its role as a mediator of peptide exchange, because peptide association to MHC class II molecules in its absence is slow and requires substantial conformational changes in the structure of MHC class II molecules [26,29]. However, characterizing DM as a typical catalyst is confounded by biochemical experiments showing that DM does not bind peptide and coprecipitates with DR only under very mild conditions. The inability of DM to bind peptide was explained when the crystal structures of DM and its mouse homolog H-2M were solved [30,31]. Whereas DM was structurally analogous to MHC class II molecules, the two helices lining the putative ‘peptide-binding groove’ were now too close to each other, preventing peptide binding to DM. The other feature emerging from the crystal structure was the lack of an obvious catalytic domain on the molecule. In the absence of DM-MHC class II cocrystals, mutational studies suggested that the acidic face of DM might be important for mediating peptide exchange. Compelling results [32,33] suggested a lateral interaction between DM and the MHC class II molecule HLA-DR3 that involves the region binding the N terminus of the bound peptide. Currently however, the precise residues, or the ‘active site’ that could mediate peptide exchange, remain unknown.
Biophysical factors contributing to DM interaction with MHC class II
There still also exists ambiguity regarding the actual effect of DM on MHC class II molecules. The initial characterization of DM included evidence that the efficacy of DM in dissociating peptides was linked to the intrinsic rate of dissociation of the peptide from MHC class II. Jensen et al. [34] defined the so-called ‘j factor’ as the fold increase of DM-mediated peptide dissociation over the uncatalyzed dissociation reaction, i.e. the ‘DM effect’. They observed that the ‘DM effect’ stayed constant irrespective of the intrinsic off-rate of the peptide. However, there are other lines of evidence that make the case that the ‘DM effect’ varies widely depending on the off rates of the peptides [35], or even that a peptide’s susceptibility to DM is inversely proportional to its intrinsic kinetic stability [36].
The sequence dependence (or lack thereof) of the j factor is an important determinant in that it implicates two very different energy barriers that DM could affect to dissociate peptides. These are (a) a sequence-independent array of 12–15 hydrogen bonds of varying strengths that are formed between residues in the MHC class II groove and the main chain of the bound peptide, and (b) a sequence-dependent set of interactions that are determined by the fit between the side chains of the peptides bound and the pockets of the groove that accommodate them [37]. Hydrogen bonds have been shown to be crucial for the stability of the bound peptides. Mutagenesis experiments of the peptide-binding groove or derivatization of the peptide to reduce the formation of hydrogen bonds have shown to destabilize peptide [38,39]. Small molecules such as alcohols (hydrogen-bond donors) that weaken hydrogen bonds also increase peptide dissociation at high doses, although, in either case, conformational perturbations of the MHC class II molecule cannot be ruled out [40]. The involvement of hydrogen bonds in DM-mediated peptide dissociation was first postulated by Jensen and coworkers based on their studies of the j factor [34] and by Sant et al. based on the observation that mutation of His to Asn at position 81 of the β chain of MHC class II I-Ad generated MHC molecules that bound peptides poorly [41]. Working with soluble DM and the MHC class II molecule HLA-DR1, we have recently shown that hydrogen bonds, especially the short, strong hydrogen bond between a highly conserved residue, β81His, and the peptide main chain close to its N terminus, are the primary targets for DM [42]. We observed that mutating this residue in HLA-DR1 destabilized bound peptides independent of sequence, and that the rapid dissociation could not be further accelerated by DM. A rescue mutant with a suitable solvent-exposed His and a mediated hydrogen bond rescued DM function, suggesting that the position and accessibility to solution (and therefore presumably to DM) were important. However, there is strong evidence that the role of DM includes sequence-specific interactions. Early studies revealed that peptides that fitted suboptimally in the groove were susceptible to DM [29], and CLIP peptide variants with different ligand-binding motifs also showed variable DM susceptibility [34]. Using variants of the HA308–316 peptide, we have previously shown that, for HLA-DR1, the filling of its P1 pocket with large hydrophobic residues (i.e. Phe, Trp, Tyr) imparts resistance to DM-mediated dissociation, whereas other residues, such as Ala, Leu, Val, or Met, make the complex susceptible to DM [29]. Hence, the question of how we reconcile these seemingly contradictory bodies of evidence arises. The answer might lie in dissecting the ‘DM effect’ into two steps: recognition of a suitable MHC class II-peptide conformation, and carrying out an effector function, leading to peptide exchange.
A unifying model to explain the mechanism of action of DM
The conformation of the MHC class II molecule has long been known to be an important factor in determining its ability to participate in peptide association or dissociation [43–51]. Many groups have shown that the binding of peptide to a MHC class II molecule involves several conformational changes; amongst these is a short-lived ‘peptide-receptive’ conformation [25,26,52] to which peptide can bind with rapid and monophasic kinetics. In the absence of DM, however, the conversion of MHC class II molecules from a stable peptide nonreceptive (closed) conformation to this receptive (open) conformation is the rate-limiting step during association. We observed that by filling the P1 pocket in HLA-DR1 by mutation, the MHC class II molecule stayed in an open conformation, from which peptides could bind and unbind rapidly [53]. Importantly, DM could now no longer interact with this mutant molecule; hence, it could not accelerate peptide association or dissociation. We thus hypothesized that the primary role of DM was to generate this receptive conformation from DR, which would then lead to quick peptide association. It is noteworthy that DM generates but does not stabilize and maintain the peptide-receptive conformation of MHC class II molecules in the absence of peptides (K.N. and S.S.-N., unpublished data). In a more recent study [42], we found that disrupting a single conserved hydrogen bond caused the bound peptide to rapidly dissociate, with kinetics resembling ‘DM-mediated’ dissociation of peptide from wild-type HLA-DR1. This mutant molecule was also resistant to DM-mediated peptide dissociation, with the crucial difference that DM could recognize and interact with the molecule if it was bound with a peptide that did not fill the P1 pocket, as observed with wild-type HLA-DR1. We interpreted these data as evidence that the mutant MHC class II molecule with a perturbed hydrogen bond represented a ‘post-DM-affected’ molecule; in other words, DM perturbed the hydrogen bonds around the P1 pocket to dissociate bound peptides. We could not rule out the role of other hydrogen bonds, and we suspect that our results reflect the unusual strength of the His81 hydrogen bond and the cooperative nature of the hydrogen-bond array [42].
We suggest that these two mutants (DR1βHis81Asp and DRβGly86Tyr) reveal the basis of the recognition and the effector functions of DM, and that this two-step functionality of DM might explain some of the earlier data. Depending on sequence, various peptide side chains will fit differently into the peptide-binding groove of MHC class II molecules, resulting in slightly different MHC class II-peptide conformations. Although this might result in variable kinetic stabilities of the complexes, we propose that the actual criterion and a true predictor for DM recognition is the conformation of the complex, and that this conformation is independent of its intrinsic stability. If DM recognizes and interacts with the MHC-peptide complex, it mediates its effector function, which is to generate an open peptide-receptive conformation. This opening up of the groove weakens the hydrogen bonds between MHC class II molecules and peptide, allowing for rapid dissociation. If there is a peptide in the milieu, it can now bind rapidly; otherwise, the groove of MHC class II molecules will close, because the lifetime of this intermediate is short [26]. The molecule stays as such until another transient interaction with DM occurs. We suspect that an empty MHC class II molecule is conformationally similar to a ‘DM-susceptible’ complex [48] (Figure 2). This might explain why DM can also interact with empty MHC class II molecules and convert them into a receptive conformation, allowing for rapid peptide binding. Once a peptide binds that converts MHC class II molecules into a compact DM-insensitive conformation, DM can no longer recognize and interact with the molecule, and it is ineffective at mediating peptide dissociation.
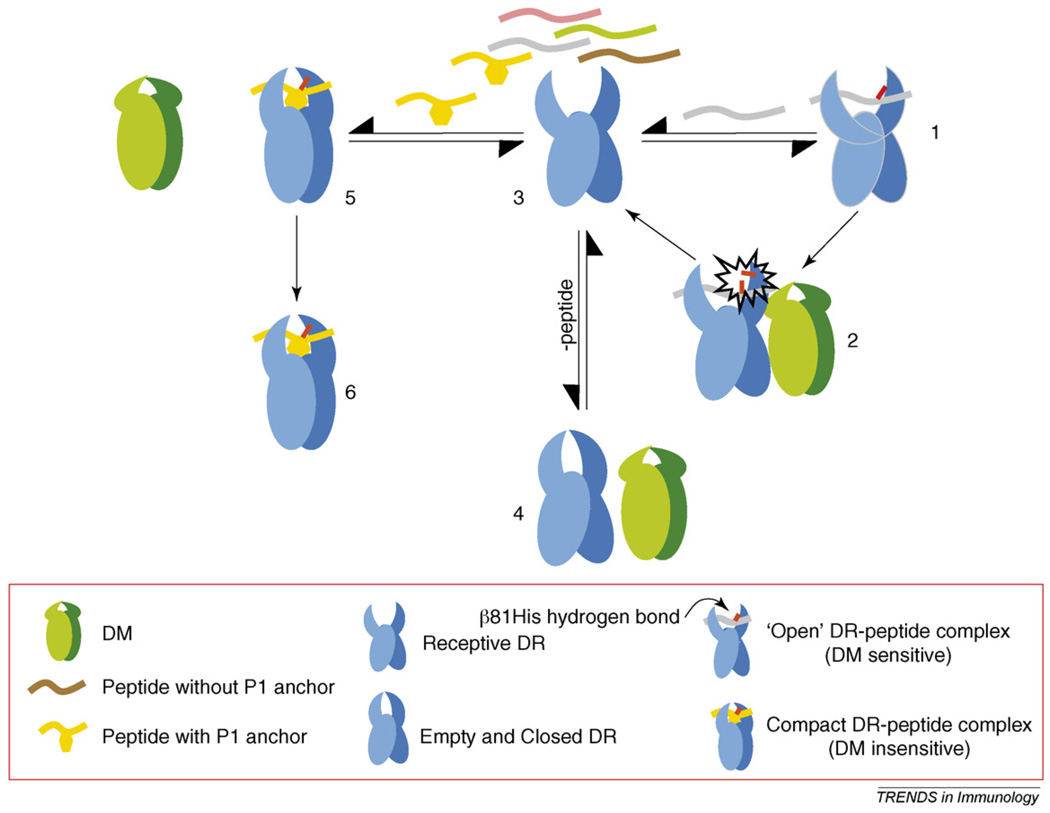
Figure 2.
A model for DM recognition of a MHC class II molecule and its effector function. A newly synthesized MHC class II, or DR1, molecule, occupied here by a peptide that does not fill Pocket 1, is in a ‘floppy’ or ‘open,’ DM-sensitive conformation (1). DM can interact transiently with the molecule by using the proposed ‘hit and run’ mechanism [42] and can break the β81His hydrogen bond between the peptide and DR1, resulting in the release of peptide (2). This generates a peptide-receptive DR1 (3), upon which several events might follow. The molecule can bind another peptide that is similar to the one described above and can then go through another round of DM-mediated dissociation (steps 1 and 2). Alternatively, in the absence of peptide, the DR1 molecule might close and become inactive over time under physiological conditions (4). This empty DR1 might now be rescued by a DM ‘hit’ to generate the peptide-receptive form again (3). Finally, if DR1 binds a peptide that fills P1, the molecule then changes to a tight, DM-insensitive conformation (5). DM cannot interact productively with this complex, and the DR1 bound to peptide is exported to the cell surface (6).
Concluding remarks
As outlined above, a comparison of DM and TPN functions has revealed striking similarities in their mechanisms of action. Both TPN and DM stabilize peptide-deficient MHC molecules. The stabilizing effect ensures that the antigen groove is maintained in a conformation receptive to capturing candidate peptides. In addition to their chaperone effects, TPN and DM act by widening the antigen groove of MHC molecules, allowing peptides to associate and dissociate at faster rates. Equally striking, TPN and DM catalyze the exchange of peptides by disrupting conserved hydrogen bonds between peptides and MHC residues; whereas TPN acts on hydrogen bonds at the C-terminal end of the groove, DM acts on those at the N-terminal end of the groove. In both cases, the targeted conserved hydrogen bonds are located in regions of the groove that crucially influence the thermal stabilities of MHC I- and II-peptide molecules. Peptide-sequence-dependent interactions were also shown to play a part in these mechanisms. Finally, once MHC class I and II molecules have captured optimal peptides, both molecules undergo conformational changes that render them insensitive to further productive interactions with TPN and DM, respectively. Thus, despite differences in structure, molecular chaperones, and maturation pathways, MHC class I and class II molecules seem to be subjected to similar and stringent peptide-selection processes that ultimately influence the repertoire presented at the cell surface. As we see it, an MHC molecule can bind many peptides of a given protein antigen, but only a small subset of these peptides can impart conformational changes in MHC-peptide complexes that render the newly formed complexes insensitive to TPN or DM. This might well explain the mechanism behind ‘immunodominance,’ a well-established phenomenon whereby a few specific peptides are selected as representative epitopes of a given protein antigen to the immune system. Clearly, epitopes derived from exogenous and endogenous antigens must undergo a necessary and rigorous process of screening to avoid possible crossreactivity and recognition of self. The editing mechanisms imposed by TPN or DM on MHC molecules might thus be nature’s elegant solution to narrow the repertoire of peptides presented and ensure the generation of a robust, noncrossreactive, and highly specific immune response, although this remains to be explicitly demonstrated.
Acknowledgements
We acknowledge support from National Institutes of Health grants R01AI063764 and R01GM53549 to S.S-.N. and R01AI045070 and R56AI065943 to M.B.
References
- 1.Ortmann B, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277:1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 2.Grandea AGK, 3rd, et al. Regulation of MHC class I heterodimer stability and interaction with TAP by tapasin. Immunogenetics. 1997;46:477–483. doi: 10.1007/s002510050308. [DOI] [PubMed] [Google Scholar]
- 3.Schoenhals GJ, et al. Retention of emptyMHCclass I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 1999;18:743–753. doi: 10.1093/emboj/18.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnden MJ, et al. Tapasin-mediated retention and optimization of peptide ligands during the assembly of class I molecules. J. Immunol. 2000;165:322–330. doi: 10.4049/jimmunol.165.1.322. [DOI] [PubMed] [Google Scholar]
- 5.Zarling AL, et al. Tapasin is a facilitator, not an editor, of class I MHC peptide binding. J. Immunol. 2003;171:5287–5295. doi: 10.4049/jimmunol.171.10.5287. [DOI] [PubMed] [Google Scholar]
- 6.Garbi N, et al. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nat. Immunol. 2006;7:93–102. doi: 10.1038/ni1288. [DOI] [PubMed] [Google Scholar]
- 7.Suh WK, et al. Interaction of MHC class I molecules with the transporter associated with antigen processing. Science. 1994;264:1322–1326. doi: 10.1126/science.8191286. [DOI] [PubMed] [Google Scholar]
- 8.Ortmann B, et al. MHC class I/β 2-microglobulin complexes associate with TAP transporters before peptide binding. Nature. 1994;368:864–867. doi: 10.1038/368864a0. [DOI] [PubMed] [Google Scholar]
- 9.Solheim JC, et al. Prominence of β 2-microglobulin, class I heavy chain conformation, and tapasin in the interactions of class I heavy chain with calreticulin and the transporter associated with antigen processing. J. Immunol. 1997;158:2236–2241. [PubMed] [Google Scholar]
- 10.Lehner PJ, et al. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line. 220. Immunity. 1998;8:221–231. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 11.Bangia N, et al. The N-terminal region of tapasin is required to stabilize the MHC class I loading complex. Eur. J. Immunol. 1999;29:1858–1870. doi: 10.1002/(SICI)1521-4141(199906)29:06<1858::AID-IMMU1858>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 12.Li S, et al. Cloning and functional characterization of a subunit of the transporter associated with antigen processing. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8708–8713. doi: 10.1073/pnas.94.16.8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sijts AJ, Pamer EG. Enhanced intracellular dissociation of major histocompatibility complex class I-associated peptides: a mechanism for optimizing the spectrum of cell surface-presented cytotoxic T lymphocyte epitopes. J. Exp. Med. 1997;185:1403–1411. doi: 10.1084/jem.185.8.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis JW, et al. HLA-A*0201 presents TAP-dependent peptide epitopes to cytotoxic T lymphocytes in the absence of tapasin. Eur. J. Immunol. 1998;28:3214–3220. doi: 10.1002/(SICI)1521-4141(199810)28:10<3214::AID-IMMU3214>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Williams AP, et al. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16:509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 16.Dick TP, et al. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 17.Howarth M, et al. Tapasin enhances MHC class I peptide presentation according to peptide half-life. Proc. Natl. Acad. Sci. U. S. A. 2004;101:11737–11742. doi: 10.1073/pnas.0306294101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright CA, et al. Tapasin and other chaperones: models of the MHC class I loading complex. Biol. Chem. 2004;385:763–778. doi: 10.1515/BC.2004.100. [DOI] [PubMed] [Google Scholar]
- 19.Kienast A, et al. Redox regulation of peptide receptivity of major histocompatibility complex class I molecules by ERp57 and tapasin. Nat. Immunol. 2007;8:864–872. doi: 10.1038/ni1483. [DOI] [PubMed] [Google Scholar]
- 20.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26:1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat. Immunol. 2007;8:873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- 22.Bouvier M. Accessory proteins and the assembly of human class I MHC molecules: a molecular and structural perspective. Mol. Immunol. 2003;39:697–706. doi: 10.1016/s0161-5890(02)00261-4. [DOI] [PubMed] [Google Scholar]
- 23.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 24.Roche PA, Cresswell P. Invariant chain association with HLA-DR molecules inhibits immunogenic peptide binding. Nature. 1990;345:615–618. doi: 10.1038/345615a0. [DOI] [PubMed] [Google Scholar]
- 25.Rabinowitz JD, et al. Formation of a highly peptide-receptive state of class II MHC. Immunity. 1998;9:699–709. doi: 10.1016/s1074-7613(00)80667-6. [DOI] [PubMed] [Google Scholar]
- 26.Natarajan SK, et al. Stable peptide binding to MHC class II molecule is rapid and is determined by a receptive conformation shaped by prior association with low affinity peptides. J. Immunol. 1999;162:4030–4036. [PubMed] [Google Scholar]
- 27.Denzin LK, Cresswell P. HLA-DM induces CLIP dissociation from MHC class II α β dimers and facilitates peptide loading. Cell. 1995;82:155–165. doi: 10.1016/0092-8674(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 28.Sloan VS, et al. Mediation by HLA-DM of dissociation of peptides from HLA-DR. Nature. 1995;375:802–806. doi: 10.1038/375802a0. [DOI] [PubMed] [Google Scholar]
- 29.Chou CL, Sadegh-Nasseri S. HLA-DM recognizes the flexible conformation of major histocompatibility complex class II. J. Exp. Med. 2000;192:1697–1706. doi: 10.1084/jem.192.12.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mosyak L, et al. The structure of HLA-DM, the peptide exchange catalyst that loads antigen onto class II MHC molecules during antigen presentation. Immunity. 1998;9:377–383. doi: 10.1016/s1074-7613(00)80620-2. [DOI] [PubMed] [Google Scholar]
- 31.Fremont DH, et al. Crystal structure of mouse H2-M. Immunity. 1998;9:385–393. doi: 10.1016/s1074-7613(00)80621-4. [DOI] [PubMed] [Google Scholar]
- 32.Pashine A, et al. Interaction of HLA-DR with an acidic face of HLA-DM disrupts sequence-dependent interactions with peptides. Immunity. 2003;19:183–192. doi: 10.1016/s1074-7613(03)00200-0. [DOI] [PubMed] [Google Scholar]
- 33.Stratikos E, et al. Identification of the lateral interaction surfaces of human histocompatibility leukocyte antigen (HLA)-DM with HLA-DR1 by formation of tethered complexes that present enhanced HLA-DM catalysis. J. Exp. Med. 2002;196:173–183. doi: 10.1084/jem.20020117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber DA, et al. Enhanced dissociation of HLA-DR-bound peptides in the presence of HLA-DM. Science. 1996;274:618–620. doi: 10.1126/science.274.5287.618. [DOI] [PubMed] [Google Scholar]
- 35.Busch R, et al. Achieving stability through editing and chaperoning: regulation of MHC class II peptide binding and expression. Immunol. Rev. 2005;207:242–260. doi: 10.1111/j.0105-2896.2005.00306.x. [DOI] [PubMed] [Google Scholar]
- 36.Sant AJ, et al. The relationship between immunodominance, DM editing, and the kinetic stability of MHC class II:peptide complexes. Immunol. Rev. 2005;207:261–278. doi: 10.1111/j.0105-2896.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- 37.Stern LJ, et al. Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature. 1994;368:215–221. doi: 10.1038/368215a0. [DOI] [PubMed] [Google Scholar]
- 38.Sant AJ, et al. Individual hydrogen bonds play a critical role in MHC class II: peptide interactions: implications for the dynamic aspects of class II trafficking and DM-mediated peptide exchange. Immunol. Rev. 1999;172:239–253. doi: 10.1111/j.1600-065x.1999.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 39.Stratikos E, et al. Enhanced catalytic action of HLA-DM on the exchange of peptides lacking backbone hydrogen bonds between their N-terminal region and the MHC class II α-chain. J. Immunol. 2004;172:1109–1117. doi: 10.4049/jimmunol.172.2.1109. [DOI] [PubMed] [Google Scholar]
- 40.Marin-Esteban V, et al. Chemical analogues of HLA-DM can induce a peptide-receptive state in HLA-DR molecules. J. Biol. Chem. 2004;279:50684–50690. doi: 10.1074/jbc.M407598200. [DOI] [PubMed] [Google Scholar]
- 41.McFarland BJ, et al. Cutting edge: a single, essential hydrogen bond controls the stability of peptide-MHC class II complexes. J. Immunol. 1999;163:3567–3571. [PubMed] [Google Scholar]
- 42.Narayan K, et al. HLA-DM targets the hydrogen bond between the histidine at position β81 and peptide to dissociate HLA-DR-peptide complexes. Nat. Immunol. 2007;8:92–100. doi: 10.1038/ni1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadegh-Nasseri S, Germain RN. How MHC class II molecules work: peptide-dependent completion of protein folding. Immunol. Today. 1992;13:43–46. doi: 10.1016/0167-5699(92)90131-P. [DOI] [PubMed] [Google Scholar]
- 44.Sadegh-Nasseri S, Germain RN. A role for peptide in determining MHC class II structure. Nature. 1991;353:167–170. doi: 10.1038/353167a0. [DOI] [PubMed] [Google Scholar]
- 45.Sadegh-Nasseri S, McConnell HM. A kinetic intermediate in the reaction of an antigenic peptide and I-Ek. Nature. 1989;337:274–276. doi: 10.1038/337274a0. [DOI] [PubMed] [Google Scholar]
- 46.Sadegh-Nasseri S, et al. MHC class II function preserved by low-affinity peptide interactions preceding stable binding. Nature. 1994;370:647–650. doi: 10.1038/370647a0. [DOI] [PubMed] [Google Scholar]
- 47.Sato AK, et al. Determinants of the peptide-induced conformational change in the human class II major histocompatibility complex protein HLA-DR1. J. Biol. Chem. 2000;275:2165–2173. doi: 10.1074/jbc.275.3.2165. [DOI] [PubMed] [Google Scholar]
- 48.Carven GJ, Stern LJ. Probing the ligand-induced conformational change in HLA-DR1 by selective chemical modification and mass spectrometric mapping. Biochemistry. 2005;44:13625–13637. doi: 10.1021/bi050972p. [DOI] [PubMed] [Google Scholar]
- 49.Dornmair K, et al. Structural intermediates in the reactions of antigenic peptides with MHC molecules. Cold Spring Harb. Symp. Quant. Biol. 1989;54:409–416. doi: 10.1101/sqb.1989.054.01.050. [DOI] [PubMed] [Google Scholar]
- 50.Witt SN, McConnell HM. Formation and dissociation of short-lived class II MHC-peptide complexes. Biochemistry. 1994;33:1861–1868. doi: 10.1021/bi00173a032. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt L, et al. Conformational isomers of a class II MHC-peptide complex in solution. J. Mol. Biol. 1999;286:207–218. doi: 10.1006/jmbi.1998.2463. [DOI] [PubMed] [Google Scholar]
- 52.Zarutskie JA, et al. The kinetic basis of peptide exchange catalysis by HLA-DM. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12450–12455. doi: 10.1073/pnas.211439398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Natarajan SK, et al. Sodium dodecyl sulfate stability of HLA-DR1 complexes correlates with burial of hydrophobic residues in pocket 1. J. Immunol. 1999;162:3463–3470. [PubMed] [Google Scholar]