Abstract
In eukaryotes, two-component regulatory systems have been demonstrated to regulate phosphorylation of mitogen-activated protein kinases (MAPKs). Here, we describe a method implementing preparation of a protein extract under denaturing conditions, followed by Western analysis using MAPK antibodies that can be used to observe the effects of components of two-component signaling pathways or other proteins on the phosphorylation status of MAPKs. The protein extraction method presented may also be used to concentrate cellular proteins for additional applications, such as metabolic labeling or analysis of other posttranslational modifications.
1. Introduction
Histidine kinases and aspartate-containing response regulator proteins are elements of two-component signaling pathways. Two-component phosphorelay systems have been identified in archaea, bacteria, protists, fungi, and plants, but to-date have not been found in any animal, including humans (Bahn, 2008; Borkovich et al., 2004; Park et al., 2009; Wolanin et al., 2002). In the simplest two-component systems, the phosphorelay begins with autophosphorylation of a conserved histidine residue in the histidine kinase in response to some environmental signal (Posas et al., 1996). This same phosphate is then transferred onto an aspartate within the response regulator domain of a different protein. The phosphorylation status of the response regulator protein determines its ability to regulate gene expression or to activate or inhibit other downstream effector proteins.
A more complex version of the pathway described above is the so-called multicomponent pathway (Borkovich et al., 2004; Catlett et al., 2003; Chang and Stewart, 1998; Wolanin et al., 2002). Such systems include hybrid histidine kinases (proteins containing both histidine kinase and response regulator domains) and histidine phosphotransfer proteins (HPTs). Multicomponent pathways begin with autophosphorylation of a histidine residue in the hybrid histidine kinase. However, instead of being transferred to a separate protein, the phosphate is passed in an intramolecular reaction to the aspartate residue on the response regulator domain in the hybrid kinase. This phosphate is subsequently transferred to a HPT protein and then to a terminal response regulator. In eukaryotic filamentous fungi such as Neurospora crassa, all histidine kinases are of the hybrid type. Neurospora possesses 11 hybrid histidine kinases, one HPT and two response regulator proteins (Borkovich et al., 2004; Galagan et al., 2003).
In Neurospora and other eukaryotes, multicomponent signaling pathways have been found to activate mitogen-activated protein (MAP) kinase cascades (Catlett et al., 2003; Chang and Stewart, 1998; West and Stock, 2001). These cascades consist of three serine/threonine protein kinases that act sequentially. The first step involves the phosphorylation of a MAP kinase kinase kinase (MAPKKK), which leads to the phosphorylation of a MAP kinase kinase (MAPKK) that triggers the phosphorylation of the down-stream MAP kinase (MAPK). The phosphorylation status of the MAPK determines its ability to regulate other proteins. Interestingly, when an upstream MAPKKK or MAPKK phosphorylates the next MAPKK or MAPK in the pathway, it is not the same phosphate (as is the case in histidine kinase signaling pathways), but a new ATP that is hydrolyzed for each phosphorylation event (Choi et al., 2008).
Neurospora possesses nine MAPKKK/MAPKK/MAPK proteins (three of each type) that are thought to make up three MAPK modules. These three modules correspond to (1) the osmosensing (OS-2 MAPK cascade; see below), (2) the cell integrity (MAK-1 MAPK cascade), and (3) the pheromone response/filamentation (MAK-2 MAPK cascade) pathways that have been well studied in yeast (Borkovich et al., 2004; Posas et al., 1998).
In Neurospora, the osmotic sensitive (os) mutants correspond to a hybrid histidine kinase (os-1/nik-1) and components of a downstream MAPK cas-cade (os-4 MAPKKK, os-5 MAPKK and os-2 p38 class MAPK; Emerson and Emerson, 1958; Livingston, 1969; Selitrennikoff et al., 1981; Zhang et al., 2002). In addition, our laboratory has demonstrated that mutants lacking the response regulator rrg-1 have many phenotypes in common with os mutants. os mutants are characterized by fragile conidia that tend to lyse and these strains are often female-sterile (Jones et al., 2007). These mutants also share the phenotypes of osmotic sensitivity and resistance to certain fungicides, traits that are believed to result from the inability to properly stimulate glycerol production or other counter solutes (Ellis et al., 1991; Fujimura et al., 2000; Kanetis et al., 2008; Pillonel and Meyer, 1997; Zhang et al., 2002).
Fludioxonil is a phenylpyrrole fungicide that is effective against Neurospora, and various genetic screens have produced several mutants (including the os mutants discussed above) that are resistant to this and other fungicides (Fujimura et al., 2000; Grindle and Temple, 1982; Ochiani et al., 2001; Selitrennikoff et al., 1981). Fludioxonil has been approved for use against a broad spectrum of fungal plant pathogens to prevent fruit spoilage (Förster, et al., 2007; Kanetis et al., 2008). Fludioxonil appears to target the same pathway as the hyperosmotic stress response pathway, leading to the activation of the OS-2 MAPK cascade (Zhang et al., 2002).
In addition to roles in osmotic stress and fungicide resistance, we have recently demonstrated that RRG-1 and the downstream OS-2 MAPK are involved in regulation of the circadian rhythm that controls asexual spore formation (conidiation) in Neurospora. Although the clock is not required to mount a rapid response during hyperosmotic conditions, Δrrg-1 mutants display a period shortening defect and a delay in the conidiation rhythm when grown on race tubes (Vitalini et al., 2007).
This chapter presents a method to assay MAPK phosphorylation after exposure of Neurospora cells to hyperosmotic stress or fungicide. The method involves freezing cells in liquid nitrogen, followed by bead lysis in an ethanol solution. Proteins are subjected to SDS–PAGE and gels electroblotted onto a membrane. MAPK antibodies are used to visualize phosphorylated and total MAPK protein. This method has been used successfully to demonstrate regulation of phosphorylation of the OS-2 MAPK by two-component signaling proteins in Neurospora (Jones et al., 2007; Vitalini et al., 2007).
2. Growth of Cultures and Exposure to Hyperosmotic Conditions or Fungicide
All strains were cultured on Vogel’s minimal medium (VM; Davis and de Serres, 1970; Vogel, 1964). Shaking liquid cultures were exposed to hyperosmotic stress induced by NaCl or the fungicide fludioxonil during the last 10 min of growth. Cells were collected by filtration and cell pads frozen in liquid nitrogen.
2.1. Reagents
- Stock solutions
- 50 μg/ml biotin in 50% ethanol (biotin solution)
- 10 mg/ml fludioxonil (dissolved in 100% DMSO)
- 4 M NaCl (dissolved in water).
- 50× concentrated VM medium salts (per liter)
- 126.8 g sodium citrate·2H2O
- 250 g KH2PO4
- 100 g NH4NO3
- 10 g MgSO4·7H2O
- 5 g CaCl2·2H2O (predissolve first in water and then added slowly)
- 5 ml biotin solution
- 5 ml trace elements solution (see below)
- After bringing solution to final volume, transfer to bottle. Add 5 ml of chloroform (acts as fungicide; sinks to bottom of bottle) to the solution. Make sure not to pipet any of the chloroform when withdrawing 50× VM solution for use.
- Trace elements solution ( for 100 ml)
- 5 g citric acid·H2O
- 5 g ZnSO4·7H2O
- 1 g Fe(NH4)2(SO4)·6H2O
- 0.25 g CuSO4·5H2O
- 0.05 g MnSO4·H2O
- 0.05 g H3BO3
- 0.05 g NaMoO4·2H2O
- Filter sterilize and store in the dark at 4 °C (Vogel, 1964).
- VM medium (per liter)
- 20 ml 50× concentrated VM salts
- 15 g sucrose
- 10 g agar (for propagation of conidia in flasks)
- Autoclave to sterilize.
2.2. Growth and collection of cultures
Inoculate a 250-ml Erlenmeyer flask containing 50 ml VM agar medium with each strain to be analyzed. Incubate flasks at 30 °C in the dark for 3 days, followed by 2–5 days at 25 °C in the light.
Suspend vegetative tissue from the VM agar cultures using 50 ml of sterile ice-cold water by swirling. The material is filtered into a sterile 250 ml Erlenmeyer flask prepared with a Handi wipe towel® (Clorox Corp., Oakland, CA) affixed to the rim with a rubberband (entire assembly autoclaved prior to use) to separate the hyphal filaments (retained by filter) from the conidia (flow through to flask).
Transfer conidial suspension to a 50-ml conical tube and centrifuge at 2500 rpm for 3–5 min at room temperature. Resuspend the pellet in 20–40 ml of ice-cold sterile water and repeat centrifugation. Resuspend the final pellet in 200–400 μl of sterile water.
Dilute the suspension (1:100) and count using a hemacytometer to quantify the number of conidia. Adjust the concentration to 1 × 108 conidia/ml. Store conidia at 4 °C in the dark for up to 1 day before use.
Sixteen-hour shaking liquid cultures were generated by inoculating 20 ml of liquid VM to a final concentration of 1 × 106 conidia/ml. Cultures were incubated in the dark at 30 °C with shaking at 200 rpm for 16 h.
During the last 10 min of growth, treat cultures with the same volume of either NaCl (final concentration 0.1 or 0.8 M) or water (control for hyperosmotic stress) or fludioxonil dissolved in dimethyl sulfoxide (DMSO; final concentration 100 μg/ml) or DMSO (control for fungicide exposure).
Collect cultures by filtration. This can be accomplished using a funnel, vacuum flask and filter paper or by filtering and then pressing out the liquid using a Handi wipe towel® (Clorox Corp.). The tissue should be quick-frozen in liquid nitrogen. The tissue may be stored at −80 °C until protein is extracted; however, lengthy storage of cell pads prior to protein extraction is not recommended and best results are obtained using freshly collected tissue.
3. Mitogen-Activated Protein Kinase Assay
A procedure for detection of MAPK phosphorylation in protein extracts is described below. This process is being described with reference to the MAPK OS-2, but it has also been used for detection of all three MAPKs in N. crassa and should be translatable to other species. The assay involves protein extraction and western blot analysis using commercially available antibodies. The antibodies have been developed against yeast Hog1p MAPK and mammalian p38 MAPK proteins, but cross-react with their highly conserved counterparts in Neurospora and other fungi, including the OS-2 homologs PdOS-2 from Penicillium digitatum (Kanetis et al., 2008), SakA from Aspergillus nidulans (Kawasaki et al., 2002), OSC-1 from Colletotrichum lagenarium (Kojima et al., 2004), and BmHog1 from Colletotrichum heterostrophus (Yoshimi et al., 2005). One antibody reacts with all forms of OS-2, whether phosphorylated or not, while the other only recognizes the phosphorylated version of the MAPK.
3.1. Reagents
- For preparation of cell extracts:
- 1% sodium dodecyl sulfate (SDS) in water
- 3 M phenylmethylsulfonyl fluoride (PMSF) in 95% ethanol. Prepare from a stock solution of 200 mM PMSF (that was dissolved in isopropanol).
- For SDS–PAGE electrophoresis:
- 2× Buffer A (per liter):
- 181.5 g Tris base
- 4 g SDS
- Adjust the pH to 8.8 (before adding SDS). Filter the solution and store at 4 °C.
- 1:3 Buffer A:
- Dilute 2× Buffer A solution with water as indicated (1:3).
- 2× Buffer B (per liter):
- 60.4 g Tris base
- 4 g SDS
- Adjust pH to 6.8 (before adding SDS). Filter solution and store at 4 °C.
- Premixed acrylamide/bisacrylamide 37.5:1, 40% solution (#1490, EMD Chemicals, Madison, WI).
- 10% ammonium persulfate (APS; diluted in water). Make fresh before use.
- Tetramethylethylenediamine (TEMED; #BP150, Fisher BioReagents, Pittsburgh, PA).
- 10% SDS (w/v in water)
- SDS sample buffer (per 10 ml):
- 2 ml glycerol
- 2 ml 10% SDS
- 1.25 ml 2 Buffer B
- 6 mg bromphenol blue
- 4.25 ml water
- Store 10× SDS sample buffer at room temperature. Add 1/20 volume of β-mercaptoethanol to an aliquot of this solution immediately before use.
- 10% SDS–PAGE resolving gel (for 12.5 ml):
- 3.1 ml water
- 3.13 ml acrylamide/bisacrylamide 37.5:1, 40% solution
- 6.25 ml 2× Buffer A
- 60 μl 10% APS
- 7 μl TEMED
- SDS–PAGE stacking gel ( for 5 ml):
- 2 ml water
- 500 μl acrylamide/bisacrylamide 37.5:1, 40% solution
- 2.5 ml 2× Buffer B
- 60 μl 10% APS
- 7 μl TEMED
- 10× running buffer (per liter):
- 30 g Tris base
- 143.75 g glycine
- 5 g SDS
- Dilute 1:10 with water to make 1× running buffer.
- For Western blot analysis:
- Transfer buffer (per liter):
- 3 g Tris base
- 14.1 g glycine
- 200 ml methanol
- Bring to final volume with water. This solution can be reused three to four times. Store in an airtight container in the dark.
- 10× Tris-buffered saline (TBS) (per liter):
- 24.2 g Tris base
- 80 g NaCl
- Adjust pH to 7.6 with HCl, then bring to final volume. Autoclave to sterilize for long-term storage. Store solution at room temperature. Dilute 1:10 with water to make 1× TBS solution for washes of the membranes to be probed with the phospho-p38 antibody.
- TBSM:
- Prepare by dissolving 5 g dry milk in 100 ml TBS. Use immediately to block membrane/dilute secondary antibody (for phospho-p38 antibody probed membranes) or store overnight at 4 °C.
- TBST:
- Add 50 μl of Tween-20 for every 100 ml of TBS. Use this solution to prepare TBSTB.
- TBSTB:
- Prepare by dissolving 5 g bovine serum albumin (BSA; Fraction V) in 100 ml TBST. Use immediately to dilute the phospho-p38 primary antibody or store overnight at 4 °C.
- 5× phosphate-buffered saline (PBS) (per liter):
- 41.2 g Na2HPO4
- 10.2 g NaH2PO4
- 20 g NaCl
- Adjust pH to 7.4 with NaOH then bring to final volume. Autoclave to sterilize for long-term storage. Store solution at room temperature. Dilute 1:5 with water to make 1× PBS solution for washes of the membranes to be probed with the Hog1p antibody.
- PBSM:
- Prepare by dissolving 5 g dry milk in 100 ml PBS. Use immediately to block the membrane/dilute secondary antibody (for Hog1p antibody probed membranes) or store overnight at 4 °C.
- PBST:
- Add 50 μl of Tween-20 for every 100 ml of PBS. Use this solution to prepare PBSTB.
- PBSTB:
- Prepare by dissolving 5 g BSA (Fraction V) in 100 ml PBST. Use immediately to dilute the Hog1p primary antibody or store overnight at 4 °C.
- Phospho-p38 rabbit primary antibody (#9211; Cell Signaling Technology, Inc., Beverly, MA), diluted 1:600 in TBSTB just before use.
- Hog1p rabbit primary antibody (#sc-9079; Santa Cruz Biotechnology, Inc., Santa Cruz, CA), diluted 1:600 in PBSTB just before use.
- Peroxidase-conjugated goat antirabbit IgG secondary antibody (Sigma Chemical Company, St. Louis, MO), diluted 1:6000 in TBSM or PBSM (depending on primary antibody diluent).
3.2. Preparation of protein extracts
Grind (~1 min) the frozen Neurospora tissue into a fine powder in liquid nitrogen using a mortar and pestle. Add additional liquid nitrogen to the mortar as needed.
Transfer 20–80 mg of ground tissue into a plastic 2-ml screw-top centrifuge tube containing 1 ml of chilled 3 mM PMSF in 95% ethanol and 0.2 g of 0.5 mm glass beads. Vortex briefly to suspend the tissue in solution. Store samples on ice briefly until several have been processed.
Vortex samples three times for 60 s with 60 s rests on ice in between.
Chill extracts at −20 °C for at least 16 h (it is fine to leave the samples for several days in the freezer). This step causes the protein to precipitate out of solution.
Centrifuge the samples at 14,000 rpm for 10 min at 4 °C in a microcentrifuge.
Remove as much of the supernatant as possible without disturbing the pellet. Evaporate the remaining ethanol by spinning the tubes for 30 min in a vacuum concentrator (Eppendorf, Westbury, NY). This step precipitates the proteins on the beads. At this point, the samples may be reconstituted (see next step) or stored at −20 °C up to several days before continuing.
Reconstitute samples in 200–250 μl of 1% SDS and heat at 85 °C for 5 min (vortex every 1–2 min). Centrifuge tubes at 14,000 rpm for 3–5 min at room temperature in a microcentrifuge.
Remove and save the protein supernatant in a new microcentrifuge tube.
Reconstitution (steps 7 and 8) may be repeated once and the supernatants can be combined.
Subject the combined supernatants to a final heating, centrifugation, and supernatant recovery as described above to remove residual cellular debris.
To ensure protein stability, add SDS sample buffer at a ratio of one volume sample buffer to two volumes protein as soon as possible. We typically add sample buffer to the majority of the extract, only leaving enough extract to successfully perform the protein assay.
Determine the concentration of protein in extracts using the BCA micro protein assay (Thermo Scientific, Pierce, Rockford, IL) according to the manufacturer’s instructions. The absorbance of assay tubes is measured at 562 nm. The standard curve consists of 0, 2, 4, 6, 8, and 10 μg BSA. Make all protein dilutions in 1% SDS.
3.3. SDS–PAGE electrophoresis and transfer of protein to membrane
Pour the 10% resolving gel between two glass plates, leaving approximately the top 3 cm empty. Using a Pasteur pipet, overlay the resolving gel with bubbles generated by shaking the 1:3 Buffer A solution. Follow by overlaying the 1:3 Buffer A solution itself. This helps keep the gel moist and yields a sharp interface at the top edge of the gel. Let the resolving gel polymerize for at least 45 min.
Rinse away any residue (such as unpolymerized acrylamide) with water. Remove excess water from above resolving gel by blotting with a piece of filter paper.
Pour the stacking gel and insert a comb. Allow the stacking gel to polymerize 5–15 min.
Rinse wells that were generated in the stacking gel by the comb with water several times to remove any unpolymerized acrylamide and then rinse the wells with 1× running buffer. Assemble gel in electrophoresis apparatus.
Protein samples (prepared as described above) should be heated at 85–100 °C and then centrifuged at room temperature for 5 min at 14,000 rpm before loading onto a 10% SDS–PAGE gel. If the gel is not run the same day, store the samples at −20 °C or lower until use. Reheat and centrifuge the samples before use. Do not use extracted protein that has not been stored in sample buffer, as the sample buffer inhibits degradation of the OS-2 MAPK protein.
Duplicate gels can be run at 15–70 mA (gels usually take 3–16 h to run depending on the voltage; never exceed 250 V), as previously described (Krystofova and Borkovich, 2005) in 1× running buffer.
The protein is transferred from the SDS–PAGE gel to a nitrocellulose membrane (0.45 μm, GE Water and Process Technologies, Trevose, PA) in a Trans-blot Cell Western blot apparatus (Bio-Rad, Hercules, CA) at 215 mA for 2–3 h in transfer buffer.
3.4. Western blot analysis using MAPK antibodies
After transfer of the protein onto the nitrocellulose membranes is complete allow the duplicate membranes to dry. One membrane will be used to determine whether the OS-2 protein is equally expressed in all samples, while the other will be used to measure the level of phosphorylated OS-2.
The membranes can be washed and probed immediately after drying, or stored for up to several weeks. Wash the membranes three times for 5 min each in TBS (for the phospho-p38 antibody probed membranes) or PBS (for the Hog1p antibody probed membranes) with shaking at room temperature.
Block the membranes in TBSM (phospho-p38 antibody-probed membranes) or PBSM (Hog1p antibody-probed membranes) for 1 h with shaking at room temperature. Blocking inhibits nonspecific binding of antibody to the membrane later in the procedure.
Wash the membranes again 3× 5 min each in TBS or PBS wash buffer, as appropriate, with shaking at room temperature.
Primary antibodies should be diluted as indicated above, in TBSTB or PBSTB. These antibodies are commercially available and were generated against mammalian (p38) or S. cerevisiae (Hog1p) MAPKs, but recognize forms of the 42 kDa OS-2 protein. The anti-phospho-p38 antibody can be used to detect phospho-OS-2. Anti-Hog1p reacts with both unphosphorylated and phosphorylated forms of OS-2, and serves as a control for the total amount of OS-2 protein in the cell. Incubate membranes overnight at 4 °C in the primary antibodies with gentle shaking.
Rinse away the excess antibody with three 5 min washes (with shaking at room temperature) in TBS or PBS, depending on primary antibody diluent.
Incubate the membranes for 1 h with the peroxidase-conjugated goat antirabbit IgG secondary antibody diluted in TBSM or PBSM (as appropriate) with shaking at room temperature.
Wash the membranes three times in wash buffer as described above (step 4) to remove excess secondary antibody.
Place the membranes on a clean transparency and treated with chemiluminescent chemicals. Chemiluminescent detection may be performed using the SuperSignal West Pico kit (Thermo Scientific, Pierce), mixing equal volumes of the Peroxide and Luminol Enhancer solutions (as previously described; Krystofova and Borkovich, 2005). Make sure the chemicals cover the membranes evenly. Rotate the transparency by hand for one minute to allow the mixture to flow evenly over the membrane and then place the transparency in the dark for 4 additional minutes. Briefly allow the excess chemiluminescent regents to drain off the membrane and then cover with a clear transparency before imaging.
3.5. Results
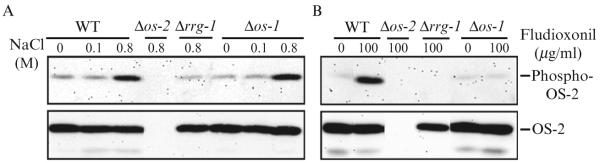
The results for a typical OS-2 phosphorylation assay are shown in Fig. 17.1. Levels of OS-2 protein are similar under all conditions in all strains, except in Δos-2 mutants, which serve as a negative control for both OS-2 protein and phospho-OS-2. Phosphorylation of OS-2 is stimulated in cultures treated with 0.8 M NaCl or 100 μg/ml fludioxonil (fungicide) in wildtype cells. Δrrg-1 (response regulator) mutants are unable to induce OS-2-phosphate levels in response to NaCl or fludioxonil above the basal level observed in wild type (Jones et al., 2007). Δos-1 (hybrid histidine kinase) mutants exhibit a hybrid response, with an increase in response to NaCl, but not to fungicide. Together, these results show that a divergence exists between the hyperosmotic stress and fungicide response pathways. The hyperosmotic stress response pathway is likely more complex than the fungicide response pathway, possibly involving an additional osmotic sensitive factor. Of note, the ability of rrg-1D921N mutants (mutated at the predicted site of phosphorylation) to induce OS-2 phosphorylation in a manner similar to wild type suggests that the unphosphorylated form of RRG-1 is the one that activates the OS-2 MAPK cascade (Jones and Borkovich, unpublished data; Jones et al., 2007).
Figure 17.1.
OS-2 phosphate levels of the Δos-1 and Δrrg-1 gene replacement mutants. (A) Analysis of OS-2 protein levels and phosphorylation in response to hyperosmotic conditions. Wild-type (74A), Δos-1, Δrrg-1, and Δos-2 strains were grown for 16 h in liquid submerged VM cultures. The cultures were brought to 0, 0.1, or 0.8 M NaCl and incubated with shaking for the last 10 min of growth. The cells were collected, whole cell extracts were prepared and then subjected to Western analysis. The top panel is a Western blot using anti-p38 that recognizes only phosphorylated OS-2, while the bottom panel is a Western blot using anti-Hog1p that reacts with all forms of the OS-2 protein. (B) Analysis of OS-2 protein levels and phosphorylation during fungicide exposure. Sixteen-hour liquid submerged VM cultures of the strains indicated in (A) were treated with 0 or 100 μg/ml fludioxonil for the last 10 min of incubation. Whole cell extracts were prepared and subjected to Western analysis with the antibodies described in (A).
4. Adapting the MAPK Assay
The MAPK detection method described above for the p38-class Neurospora MAPK OS-2 has also been used to detect the OS-2 homolog PdOS-2 from P. digitatum (Kanetis et al., 2008). A similar MAPK assay has been implemented to observe the level of phosphorylated MAK-1 and MAK-2, the other two MAPKs found in Neurospora (Pandey et al., 2004; Park et al., 2008). MAK-1 (50 kDa) and MAK-2 (39 kDa) are p44/42/Erk class MAPKs. These two proteins are recognized by commercially available antibodies that were generated against mammalian (p44/42) MAPK sequences. Both phosphorylated and nonphosphorylated MAK-1 and MAK-2 react with one antibody (anti-p44/42 #9102; 1:200 dilution in TBSTB; Cell Signaling Technology, Inc.), while another antibody recognizes only the phosphorylated forms of the two proteins (antiphospho-p44/42 #9101; 1:200 dilution in TBSTB; Cell Signaling Technology, Inc.). An advantage of this assay for MAK-1 and MAK-2 is that both proteins can be detected on the same Western blot, due to their large difference in molecular mass.
5. Discussion
The method discussed in this paper may theoretically be modified to observe the phosphorylation status of any MAPK or the level of any cellular protein for which an antibody is available. We have used this technique to detect epitope-tagged proteins from lysed cells of Neurospora strains that have been transformed with a tagged gene construct (Jones and Borkovich, unpublished results). Furthermore, the protein extraction method discussed above can be used to generate extremely concentrated protein solutions. The concentration of the extract can be controlled by adjusting the volume of 1% SDS used to reconstitute the proteins.
The cell lysis method has been used to extract proteins from many types of Neurospora tissues, including various plate and shaking liquid cultures. We commonly use tissues harvested from overnight shaking liquid VM cultures. A 20-ml culture generates an adequate quantity of tissue from strains that grow similarly to wild type. However, larger cultures were sometimes needed to obtain enough tissue from slow-growing mutants.
Some Western blot procedures will tolerate stripping of a membrane after probing with the first antibody and then reprobing with a second antibody. Such a procedure would reduce the number of blots needed. However, in our experience, probing the same blot with the phospho and protein MAPK antibodies, as opposed to use of duplicate blots, is not recommended, due to issues with incomplete removal of the first antibody and/or removal of protein from the blot during the stripping procedure. In particular, we have found that the Hog1p antibody is difficult to remove completely.
The primary antibody solution can be reused two to three times to probe a membrane. Although this practice can be used to conserve relatively expensive primary antibody during preliminary studies, freshly prepared antibody solutions are preferred for published experiments, as their actual titer is known. If reusing a primary antibody solution, it is important that it be diluted in PBSTB or TBSTB (not a milk solution) and stored at −20 °C between uses. Thaw the antibody solution at room temperature before reuse.
The results in Fig. 17.1 show that levels of the OS-2 MAPK protein do not fluctuate greatly over time or under different experimental conditions. However, the phosphorylation status of OS-2 is altered in response to fungicide treatment or a hyperosmotic environment (see Fig. 17.1; Jones et al., 2007) and regularly oscillates throughout the day (Vitalini et al., 2007) in wild-type strains. Future work will focus on analysis of additional upstream regulators, including additional hybrid histidine kinases, that regulate OS-2 phosphorylation in Neurospora.
ACKNOWLEDGMENTS
We thank Dr. Gyungsoon Park for helpful discussions and assistance with fine-tuning the MAK-1 and MAK-2 detection protocol.
REFERENCES
- Bahn YS. Master and commander in fungal pathogens: The two-component system and the HOG signaling pathway. Eukaryot. Cell. 2008;7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich KA, Alex LA, Yarden O, Freitag M, Turner GE, Read ND, Seiler S, Bell-Pedersen D, Paietta J, Plesofsky N, Plamann M, Goodrich-Tanrikulu M, et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 2004;68:1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Yoder OC, Turgeon BG. Whole-genome analysis of two-component signal transduction genes in fungal pathogens. Eukaryot. Cell. 2003;2:1151–1161. doi: 10.1128/EC.2.6.1151-1161.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Stewart RC. The two-component system. Regulation of diverse signaling pathways in prokaryotes and eukaryotes. Plant Physiol. 1998;117:723–731. doi: 10.1104/pp.117.3.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HS, Kim JR, Lee SW, Cho KH. Why have serine/tyrosine kinases been evolutionarily selected in eukaryotic signaling cascades? Comput. Biol. Chem. 2008;32:218–221. doi: 10.1016/j.compbiolchem.2008.02.005. [DOI] [PubMed] [Google Scholar]
- Davis RH, de Serres FJ. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17A:79–143. [Google Scholar]
- Ellis SW, Grindle M, Lewis DH. Effect of osmotic stress on yield and polyol content of dicarboximide-sensitive and -resistant strains of Neurospora crassa. Mycol. Res. 1991;95:457–464. [Google Scholar]
- Emerson S, Emerson MR. Production, reproduction, and reversion of protoplast-like structures in the osmotic strain of Neurospora crassa. Proc. Natl. Acad. Sci. USA. 1958;44:668–671. doi: 10.1073/pnas.44.7.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster H, Driever GF, Thompson DC, Adaskaveg JE. Postharvest decay management for stone fruit crops in California using the “reduced-risk” fungicides fludioxonil and fenhexamid. Plant Dis. 2007;91:209–215. doi: 10.1094/PDIS-91-2-0209. [DOI] [PubMed] [Google Scholar]
- Fujimura M, Ochiai N, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. Sensitivity to phenylpyrrole fungicides and abnormal glycerol accumulation in os and cut mutant strains of Neurospora crassa. J. Pestic. Sci. 2000;25:31–36. [Google Scholar]
- Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- Grindle M, Temple W. Fungicide-resistance of os mutants of Neurospora crassa. Neurospora Newslett. 1982;29:16–17. [Google Scholar]
- Jones CA, Greer-Phillips SE, Borkovich KA. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell. 2007;18:2123–2136. doi: 10.1091/mbc.E06-03-0226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanetis L, Forster H, Jones CA, Borkovich KA, Adaskaveg JE. Characterization of genetic and biochemical mechanisms of fludioxonil and pyrimethanil resistance if field isolates of Penicillium digitatum. Phytopathology. 2008;98:205–214. doi: 10.1094/PHYTO-98-2-0205. [DOI] [PubMed] [Google Scholar]
- Kawasaki L, Sanchez O, Shiozaki K, Aguirre J. SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol. Microbiol. 2002;45:1153–1163. doi: 10.1046/j.1365-2958.2002.03087.x. [DOI] [PubMed] [Google Scholar]
- Kojima K, Takano Y, Yoshimi A, Tanaka C, Kikuchi T, Okuno T. Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 2004;53:1785–1796. doi: 10.1111/j.1365-2958.2004.04244.x. [DOI] [PubMed] [Google Scholar]
- Krystofova S, Borkovich KA. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gbetagamma dimer required for normal female fertility, asexual development, and Galpha protein levels in Neurospora crassa. Eukaryot. Cell. 2005;4:365–378. doi: 10.1128/EC.4.2.365-378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingston LR. Locus-specific changes in cell wall composition characteristic of osmotic mutants of Neurospora crassa. J. Bacteriol. 1969;99:85–90. doi: 10.1128/jb.99.1.85-90.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai N, Fujimura M, Motoyama T, Ichiishi A, Usami R, Horikoshi K, Yamaguchi I. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest Manag. Sci. 2001;57:437–442. doi: 10.1002/ps.302. [DOI] [PubMed] [Google Scholar]
- Pandey A, Roca MG, Read ND, Glass NL. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryot. Cell. 2004;3:348–358. doi: 10.1128/EC.3.2.348-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G, Pan S, Borkovich KA. Mitogen-activated protein kinase cascade required for regulation of developmental and secondary metabolism in Neurospora crassa. Eukaryot. Cell. 2008;7:2113–2122. doi: 10.1128/EC.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G, Jones C, Borkovich K. Signal transduction pathways. In: Borkovich KA, Ebbole DJ, editors. Cellular and Molecular Biology of Filamentous Fungi. 2009. in press. [Google Scholar]
- Pillonel C, Meyer T. Effect of phenylpyrroles on glycerol accumulation and protein kinase activity of Neurospora crassa. Pest Manag. Sci. 1997;49:229–236. [Google Scholar]
- Posas F, Wurgler-Murphy SM, Maeda T, Witten EA, Thai TC, Saito H. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr. Opin. Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- Selitrennikoff C, Lilley B, Zucker R. Formation and regeneration of protoplasts derived from a temperature-sensitive osmotic strain of Neurospora crassa. Exp. Mycol. 1981;5:155–161. [Google Scholar]
- Vitalini MW, de Paula RM, Goldsmith CS, Jones CA, Borkovich KA, Bell-Pedersen D. Circadian rhythmicity mediated by temporal regulation of the activity of p38 MAPK. Proc. Natl. Acad. Sci. USA. 2007;104:18223–18228. doi: 10.1073/pnas.0704900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ. Distribution of lysine pathways among fungi: Evolutionary implications. Am. Nat. 1964;98:435–446. [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Wolanin PM, Thomason PA, Stock JB. Histidine protein kinases: Key signal transducers outside the animal kingdom. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-reviews3013. REVIEWS3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi A, Kojima K, Takano Y, Tanaka C. Group III histidine kinase is a positive regulator of Hog1-type mitogen-activated protein kinase in filamentous fungi. Eukaryot. Cell. 2005;4:1820–1828. doi: 10.1128/EC.4.11.1820-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lamm R, Pillonel C, Lam S, Xu J-R. Osmoregulation and fungicide resistance: The Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Mircobiol. 2002;68:532–538. doi: 10.1128/AEM.68.2.532-538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]