Abstract
Background
Polymorphisms in chemokine (C-C motif) receptors 2 and 5 genes (CCR2 and CCR5) have been associated with HIV-1 infection and disease progression. We investigated the impact of CCR2-CCR5 haplotypes on HIV-1 viral load (VL) and heterosexual transmission in an African cohort. Between 1995 and 2006, cohabiting Zambian couples discordant for HIV-1 (index seropositive and HIV-1 exposed seronegative {HESN}) were monitored prospectively to determine the role of host genetic factors in HIV-1 control and heterosexual transmission. Genotyping for eight CCR2 and CCR5 variants resolved nine previously recognized haplotypes. By regression and survival analytic techniques, controlling for non-genetic factors, we estimated the effects of these haplotypic variants on a) index partner VL, b) seroconverter VL, c) HIV-1 transmission by index partners, d) HIV-1 acquisition by HESN partners.
Results
Among 567 couples, 240 virologically linked transmission events had occurred through 2006. HHF*2 homozygosity was associated with significantly lower VL in seroconverters (mean beta = -0.58, log10 P = 0.027) and the HHD/HHE diplotype was associated with significantly higher VL in the seroconverters (mean beta = 0.54, log10 P = 0.014) adjusted for age and gender in multivariable model. HHD/HHE was associated with more rapid acquisition of infection by the HESNs (HR = 2.0, 95% CI = 1.20-3.43, P = 0.008), after adjustments for index partner VL and the presence of genital ulcer or inflammation in either partner in Cox multivariable models. The HHD/HHE effect was stronger in exposed females (HR = 2.1, 95% CI = 1.14-3.95, P = 0.018).
Conclusions
Among Zambian discordant couples, HIV-1 coreceptor gene haplotypes and diplotypes appear to modulate HIV-1 VL in seroconverters and alter the rate of HIV-1 acquisition by HESNs. These associations replicate or resemble findings reported in other African and European populations.
Background
Sub-Saharan Africa is home to about 10% of the world's population but bears nearly 64% of all HIV-1 infections [1], with most HIV-1 transmission occurring heterosexually. In Zambia, about one in five cohabiting couples involves an HIV-1 seropositive (index) and a seronegative (exposed) partner; these serodiscordant couples are at high risk of heterosexual transmission, with an estimated rate of eight transmission events per 100 person-years of follow-up [2].
The rate of within-couple heterosexual HIV-1 transmission is highly variable, and a number of viral, host and environmental factors may modify transmission (infectiousness), acquisition (susceptibility) or both [3]. Donor HIV-1 viral load (VL), age, sex, history of sexually transmitted infection (STI), unprotected sex, and possible HIV-1 subtype are among the major factors implicated [4,5]. In southern Africa, unusual biological features of the predominant C subtype of HIV-1 [5] and absence of the human CC chemokine receptor 5 gene (CCR5) 32-bp deletion (Δ32) as a resistance factor may contribute to relatively high transmission rate.
The recognition that Caucasians who are homozygous for CCR5-Δ32 are highly resistant to HIV-1 infection was a landmark finding in research on HIV-1 transmission [6-9]. It stimulated a concerted effort to elucidate the impact of other genetic variations in CCR5 and the adjacent gene CCR2 on HIV-1 transmission and disease progression [10-12]. Research on the association of these variants with transmission has been largely cross-sectional or retrospective; the few prospective studies have focused on vertical (mother-to-child) transmission [13] and on HIV-1 exposed seronegatives (HESNs), in part because of the difficulty in enrolling and following HIV-1-discordant couples.
In Zambia, thousands of cohabiting and HIV-1 discordant couples have been offered voluntary counseling and testing (VCT) services since 1995 [2], and some of them have been followed for more than 10 years [14]. Despite counseling and behavioral interventions, the rate of HIV-1 transmission among these couples has remained high [15]. This circumstance permitted us to investigate the association of polymorphism in CCR2 and CCR5 with heterosexual transmission of phylogenetically related [16] HIV-1 within discordant partners.
The major published studies [11,13,17-20] examining the effects of CCR2 and CCR5 SNPs/haplotypes/diplotypes on HIV-1 infection or disease progression have shown a wide spectrum of effects in various populations (See Additional File 1; Table S1). We systematically tested hypotheses on these as well as other markers that occurred frequently enough in our population to permit meaningful inferences, especially in confirmation of earlier findings.
Results
General characteristics of Zambian couples with linked HIV-1 viruses
During the study period 567 couples were eligible for analysis. Linked transmission occurred in 240 of the 567; (Table 1). nearly all (> 95%) of the HIV-1 sequences from these transmission pairs corresponded to viral subtype C (HIV-1C) [16]. Male-to-female transmission accounted for nearly three-fifths of the incident infections (Table 1). The younger age of exposed women and, to a lesser extent, exposed men was associated with seroconversion. Certain non-genetic characteristics of the partners were also independently associated with increased transmission: genital ulcers or genital inflammation in any partner (HR = 3.62, 95% CI: 2.65-4.93, P < 0.0001) and high VL in the index partner (HR = 1.59, 95% CI: 1.32-1.91, P < 0.0001). These factors were retained in subsequent models that tested the impact of genetic markers.
Table 1.
Demographic, epidemiologic and virologic characteristics of the HIV-1 nontransmission and transmission serodiscordant Zambian couples
| Nontransmission couples | Transmission couples | P | |
|---|---|---|---|
| Characteristic | |||
| Number of couples | 327 | 240 | |
| Male/Female (index partner) | 148/179 | 147/93 | 0.0002 |
| Age of partners (yrs) | |||
| Index | 31.5 ± 7.9 | 30.6 ± 7.8 | 0.170 |
| Exposed | 32.0 ± 8.3 | 28.6 ± 7.3 | < 0.0001 |
| Follow-up time (median [IQR], months) | 31.5 [17.0-56.1] | 17.7 [8.8-36.2] | < 0.0001 |
| Male circumcised | |||
| Index | 8.8% | 9.0% | 0.970 |
| Exposed | 19.5% | 9.9% | 0.053 |
| Genital ulcers | |||
| Index | 12.8% | 36.6% | < 0.0001 |
| Exposed | 4.1% | 26.3% | < 0.0001 |
| Genital inflammation a | |||
| Index | 10.3% | 26.3% | < 0.0001 |
| Exposed | 6.6% | 29.1% | < 0.0001 |
| Any sexually transmitted disease | |||
| Index | 20.7% | 50.2% | < 0.0001 |
| Exposed | 10.0% | 46.0% | < 0.0001 |
| HIV-1 RNA level (log10) in index partner | 4.47 ± 0.90 | 4.96 ± 0.70 | < 0.0001 |
| HIV-1 RNA level (log10) in seroconverted partner | 4.50 ± 0.80 | NA | |
a In the 3-6 months before HIV-1 transmission (transmission couples) or latest follow-up visit (nontransmission couples).
Distribution of CCR2-CCR5 haplotypes in Zambian couples
Eight CCR2-CCR5 haplotypes were observed in the frequency distribution shown in Table 2. Nearly 50% of all haplotypes were HHA or HHF*2. Haplotype HHB was rarely seen, and the Δ32-containing haplotype HHG*2 was not observed at all. The most common genotypes (diplotypes) were HHA/HHF*2, HHA/HHD, HHA/HHA, HHA/HHE, HHD/HHF*2, and HHE/HHF*2 (See Additional File 2; Table S2). The overall distribution of CCR2-CCR5 haplotypes did not conform to HWE (Table 2). After stratification of the cohort into transmission and nontransmission index partners, seroconverters, and exposed uninfected partners, the haplotype distribution deviated significantly from HWE in all three seropositive groups, but not in the HESNs.
Table 2.
Frequencies of CCR2-CCR5 polymorphisms among HIV-1 serodiscordant couples, index partners, and HIV-1 exposed seronegative partners
| HESNa partners | |||||
|---|---|---|---|---|---|
| All (N = 1134) | Index partners (N = 567) | All (N = 567) | Seroconverters (N = 240) | Uninfected (N = 327) | |
| Haplotype | N (%) | N (%) | N (%) | N (%) | N (%) |
| HHA | 601 (26.5) | 305 (26.9) | 296 (26.1) | 133 (27.7) | 163 (24.9) |
| HHB | 44 (1.9) | 25 (2.2) | 19 (1.7) | 8 (1.7) | 11 (1.7) |
| HHC | 178 (7.9) | 69 (6.1) | 109 (9.6) | 42 (8.8) | 67 (10.2) |
| HHD | 370 (16.3 | 190 (16.8) | 180 (15.9) | 75 (15.6) | 105 (16.1) |
| HHE | 310 (13.7) | 138 (12.2) | 172 (15.2) | 61 (12.7) | 111 (17.0) |
| HHF*1 | 128 (5.6) | 76 (6.7) | 52 (4.6) | 24 (5.0) | 28 (4.3) |
| HHF*2 | 480 (21.2) | 259 (22.8) | 221 (19.5) | 99 (20.6) | 122 (18.7) |
| HHG*1 | 157 (6.9) | 72 (6.4) | 85 (7.5) | 38 (7.9) | 47 (7.2) |
| HWE:Pb | 0.0001 | 0.0001 | 0.267 | 0.024 | 0.160 |
a HESN = HIV-1 exposed seronegative
b P values for tests of Hardy-Weinberg equilibrium in each of the patient groups.
CCR2-CCR5 determinants of VL
Although HHA and HHC have previously shown protective effects in the form of associations with lower VL in a mixed population[21], we did not observe such an effect on VL in Zambians with either haplotype overall or with any specific diplotypes containing either of them.
In prior studies, HHF*2 has shown somewhat inconsistent associations with VL and disease control [11,17,19,20,22,23]. In our Zambian study population HHF*2 showed a weak association with lower VL in both index partners (β = -0.21, log10 P = 0.024) and seroconverters (β = -0.10, log10 P = 0.089). When the index partners and seroconverters were stratified by HHF*2 genotype, a stronger association in the latter group was largely attributable to HHF*2 homozygosity (β = -0.70, log10 P = 0.007) (Table 3).
Table 3.
The impact of CCR2-CCR5 haplotypes on HIV-1 viral load in Zambian index partners and seroconverters
| Index partners | Recent seroconverters | |||||
|---|---|---|---|---|---|---|
| (N = 567) | (N = 240) | |||||
| Viral Load Table for CCR5 haplotype/diplotype | ||||||
| Haplotype/diplotype | N | β ± SEa | P* | N | β ± SEa | P* |
| HHF*2 | 232 | -0.21 ± 0.09 | 0.024 | 89 | -0.10 ± 0.58 | 0.089 |
| HHF*2/HHF*2 | 27 | -0.08 ± 0.16 | 0.625 | 10 | -0.70 ± 0.26 | 0.007 |
| HHD (All) | 164 | -0.12 ± 0.06 | 0.052 | 69 | 0.24 ± 0.10 | 0.021 |
| HHE (All) | 131 | 0.13 ± 0.08 | 0.096 | 61 | 0.12 ± 0.12 | 0.339 |
| HHD/HHE | 18 | -0.22 ± 0.20 | 0.270 | 16 | 0.49 ± 0.21 | 0.020 |
| HHD/X (No HHE) | 146 | -0.11 ± 0.08 | 0.157 | 53 | 0.14 ± 0.13 | 0.284 |
| HHE/X (No HHD) | 113 | 0.19 ± 0.09 | 0.026 | 45 | -0.06 ± 0.13 | 0.673 |
| Multivariable Model for Interaction** | ||||||
| Haplotype/diplotype | N | β ± SEa | P* | N | β ± SEa | P* |
| HHD/X (No HHE or HHF*2) | 109 | -0.16 ± 0.09 | 0.560 | 37 | 0.24 ± 0.16 | 0.118 |
| HHE/X (No HHD or HHF*2) | 69 | 0.36 ± 0.11 | 0.002 | 31 | 0.10 ± 0.16 | 0.556 |
| HHD/HHE | 18 | -0.16 ± 0.20 | 0.444 | 16 | 0.54 ± 0.22 | 0.014 |
| HHF*2/HHF*2 | 27 | -0.02 ± 0.16 | 0.918 | 10 | -0.58 ± 0.26 | 0.027 |
| HHF2/X (No HHD, HHE or HHF*2) | 124 | 0.05 ± 0.09 | 0.602 | 49 | 0.10 ± 0.14 | 0.496 |
a SE, standard error of the β estimate according to linear regression models.
* P value adjusted for the sex and age at VL for all the individuals.
** Individuals with HHD/HHF*2, HHE/HHF*2 diplotypes are in the reference group in the multivariable model for interaction.
Both HHD and HHE have been associated with higher VL in several studies [11,13,18,24,25]. In our Zambian cohort, dominant models, including each haplotype plus non-genetic factors analyzed by GLM, indicated that HHD was associated with higher VL (β = 0.24, log10 P = 0.021) in the seroconverters, but a modest effect in the opposite direction was observed in index partners. HHE showed a trend toward association with higher VL in index partners and a similar non-significant association with higher VL in seroconverters adjusted for age and gender (Table 3). Because this pattern of association could be explained by combinations of haplotypes carried, we explored the effect of diplotypes further. Among all diplotypes of frequency > 0.05, HHD/HHE showed the strongest association with higher VL (β = 0.49, log10 P = 0.02)
We next constructed a multivariable model with all the haplotypes and diplotypes that showed a trend toward association (log10 P < 0.10) with higher or lower VL in either index partners or seroconverters to test their independent influences on VL (Table 3). In this model, by including uninformative diplotypes (HHD/HHF*2 and HHE/HHF*2) in the reference group, each diplotype implicated could be tested independently of the others. In the index partners, HHE/X shows a strong association with higher VL. In seroconverters the HHD/HHE and HHF*2/HHF*2 diplotypes remained significantly and independently associated with VL after controlling for individual haplotype effects (Table 3).
CCR2-CCR5 determinants of transmission from index partners and of seroconversion in HESNs
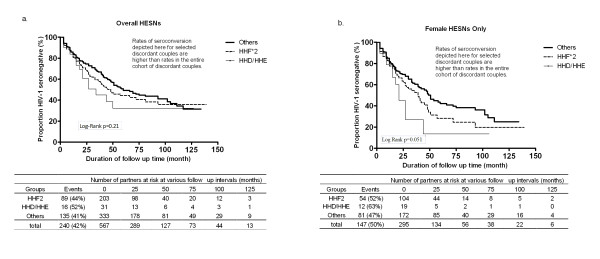
The few studies that have attempted to assess the role of the receptor polymorphism in transmission and susceptibility have shown rather diverse associations of common CCR5 haplotypes, without any discernible pattern (See Additional File 1, Table S1). No SNP or haplotype carried by Zambian index partners was significantly associated with transmission (data not shown). In the survival analysis, HESNs with the HHD/HHE diplotypes showed significantly more rapid seroconversion than HESNs with other haplotypes (Table 4 and Figure 1a) after adjustments for index partner VL and the presence of genital ulcer or inflammation in either partner. Although HHF*2 did not show statistically significant association with faster HIV-1 acquisition, we assigned it to a separate stratum in the Kaplan-Meier plot because aggregating it in the reference group would have given the appearance of a weaker HHD/HHE effect (Multivariable Cox model HR = 2.0, 95% CI = 1.20-3.43, P = 0.008). Stratification by gender revealed a stronger impact of HHD/HHE on HESN women than men (Table 4 and Figure 1b) (Multivariable Cox model HR = 2.1, 95% CI = 1.14-3.95, P = 0.018).
Table 4.
Proportional hazards analysis of the effect of CCR2-CCR5 haplotype or diplotype on HIV-1 acquisition.
| Overall | Male-to-Female | Female-to-Male | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (567 couples) | (295 couples) | (272 couples) | ||||||||||
| Cox model for individual CCR2-CCR5 haplotype or diplotype. | ||||||||||||
| Haplotype/diplotype | N* | HR | 95% CI | P** | N* | HR | 95% CI | P** | N* | HR | 95% CI | P** |
| HHF*2 | 203 | 1.1 | 0.85-1.46 | 0.417 | 104 | 1.1 | 0.77-1.56 | 0.61 | 99 | 1.2 | 0.75-1.75 | 0.531 |
| HHD | 167 | 1.0 | 0.74-1.32 | 0.983 | 94 | 0.9 | 0.59-1.23 | 0.385 | 73 | 1.2 | 0.75-1.92 | 0.442 |
| HHE | 158 | 1.1 | 0.82-1.51 | 0.495 | 85 | 1.3 | 0.86-1.87 | 0.234 | 73 | 0.9 | 0.57-1.53 | 0.781 |
| HHD/HHE | 31 | 1.9 | 1.14-3.16 | 0.015 | 19 | 2.0 | 1.08-3.57 | 0.028 | 12 | 1.7 | 0.60-4.66 | 0.321 |
| Multivariable model for CCR2-CCR5 HHD/HHE diplotype and HHF*2 haplotype. | ||||||||||||
| Genetic factors | N* | HR | 95% CI | P** | N* | HR | 95% CI | P** | N* | HR | 95% CI | P** |
| HHD/HHE | 31 | 2.0 | 1.20-3.43 | 0.008 | 19 | 2.1 | 1.14-3.95 | 0.018 | 12 | 1.8 | 0.64-5.08 | 0.267 |
| HHF*2 | 203 | 1.2 | 0.90-1.57 | 0.222 | 104 | 1.2 | 0.83-1.72 | 0.337 | 99 | 1.2 | 0.77-1.82 | 0.439 |
| Any genital ulcer or inflammation | 299 | 3.6 | 2.65-4.93 | < .0001 | 162 | 3.0 | 2.04-4.51 | < .0001 | 137 | 4.6 | 2.79-7.65 | < .0001 |
| Donor VL (per 1.0 log10 unit) | 523 | 1.6 | 1.32-1.91 | < .0001 | 263 | 1.3 | 1.03-1.74 | 0.028 | 260 | 1.8 | 1.35-2.47 | < .0001 |
*N represents the number of HESNs with each genotype.
**P values adjusted for genital ulcer, genital inflammation in either partner, and index partner log10 VL.
Figure 1.
Association of two CCR2-CCR5 diplotype (HHD/HHE and HHF*2) with time to HIV-1 acquisition among initially seronegative partners of HIV-1 discordant Zambian couples. Analysis was based on 567 initially seronegative HESNs (panel a) or 295 seronegative female HESNs only (panel b). Vertical lines on each Kaplan-Meier curve represent subjects censored at the last follow up visit (before December 2006).
Discussion
Many investigations into genetic determinants of HIV/AIDS have evaluated the effects of Δ32, selected SNPs, and haplotypes across CCR2-CCR5 on disease progression in a variety of infected populations. Studies of these markers as determinants of acquisition have usually been conducted in pairs of mothers and infants or in exposed men of European ancestry whose male sexual contacts are largely unknown [13,17,25-27]. Our relatively large prospective study of heterosexual discordant African couples has produced further evidence for involvement of variants in these genes in both control and occurrence of HIV-1 infection.
HHE was associated with slightly higher VL than was seen with other haplotypes, a finding consistent with observations in a number of other studies on different ethnic groups and various modes of transmission [11,17,28,29]. Further confirmation of the effect of HHE highlights its potential impact on clinical HIV-1 disease control in diverse populations, in contrast to that of the protective Δ32 variant whose distribution is confined to individuals of European ancestry. We detected an association of homozygous HHF*2 (containing CCR2-64I) with lower VL in recent seroconverters but found less certain effects of heterozygous HHF*2. This finding is consistent with previous reports [11,17,19,20,22]. Although an early meta-analysis persuasively documented modest protection by the 64I allele against progression of HIV-1 subtype B infection [22], results in subsequent studies have been less consistent--showing association with slow progression either among Europeans, but not African-Americans [19,20] or among African-Americans but not Europeans [23,29]. For populations with subtype C infection, however, no previous study is available as a basis for comparison.
As for the influence of CCR2-CCR5 alleles or haplotypes on transmission and acquisition of infection, the highly significant deviation of the distribution of haplotypes from HWE among the index, but not the exposed partners, was strong evidence of a selective effect, and the differential deviation of the seroconverters, but not the persistently seronegatives, corroborated the difference. Neither chance nor systematic selection of couples into the study cohort by their CCR2-CCR5 profile unrelated to infection seems as plausible an explanation as the direct effect on acquisition of HIV-1 infection proposed here.
No CCR2-CCR5 variant carried by index partners was associated with an appreciable difference in transmission--not even the diplotype HHD/HHE associated with a statistically significant higher mean VL. This relative deviation in level of viremia was apparently not equivalent to the larger deviation conferred by index partner HLA-B*57, a genetic marker associated with a significantly lower transmission rate in this population [14]. Such differential impact of the different genetic markers may reflect a threshold effect by which a deviation of VL greater than a certain level overrides any genetic influence, but the number of subjects in our cohort was insufficient to assess that possibility.
We observed a trend toward an increased rate of acquisition among the exposed partners carrying HHF*2. In another African population (Cameroon), the frequency of CCR2-64I (HHF*2) was higher in the HIV-1 seropositives (most likely of mixed viral subtype) than in the seronegatives [30]. However, we remain skeptical about the importance of these findings for several reasons. First, the association and its significance in Zambians diminished in the multivariable analysis. Second, previous evidence for a role of HHF*2 in occurrence of infection is sparse, and there is no other report from a prospective study. Third, considerable uncertainty remains about the functional relevance of the HHF*2 polymorphism and CCR2 itself to HIV-1 infection [31]. Further population studies alone are unlikely to clarify more precisely the true nature of this genetic contribution.
More rapid HIV-1 acquisition among exposed seronegatives occurred in association with the HHD/HHE diplotype, and the association was stronger in exposed women than men. An association with this diplotype has not been reported before, most likely because the single SNP allele that distinguishes HHD from other haplotypes is only frequent enough in persons of African ancestry. The relatively higher frequency (7%) of HHD/HHE in our population than in Caucasians or other smaller groups of Africans may have facilitated detection of its effect. Associations with higher risk of mother-to-child transmission have been reported for HHD in Africans [32] and with homozygous HHD in African Americans [33]. HHE has also been reported to be detrimental for HIV infection as well as disease progression, but HHD/HHE has not been studied previously as a diplotype. Although our findings do not constitute exact replication of previous work, they appear to indicate consistent effects of the two haplotypes across populations with different viral subtypes.
The effects of HHD/HHE appeared stronger in male-to-female transmission. Differences in VL among the donor groups did not explain this difference according to direction of transmission. Nor did the difference arise from any obvious difference in age or sexual exposure of the two groups. For each subgroup stratified by gender, the number of seronegative subjects carrying these genotypes (diplotypes) was relatively small. Analysis based on larger samples will be necessary to reach a reliable conclusion about such gender-specific associations.
One feature of our study worth noting is the advantage of survival analysis of time to transmission/acquisition in detecting relationships that may be weaker in the cross-sectional or case-control approach often used to assess genetic influences on HIV-1 infection. Survival methods may be more sensitive in capturing time-dependent genetic effects on infection just as they have been in the analysis of disease progression.
We did not adjust statistically for the number of genetic polymorphisms tested. Rather we have emphasized those nominally significant associations with CCR2-CCR5 variants that have previously been implicated in HIV/AIDS and de-emphasized those whose involvement was less predictable from earlier studies. The previously documented HHE association with higher VL [11,17,28,29] provided ample rationale for interpreting our results as confirmatory without treating all haplotypes as equally likely to be involved. The impact of HHD/HHE on seroconversion was predicted somewhat less directly by earlier work associating HHD with a higher frequency of neonatal infection [33]. An even more important reason why these relationships cannot be readily dismissed as chance findings is that they were observed in the context of significant deviations from HWE of the haplotype distributions in each of the seropositive groups but not the seronegative group.
A consistent effect of the frequent HHE with higher VL in subtype C HIV-1-infected Africans as well as subtype B-infected Europeans and a stronger effect of HHD/HHE could have further ramifications. Since the response to antiretroviral treatment in Europeans may be modified by (Δ32) [34-36] and perhaps by other receptor variants [37,38], investigators in African settings should consider whether similar studies of CCR2-CCR5 polymorphism might provide epidemiologically or clinically useful prognostic information.
Conclusions
In summary, our analysis of CCR2-CCR5 haplotypes consisting of common combinations of SNP alleles spanning those two genes has confirmed a previously reported association of haplotype HHF*2 with favorable response to HIV-1 infection; and our longitudinal analysis of seroconversion in HESN African heterosexual partners has detected probable contributions by the HHD/HHE diplotype to acquisition of infection [11,17,39]. Further insight into these relationships will be gained from studies of correlation between gene variation and gene function, as well as investigation of other representative and informative populations of infected and uninfected Africans.
Methods
Study population
Our study population comprised HIV-1 serodiscordant, cohabiting heterosexual couples enrolled in the Zambia-Emory HIV Research Project between 1995 and 2006. The procedures for screening, recruitment, counseling, follow-up visits and laboratory testing have been described elsewhere [15,40]. All couples whose HESN partner acquired virologically linked HIV-1 from the index partner during follow-up were included in this study. For closer comparability to the transmitters, nontransmission couples were selected from a large number based on self-reported behavioral or clinical measures of unprotected sex. Virologically linked HIV-1 transmission was defined as identity between viruses from index and seroconverting partners, according to phylogenetic analysis of sub-genomic sequences of gag, env (gp120 and gp41), and long terminal repeat regions [16,40]. Participant characteristics have previously been thoroughly examined as potential risk factors for transmission in this cohort [15,16,40,41]. Risk factors considered here include index partner (donor) viral load (VL), age of each partner, and genital ulceration/inflammation in each partner. The study population consisted of 567 couples with: a) adequate data and biologic material for both partners, b) observation of nontransmission couples for at least nine months, c) intra-couple virologic linkage when transmission occurred, and d) none of the partners on anti-retroviral treatment.
Non-genetic factors
VL was quantified as the number of HIV-1 RNA copies per ml of plasma using Roche Amplicor 1.0 assay (Roche diagnostic Systems Inc., Branchburg, NJ) in a laboratory certified by the virology quality assurance program of the AIDS Clinical Trials Group (ACTG). The lower detection limit was 400 copies/mL of plasma. For this work, VL was transformed to log10 and treated as a continuous variable. Previous analyses [40] indicated that index partners with a medium number of HIV-1 RNA copies/mL (104-105, log10 = 4-5) or a high number of copies/mL (> 105, log10 > 5) were more likely to transmit the virus than those with a low number (< 104, log10 < 4).
Genotyping
Genomic DNA was extracted from whole blood and buffy coats using the QIAamp blood kit and protocols recommended by the manufacturer (QIAGEN Inc., Valencia, CA). PCR-based typing differentiated the dimorphic variants at eight sites--one in CCR2 (the SNP encoding V64I--rs1799864) and seven in CCR5 [six SNPs in or adjacent to the cis-regulatory or promoter region (A29G--rs2856758, G303A--rs1799987, T627C--rs1799988, C630T--rs41469351, A676G--rs1800023 and C927T--rs1800024)] and the 32-bp deletion (Δ32--rs333). CCR5 haplotypes were typed by a combination of two methods: a PCR typing scheme and a TaqMan SNP typing scheme. The PCR typing scheme used 12 combinations of sequence-specific primers (SSP) plus four additional SSP reactions in conjunction with T627C-specific primers to define the A29G variant as described for previous work [11,13,17-20]. Combination of variants at the eight sites form nine relatively frequent CCR2-CCR5 haplotypes (HHA-HHE, HHF*1, HHF*2, HHG*1 and HHG*2) according to the nomenclature of the Tri-Service HIV-1 Natural History Study (TSS) [42]. HHF*2 is the only haplotype carrying the V64I mutation. A TaqMan genotyping assay was used to confirm the PCR-based SNP typing and assign CCR5 haplotypes for 126 individuals. TaqMan assays were performed using customized TaqMan probes for 7 SNP sites; SNP alleles were assigned after real-time PCR using the ABI 7500 Fast System (Applied Biosystems) according to procedures recommended by the manufacturer.
Statistical analysis
Non-genetic factors (VL, age, gender, genital ulcer, genital inflammation, circumcision, and presence of sperm) were compared between seroconverting and non-seroconverting exposed partners using χ2 and t-tests. Hardy-Weinberg equilibrium (HWE) for each SNP and CCR haplotype distribution was assessed using SAS Genetics (see below). HWE was calculated for the entire cohort and for four separate partner groups: transmission index, nontransmission index, seroconverting, and exposed uninfected partners. Associations of frequent haplotypes/genotypes with HIV-1 VL among the index partners and seroconverters were tested using general linear model (GLM) statistics with adjustment for age and gender.
For analysis of time-to-infection (transmission and acquisition), follow-up time for each couple was measured from the date of their enrollment into the cohort to 1) the date of HIV-1 infection (first seropositive visit) of the initially uninfected exposed partner or 2) the most recent seronegative visit prior to administrative censoring date (December 31, 2006). Time-to-infection was displayed in Kaplan-Meier plots, and comparisons between genetically distinctive groups were evaluated with Wilcoxon and log-rank tests. These plots illustrate differences in transmission associated with specific genetic markers; they do not reflect transmission rates in the entire prospectively observed discordant couple population. The overall annual HIV-1 seroincidence (7-8/100 PY) represents a one-half to two-thirds reduction in transmission following joint testing and counseling.
Statistical analysis of genetic variants of CCR2 and CCR5 consisted of testing hypotheses derived from earlier work on acquisition or progression of infection (See Additional File 1; Table S1) followed by systematic search for novel associations in our study population. Multivariable Cox proportional hazards models were used to control for non-genetic covariates. We estimated the hazard ratios (HR), its 95% confidence interval (CI), and the corresponding two-sided P-values. For hypotheses on genetic markers consistent with previously reported associations, statistical testing was performed without correction for multiple comparisons. All statistical analyses were done using SAS® 9.2 including SAS/Genetics™ (SAS Institute Inc., Cary, NC).
List of Abbreviations
CCR5: C-C chemokine receptor 5; CCR2: C-C chemokine receptor 2; AIDS: acquired immunodeficiency syndrome; HIV: human immunodeficiency virus; HR: hazard ratio; VL: viral load; SSP: sequence-specific primers; SNP: single nucleotide polymorphism; GLM: general linear model; HWE: Hardy-Weinberg equilibrium; HHA, etc: human haplotype A, etc.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RM* performed the statistical analyses and participated in the preparation of multiple drafts of the manuscript. LH* performed the laboratory work, participated in the statistical analyses and participated in the preparation of multiple drafts of the manuscript. WS assisted in planning the laboratory work, performed the assays, and reviewed the manuscript. IB prepared analytic data sets of the clinical, epidemiologic, and genetic data; and assisted in editing the manuscript. JM organized the cohort studies, supervised the data collection in the field and reviewed the manuscript. SA conceived the cohort studies, participated in the design of the genetic substudies, and reviewed the analyses and the manuscript. EH participated in the design of the genetic substudies, supervised the performance of the viral sequencing and viral load measurements, and reviewed the manuscript. SS participated in the analyses and in the editing of the manuscript. JT participated in the design of the genetic substudies, supervised all aspects of the genotyping, participated in the analyses and reviewed multiple drafts of the manuscript. RAK conceived the genetics studies, supervised the statistical analyses, and reviewed and edited all drafts of the manuscript.
Supplementary Material
Table S1: Studies of associations between polymorphisms in CCR2 and CCR5 and acquisition or progression of HIV-1 infection. Summary of the recent publications on CCR2-CCR5 haplotypes and association with HIV-1 acquisition or disease progression. Includes references [43-45]
Table S2: CCR2-CCR5 haplotypes and diplotypes as observed in HIV-1 discordant Zambian couples. Frequency of CCR2-CCR5 haplotypes and common diplotypes in overall Zambia cohort and subgroups. Rare diplotypes with count less than 12 in overall cohort are not shown.
Contributor Information
Rakhi Malhotra, Email: rakhi_shai@uab.edu.
Liangyuan Hu, Email: lyhu@uab.edu.
Wei Song, Email: wsong@uab.edu.
Ilene Brill, Email: ibrill@uab.edu.
Joseph Mulenga, Email: joseph.mulenga@znbts.gov.zm.
Susan Allen, Email: sallen5@sph.emory.edu.
Eric Hunter, Email: eric.hunter2@emory.edu.
Sadeep Shrestha, Email: sshresth@uab.edu.
Jianming Tang, Email: jtang@uab.edu.
Richard A Kaslow, Email: rkaslow@uab.edu.
Acknowledgements
We thank study participants, staff, interns, and Project Management Group members of the Zambia-Emory HIV Research Project in Lusaka, Zambia; technical staff and students at the virology laboratory at the University Teaching Hospital, Lusaka, the immunogenetics laboratory at UAB School of Public Health; and the data analysis group at UAB school of Public Health.
References
- UNAIDS. Book UNAIDS/WHO "AIDS Epidemic Update: December 2006". City; 2006. UNAIDS/WHO "AIDS Epidemic Update: December 2006". (Editor ed.^eds.) [Google Scholar]
- Dunkle KL, Stephenson R, Karita E, Chomba E, Kayitenkore K, Vwalika C, Greenberg L, Allen S. New heterosexually transmitted HIV infections in married or cohabiting couples in urban Zambia and Rwanda: an analysis of survey and clinical data. Lancet. 2008;371:2183–2191. doi: 10.1016/S0140-6736(08)60953-8. [DOI] [PubMed] [Google Scholar]
- Borrow P, Shattock RJ, Vyakarnam A. Innate immunity against HIV: a priority target for HIV prevention research. Retrovirology. 2010;7:84. doi: 10.1186/1742-4690-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce RA, Sena A, Cates W Jr, Cohen MS. Sexual transmission of HIV. N Engl J Med. 1997;336:1072–1078. doi: 10.1056/NEJM199704103361507. [DOI] [PubMed] [Google Scholar]
- Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D. et al. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Paxton WA, Choe S, Ceradini D, Martin SR, Horuk R, MacDonald ME, Stuhlmann H, Koup RA, Landau NR. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/S0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C. et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- Dean M, Carrington M, Winkler C, Huttley GA, Smith MW, Allikmets R, Goedert JJ, Buchbinder SP, Vittinghoff E, Gomperts E. et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- Zimmerman PA, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G. et al. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]
- McDermott DH, Zimmerman PA, Guignard F, Kleeberger CA, Leitman SF, Murphy PM. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/S0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- Tang J, Shelton B, Makhatadze NJ, Zhang Y, Schaen M, Louie LG, Goedert JJ, Seaberg EC, Margolick JB, Mellors J, Kaslow RA. Distribution of chemokine receptor CCR2 and CCR5 genotypes and their relative contribution to human immunodeficiency virus type 1 (HIV-1) seroconversion, early HIV-1 RNA concentration in plasma, and later disease progression. J Virol. 2002;76:662–672. doi: 10.1128/JVI.76.2.662-672.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CA, Hendel H, Carrington M, Smith MW, Nelson GW, O'Brien SJ, Phair J, Vlahov D, Jacobson LP, Rappaport J. et al. Dominant effects of CCR2-CCR5 haplotypes in HIV-1 disease progression. J Acquir Immune Defic Syndr. 2004;37:1534–1538. doi: 10.1097/01.qai.0000127353.01578.63. [DOI] [PubMed] [Google Scholar]
- Ometto L, Bertorelle R, Mainardi M, Zanchetta M, Tognazzo S, Rampon O, Ruga E, Chieco-Bianchi L, De Rossi A. Polymorphisms in the CCR5 promoter region influence disease progression in perinatally human immunodeficiency virus type 1-infected children. J Infect Dis. 2001;183:814–818. doi: 10.1086/318828. [DOI] [PubMed] [Google Scholar]
- Tang J, Shao W, Yoo YJ, Brill I, Mulenga J, Allen S, Hunter E, Kaslow RA. Human leukocyte antigen class I genotypes in relation to heterosexual HIV type 1 transmission within discordant couples. J Immunol. 2008;181:2626–2635. doi: 10.4049/jimmunol.181.4.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S, Meinzen-Derr J, Kautzman M, Zulu I, Trask S, Fideli U, Musonda R, Kasolo F, Gao F, Haworth A. Sexual behavior of HIV discordant couples after HIV counseling and testing. Aids. 2003;17:733–740. doi: 10.1097/00002030-200303280-00012. [DOI] [PubMed] [Google Scholar]
- Trask SA, Derdeyn CA, Fideli U, Chen Y, Meleth S, Kasolo F, Musonda R, Hunter E, Gao F, Allen S, Hahn BH. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J Virol. 2002;76:397–405. doi: 10.1128/JVI.76.1.397-405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G, Cabrera S, McBride M, Cao XH, Merrill G. et al. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci USA. 1999;96:12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez E, Kulkarni H, Bolivar H, Mangano A, Sanchez R, Catano G, Nibbs RJ, Freedman BI, Quinones MP, Bamshad MJ. et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- Smith MW, Dean M, Carrington M, Winkler C, Huttley GA, Lomb DA, Goedert JJ, O'Brien TR, Jacobson LP, Kaslow R. et al. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- Easterbrook PJ, Rostron T, Ives N, Troop M, Gazzard BG, Rowland-Jones SL. Chemokine receptor polymorphisms and human immunodeficiency virus disease progression. J Infect Dis. 1999;180:1096–1105. doi: 10.1086/314997. [DOI] [PubMed] [Google Scholar]
- Kageyama S, Mimaya J, Yamada K, Kurimura T, Shiraki K. Polymorphism of CCR5 affecting HIV disease progression in the Japanese population. AIDS Res Hum Retroviruses. 2001;17:991–995. doi: 10.1089/088922201300343663. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Rosenberg PS, Goedert JJ, Ashton LJ, Benfield TL, Buchbinder SP, Coutinho RA, Eugen-Olsen J, Gallart T, Katzenstein TL. et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: An international meta-analysis of individual-patient data. Ann Intern Med. 2001;135:782–795. doi: 10.7326/0003-4819-135-9-200111060-00008. [DOI] [PubMed] [Google Scholar]
- Schinkel J, Langendam MW, Coutinho RA, Krol A, Brouwer M, Schuitemaker H. No evidence for an effect of the CCR5 delta32/+ and CCR2b 64I/+ mutations on human immunodeficiency virus (HIV)-1 disease progression among HIV-1-infected injecting drug users. J Infect Dis. 1999;179:825–831. doi: 10.1086/314658. [DOI] [PubMed] [Google Scholar]
- Dorak MT, Tang J, Penman-Aguilar A, Westfall AO, Zulu I, Lobashevsky ES, Kancheya NG, Schaen MM, Allen SA, Kaslow RA. Transmission of HIV-1 and HLA-B allele-sharing within serodiscordant heterosexual Zambian couples. Lancet. 2004;363:2137–2139. doi: 10.1016/S0140-6736(04)16505-7. [DOI] [PubMed] [Google Scholar]
- Nguyen L, Li M, Chaowanachan T, Hu DJ, Vanichseni S, Mock PA, van Griensven F, Martin M, Sangkum U, Choopanya K. et al. CCR5 promoter human haplogroups associated with HIV-1 disease progression in Thai injection drug users. Aids. 2004;18:1327–1333. doi: 10.1097/00002030-200406180-00012. [DOI] [PubMed] [Google Scholar]
- An P, Martin MP, Nelson GW, Carrington M, Smith MW, Gong K, Vlahov D, O'Brien SJ, Winkler CA. Influence of CCR5 promoter haplotypes on AIDS progression in African-Americans. Aids. 2000;14:2117–2122. doi: 10.1097/00002030-200009290-00007. [DOI] [PubMed] [Google Scholar]
- O'Brien SJ, Moore JP. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol Rev. 2000;177:99–111. doi: 10.1034/j.1600-065x.2000.17710.x. [DOI] [PubMed] [Google Scholar]
- Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, Lee B, Doms RW, Margolick J, Buchbinder S. et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, Craig FE, O'Connell P, Tryon V, Clark RA. et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- Ma L, Marmor M, Zhong P, Ewane L, Su B, Nyambi P. Distribution of CCR2-64I and SDF1-3'A alleles and HIV status in 7 ethnic populations of Cameroon. J Acquir Immune Defic Syndr. 2005;40:89–95. doi: 10.1097/01.qai.0000157008.66584.d6. [DOI] [PubMed] [Google Scholar]
- Arenzana-Seisdedos F, Parmentier M. Genetics of resistance to HIV infection: Role of co-receptors and co-receptor ligands. Semin Immunol. 2006;18:387–403. doi: 10.1016/j.smim.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Franceschini N, Mwapasa V, North KE, Meshnick SR. Regulation of CCR5 expression in human placenta: insights from a study of mother-to-child transmission of HIV in Malawi. PLoS One. 2010;5:e9212. doi: 10.1371/journal.pone.0009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrikis LG, Neumann AU, Thomson B, Korber BT, McHardy P, Karanicolas R, Deutsch L, Huang Y, Lew JF, McIntosh K. et al. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J Virol. 1999;73:10264–10271. doi: 10.1128/jvi.73.12.10264-10271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdez H, Purvis SF, Lederman MM, Fillingame M, Zimmerman PA. Association of the CCR5delta32 mutation with improved response to antiretroviral therapy. Jama. 1999;282:734. doi: 10.1001/jama.282.8.734. [DOI] [PubMed] [Google Scholar]
- Kasten S, Goldwich A, Schmitt M, Rascu A, Grunke M, Dechant C, Kalden JR, Harrer T. Positive influence of the Delta32CCR5 allele on response to highly active antiretroviral therapy (HAART) in HIV-1 infected patients. Eur J Med Res. 2000;5:323–328. [PubMed] [Google Scholar]
- Barroga CF, Raskino C, Fangon MC, Palumbo PE, Baker CJ, Englund JA, Spector SA. The CCR5Delta32 allele slows disease progression of human immunodeficiency virus-1-infected children receiving antiretroviral treatment. J Infect Dis. 2000;182:413–419. doi: 10.1086/315704. [DOI] [PubMed] [Google Scholar]
- Wang YM, Wang B, Dyer WB, Lachireddy K, Peng NK, Saksena NK. Evidence for predominance of CCR5-using HIV-1 strains during highly active antiretroviral therapy. Curr HIV Res. 2007;5:221–234. doi: 10.2174/157016207780077048. [DOI] [PubMed] [Google Scholar]
- Puissant B, Roubinet F, Massip P, Sandres-Saune K, Apoil PA, Abbal M, Pasquier C, Izopet J, Blancher A. Analysis of CCR5, CCR2, CX3CR1, and SDF1 polymorphisms in HIV-positive treated patients: impact on response to HAART and on peripheral T lymphocyte counts. AIDS Res Hum Retroviruses. 2006;22:153–162. doi: 10.1089/aid.2006.22.153. [DOI] [PubMed] [Google Scholar]
- Catano G, Chykarenko ZA, Mangano A, Anaya JM, He W, Smith A, Bologna R, Sen L, Clark RA, Lloyd A. et al. Concordance of CCR5 genotypes that influence cell-mediated immunity and HIV-1 disease progression rates. J Infect Dis. 2011;203:263–272. doi: 10.1093/infdis/jiq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fideli US, Allen SA, Musonda R, Trask S, Hahn BH, Weiss H, Mulenga J, Kasolo F, Vermund SH, Aldrovandi GM. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res Hum Retroviruses. 2001;17:901–910. doi: 10.1089/088922201750290023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Tang S, Lobashevsky E, Zulu I, Aldrovandi G, Allen S, Kaslow RA. HLA allele sharing and HIV type 1 viremia in seroconverting Zambians with known transmitting partners. AIDS Res Hum Retroviruses. 2004;20:19–25. doi: 10.1089/088922204322749468. [DOI] [PubMed] [Google Scholar]
- Mangano A, Gonzalez E, Dhanda R, Catano G, Bamshad M, Bock A, Duggirala R, Williams K, Mummidi S, Clark RA. et al. Concordance between the CC chemokine receptor 5 genetic determinants that alter risks of transmission and disease progression in children exposed perinatally to human immunodeficiency virus. J Infect Dis. 2001;183:1574–1585. doi: 10.1086/320705. [DOI] [PubMed] [Google Scholar]
- Pedersen BR, Kamwendo D, Blood M, Mwapasa V, Molyneux M, North K, Rogerson SJ, Zimmerman P, Meshnick SR. CCR5 haplotypes and mother-to-child HIV transmission in Malawi. PLoS One. 2007;2:e838. doi: 10.1371/journal.pone.0000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John GC, Bird T, Overbaugh J, Nduati R, Mbori-Ngacha D, Rostron T, Dong T, Kostrikis L, Richardson B, Rowland-Jones SL. CCR5 promoter polymorphisms in a Kenyan perinatal human immunodeficiency virus type 1 cohort: association with increased 2-year maternal mortality. J Infect Dis. 2001;184:89–92. doi: 10.1086/321006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaley PA, French N, Kaleebu P, Gilks C, Whitworth J, Hill AV. HIV in Africa (Communication arising): chemokine-receptor genes and AIDS risk. Nature. 2002;417:140. doi: 10.1038/417140a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Studies of associations between polymorphisms in CCR2 and CCR5 and acquisition or progression of HIV-1 infection. Summary of the recent publications on CCR2-CCR5 haplotypes and association with HIV-1 acquisition or disease progression. Includes references [43-45]
Table S2: CCR2-CCR5 haplotypes and diplotypes as observed in HIV-1 discordant Zambian couples. Frequency of CCR2-CCR5 haplotypes and common diplotypes in overall Zambia cohort and subgroups. Rare diplotypes with count less than 12 in overall cohort are not shown.