Abstract
Three meiosis-specific chromosomal components in budding yeast, Mek1, Red1, and Hop1, are required for recombination, proper segregation of homologs, and the meiotic recombination checkpoint. Mek1 is a protein kinase. Mutations that increase the size of the ATP binding pocket of Mek1 (mek1-as1) sensitize the kinase to specific small molecule inhibitors. Experiments using mek1-as1 demonstrate that the requirement for Mek1 kinase activity coincides with the formation of double strand breaks (DSBs) and that this activity is necessary after DSB formation to prevent repair by DMC1-independent pathways. Contrary to previous reports, Red1 is not a substrate for Mek1. Instead, RED1 is required for wild-type levels of Mek1 kinase activity. In addition, activation of Mek1 requires HOP1, the formation of Red1/Hop1 complexes and a functional Mek1 FHA domain. The requirement for RED1 to produce active kinase can be bypassed by a mek1 mutation that creates a constitutively active Mek1 kinase. We propose that Red1 is phosphorylated by a kinase other than MEK1 and that phosphothreonines on Red1 then interact with the Mek1 FHA domain to recruit the kinase to sites of DSBs where Mek1 is activated to prevent DMC1-independent DSB repair.
INTRODUCTION
Meiosis is a specialized cell division that produces gametes with half the number of chromosomes as the parental cells. To accomplish this feat, two chromosomal divisions follow just a single round of chromosome replication. At the first meiotic division (MI), homologous chromosomes segregate to opposite poles. To align properly at MI, homologs must be physically connected to each other by crossovers and by sister chromatid cohesion distal to the sites of crossing over. Failure to make these connections results in high levels of chromosome nondisjunction (Bascom-Slack et al., 1997).
Meiotic recombination is initiated by the introduction of double strand breaks (DSBs). DSB formation requires a number of different genes, most notably SPO11, a highly conserved topoisomerase-like protein that catalyzes the break (Keeney, 2001). After resection of the 5′ ends, a 3′ single-stranded (ss) tail invades the homologous duplex. This step is facilitated by DMC1, a meiosis-specific recA homolog that promotes interhomolog crossovers (Bishop et al., 1992; Schwacha and Kleckner, 1997; Hunter and Kleckner, 2001). The single end invasion intermediate is then converted to a double Holliday junction, which is resolved to produce recombinant chromosomes (Allers and Lichten, 2001). Mutations that block at steps subsequent to DSB formation cause a prophase arrest due to triggering the meiotic recombination checkpoint (Roeder and Bailis, 2000).
For crossovers to be functional in promoting MI segregation in budding yeast, they must occur in the context of a proteinaceous structure formed between homologous chromosomes called the synaptonemal complex (SC) (Engebrecht et al., 1990). The SC is formed by condensation of sister chromatids along protein cores called axial elements (AEs). Synapsis is complete when AEs are connected by proteins in the central region such as Zip1 or Scp1 (Meuwissen et al., 1992; Sym et al., 1993).
Genetic studies from budding yeast indicate that meiosis-specific components of the SC play important roles in recombination, chromosome segregation and the meiotic recombination checkpoint. Mek1, Hop1, and Red1 are meiosis-specific proteins that localize to AEs (Smith and Roeder, 1997; Bailis and Roeder, 1998). Recombination is reduced, but not eliminated in mek1/hop1/red1 mutants, and those crossovers that do occur are not effective for disjunction (Hollingsworth and Byers, 1989; Rockmill and Roeder, 1990, 1991; Leem and Ogawa, 1992; Baumgartner and Hollingsworth, unpublished data). The stoichiometry between the three proteins is important for effective chromosome segregation and for the meiotic recombination checkpoint (Hollingsworth and Ponte, 1997; Bailis et al., 2000). Both homo and hetero-oligomers have been observed with Hop1 and Red1 (Kironmai et al., 1998; Woltering et al., 2000) and Mek1 interacts with Hop1 in a RED1-dependent manner (Bailis and Roeder, 1998). All three genes are important in ensuring that crossovers occur between homologs and not between sister chromatids (Hollingsworth et al., 1995; Schwacha and Kleckner, 1997; Thompson and Stahl, 1999). Finally, red1 and mek1 mutants have a partial defect in sister chromatid cohesion (Bailis and Roeder, 1998).
Mek1 is a serine/threonine protein kinase (Bailis and Roeder, 1998; de los Santos and Hollingsworth, 1999). Recently, a chemical genetic approach has been developed in which the ATP binding pocket of a kinase is enlarged by mutation, thereby making the kinase sensitive to small molecule inhibitors (Bishop et al., 2001). In addition, the direct substrates of the kinase can be identified using radioactively labeled ATP analogs (Shah and Shokat, 2002). Importantly, analog-sensitive (as) kinases exhibit the same specificity for target substrates as their wild-type counterparts (Witucki et al., 2002). We have applied this approach to Mek1 (mek1-as1) and discovered that Mek1 kinase activity is required after DSB formation for preventing DMC1-independent DSB repair.
The mek1-as1 allele was also used to test the hypothesis that Red1 is a direct target of Mek1 (Bailis and Roeder, 1998; de los Santos and Hollingsworth, 1999). Our work demonstrates that, rather than being a substrate of Mek1, Red1 is necessary for maximum levels of Mek1 kinase activity. Based on these data, we present a model for how Mek1, Red1, and Hop1 work together during meiosis to regulate the pathway of DSB repair.
MATERIALS AND METHODS
Plasmids
Plasmids for this study were made by standard procedures by using the Escherichia coli strain BSJ72 (Maniatis et al., 1982). Details of plasmid constructions are available upon request. pTS30, pTS31, pTS32, and pTS33 carry MEK1, mek1-K199R, mek1-T327A, and mek1-T327D fused to MEK1p-GST, respectively, in the ADE2-integrating vector pRS402. The Q247A and Q247G mutations were introduced into pTS30 by site-directed mutagenesis by using the QuikChange kit (Stratagene, La Jolla, CA) and confirmed by DNA sequencing (our unpublished data). The R51A mutation was similarly introduced into GST-mek1-as1 and GST-MEK1. All mutant alleles conferring a phenotype were sequenced to ensure no inadvertent mutations were introduced during the mutagenesis (our unpublished data). pLW1 and pLW3 are 2μ URA3 plasmids containing GST-MEK1 and GST-mek1-as1, respectively. pDW15, 16, 17, 18, and 19 are 2μ LEU2 plasmids carrying GST-MEK1, GST-mek1-K199R, GST-mek1-T327A, GST-mek1-T327D, and GST-mek1-T327S, respectively (de los Santos and Hollingsworth, 1999). pLW19 carries GST-mek1-as1 on a URA3 integrating plasmid. pLW11 contains a C-terminal fragment of RED1 (amino acids 427–827) fused to GST for expression in Escherichia coli. pSB3 and pSB3-K348E are URA3 integrating plasmids carrying RED1 and red1-K348E, respectively (Woltering et al., 2000). pNH212 carries RED1–3HA in a 2μ URA3 vector, whereas pLP37 contains MEK1 in a URA3 integrating vector (de los Santos and Hollingsworth, 1999).
Yeast Strains and Media
Liquid and solid media were as described previously (Vershon et al., 1992; de los Santos and Hollingsworth, 1999). Inhibitors were diluted from 10 mM stocks in dimethyl sulfoxide into spo medium immediately before pouring plates or added directly to liquid sporulation medium (2% potassium acetate).
All strains were derived from the SK1 background with the exception of NH572 and NH575, which are derivatives of the BR strain background. The genotypes of each strain are listed in Table 1. Details of the SK1 strain constructions are available upon request. In all of the NH423 strains, the GST plasmids were integrated into each haploid and then mated to make diploids containing two copies of the plasmid. The BR strains were constructed by disrupting RED1 with red1::LEU2 by using pNH119 (Hollingsworth and Johnson, 1993) in BR1373–6D and BR1919–8D (Rockmill and Roeder, 1990) and then crossing the haploids to make NH572. pTS1 (de los Santos and Hollingsworth 1999) was used disrupt MEK1 in BR1373–6D. BR1373-6Dmek1 was then crossed to BR1919-8D and a MATα mek1::LEU2 segregant was backcrossed to BR1373-6Dmek1 to make NH575. To ensure isogenicity between the MEK1 and mek1 diploids, MEK1 was integrated into NH575 by using pLP37 (de los Santos and Hollingsworth 1999) and compared with NH575 transformed with the vector pRS306. mek1 and red1 disruptions were confirmed by Southern blots and tetrad dissection (our unpublished data).
Table 1.
S. cerevisiae strains
Inhibitors
Stocks (10 mM) of 4-amino-1-tert-butyl-3-(1′naphthyl)pyrazolo[3,4-d]pyrimidine (1-Na-PP1), 4-amino-1-(tert-butyl)-3-(1′-naphthylmethyl)pyrazolo [3,4-d] pyrimidine (1-NM-PP1), and 4-amino-1-tert-butyl)-3-(2′-naphthylmethyl) pyrazolo[3,4-d] pyrimdine (2-NM-PP1) were either synthesized at the University of California at San Francisco or generously provided by Cellular Genomics (New Haven, CT).
Time Courses
Cells were sporulated at 30°C as described in de los Santos and Hollingsworth (1999). Analysis of meiotic progression, sporulation, and DSBs is described in Woltering et al. (2000). To assay for sensitivity to inhibitor, 1-Na-PP1 was added to a final concentration of 1 μM to 1 ml of cells. The cells were then returned to 30°C and incubated until 48 h after transfer to sporulation medium. At this time, the asci were either counted or dissected, depending upon the experiment.
Antibodies, Immunoprecipitations, and Western Blots
Gst-Red1427–827 antigen for antibody production was obtained using BL21(DE3) bacteria transformed with pLW11 following the protocol of Kellogg et al., (1995). Antigen was sent to the Pocono Rabbit Farm where a standard protocol was used to inject rabbits for antibody production. The resulting serum contains antibodies against both Gst and Red1 and is referred to as the G/R serum. α-Hop1 antibodies are described in de los Santos and Hollingsworth (1999). α-Gst antibodies were a gift from Doug Kellogg (University of California, Santa Cruz, Santa Cruz, CA). Immunoprecipitations were performed using soluble extracts from meiotic cells as described in de los Santos and Hollingsworth (1999). For Western blots, the G/R antibodies were used at a dilution of 1:10,000, whereas the α-Gst and α-Hop1 antibodies were diluted 1:5000.
Kinase Assays
For experiments using Mek1 partially purified by glutathione precipitation, soluble extracts from 80 ml of sporulating cells were incubated with 750 μl of a slurry containing 1:1 glutathione-Sepharose:lysis buffer. Protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 2 mM benzamidine HCl, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin) and phosphatase inhibitors (10 mM NaF, 1 mM Na4P2O7) were included at all stages except the actual kinase reaction unless otherwise specified. Lysis buffer contains 200 mM NaCl, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% Triton X-100. The beads were washed twice with 5 ml of wash buffer A (500 mM NaCl, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1% Triton X-1000) and once with 5 ml of wash buffer B (200 mM NaCl, 25 mM Tris-HCl, pH 7.5, 1 mM EDTA) and twice with 5 ml of kinase buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgCl2). After resuspension in 1 ml of kinase buffer, 125 μl of slurry was aliquoted into eight tubes. Residual liquid was removed using a 30-gauge needle. A standard reaction contains 37 μl of kinase buffer, 2 μlof1mM ATP, and 1 μl of 10 mCi/ml 32P γ-labeled ATP. For reactions containing 5 μM inhibitor, 2 μl of 100 μM 1-Na-PP1 was added and the amount of kinase buffer reduced to 35 μl. For reactions using 32P γ-labeled ATP analogs (generously provided by Cellular Genomics), 1 μl of 5 mCi/ml was used. The reactions were incubated at room temperature for 30 min. After addition of 10 μl of 5× protein sample buffer, the samples were heated at 95°C for 5 min and loaded onto an 8% polyacrylamide gel. The gels were run overnight at 8 mA and transferred onto nitrocellulose and exposed for autoradiography. Afterward, the filters were probed with antibodies to detect Gst-Mek1, Hop1, and Red1 depending upon the experiment. For kinase assays using immunoprecipitates (IPs), 20 μl of protein A beads:2 μl of α-Gst antibody:antigen complexes were used in reactions as described above. Phosphatase treatment of IPs was performed as described in de los Santos and Hollingsworth (1999).
RESULTS
Mutation of a Conserved Glutamine in the Mek1 ATP Binding Pocket Creates Inhibitor-sensitive Alleles
Alignment of Mek1 with other protein kinases predicted that the Mek1 ATP binding pocket could be enlarged by mutating the glutamine at position 247 to either glycine or alanine, thereby creating two alleles of MEK1 designated mek1-as1 and mek1-as2, respectively (for more information, see http://kinase.ucsf.edu/ksd/). The mutations were also introduced into GST-MEK1, an allele encoding a functional fusion protein to facilitate detection of the kinase (de los Santos and Hollingsworth, 1999). Because there were no differences between the tagged and untagged versions of MEK1 (our unpublished data), only experiments using derivatives of GST-MEK1 (hereafter referred to as MEK1) are presented. MEK1 function was assayed by sporulating cells in the presence of varying concentrations of inhibitor and measuring spore viability by tetrad dissection. In the SK1 strain background, deletion of MEK1, as well as a mutation in the catalytic site of the kinase (mek1-K199R) results in ≤1.0% viable spores (Table 2; de los Santos and Hollingsworth, 1999). MEK1 was unaffected by the presence of inhibitors at all concentrations tested (up to 5 μM) (Table 2; our unpublished data). The mek1-as2 allele exhibited some reduction in spore viability even in the absence of the inhibitor, suggesting that the alanine substitution decreases the ability of the kinase to use cellular ATP. Consistent with this idea, overexpression of mek1-as2 rescues the reduction in spore viability (our unpublished data). In contrast, the mek1-as1 mutant is indistinguishable from wild-type in the absence of inhibitor (Table 2). Whereas no phenotype was observed with either mek1-as allele by using 2-NM-PP1 (our unpublished data), a dose response was seen for both mek1-as alleles with the 1-Na-PP1 and 1-NM-PP1 inhibitors (Table 2). Although the presence of 1-Na-PP1 decreases spore viability of mek1-as, alleles, addition of inhibitor has no effect on meiotic progression or sporulation in either MEK1 or mek1-as diploids (our unpublished data). Because mek1-as1 gives a mutant phenotype only in the presence of inhibitor, it was used for all experiments involving the use of inhibitor or ATP analogs.
Table 2.
Effects of different inhibitors on spore viability in the presence of various alleles of MEK1
| % Spore viability (no. tetrads)
|
|||||
|---|---|---|---|---|---|
| 1-Na-PP1
|
1-NM-PP1
|
||||
| Genotype | No I | 0.1 μM | 0.5 μM | 0.1 μM | 0.5 μM |
| MEK1 | 92.7 (72) | 95.5 (72) | 94.0 (96) | 92.1 (48) | 95.8 (96) |
| mek1-as1 | 93.6 (46) | 13.4 (95) | 1.0 (96) | 19.6 (120) | 5.7 (96) |
| mek1-as2 | 76.7 (94) | 14.9 (94) | 2.4 (114) | 5.2 (120) | 1.6 (96) |
| mek1::LEU2 | <1.0 (27) | ||||
| mek1-K199R | 0.9 (58) | ||||
No I, no inhibitor.
The pTS30-based, GST-MEK1-integrating plasmids were transformed into YTS1 ade2. Transformants were patched onto selective medium and then transferred to sporulation medium containing the indicated amount of inhibitor.
The Requirement for Mek1 Kinase Activity Coincides with DSB Formation and Heteroallelic Recombination
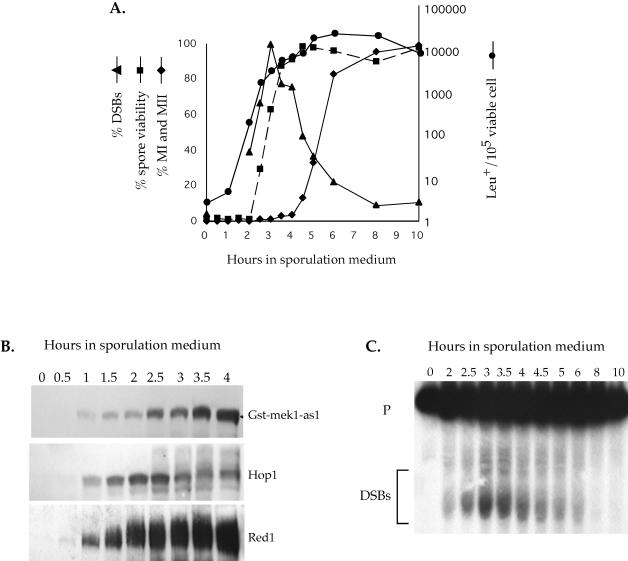
Using the ability to inhibit Mek1-as1, execution point analysis was performed to determine when during meiosis Mek1 kinase activity is required. A mek1-as1 diploid was transferred to sporulation medium and incubated at 30°C. At various time points, a final concentration of 1 μM 1-Na-PP1 was added to 1-ml aliquots, and the cells were returned to 30°C to complete sporulation. MEK1 function was assayed in these samples by tetrad dissection to determine spore viability. Addition of the inhibitor between 0 and 2 h resulted in a null mek1 phenotype (Figure 1A). After 2 h, spore viability began to rise, reaching wild-type levels by 3.5 h. This result defines the endpoint for the requirement for Mek1 kinase activity in the formation of viable spores (Figure 1A).
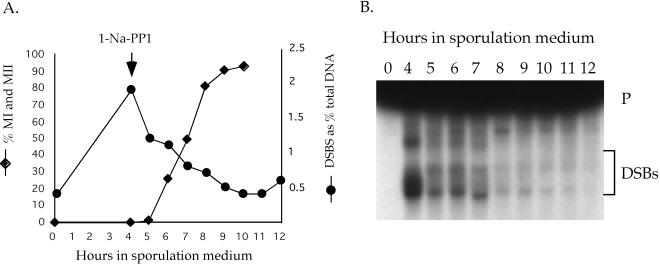
Figure 1.
Time course of mek1-as1 inhibitor sensitivity relative to other meiotic parameters. The GST-mek1-as1 diploid NH465::pLW19 was transferred to sporulation medium at 30°C and assayed for various meiotic landmarks as described in MATERIALS AND METHODS. (A) Graph of different meiotic parameters as function of time. Leu+ prototrophs, DSBs, and meiotic progression were measured in the absence of inhibitor. The DSB curve was obtained by quantitation of the data presented in C. For spore viability, a final concentration of 1 μM 1-Na-PP1 was added at various time points indicated by squares and the cells were returned to 30°C until 48 h when the asci were dissected. Spore viability is indicated by a dashed line. (B) Western blot analysis. Total extract (25 μg) from each time point was probed with α-Gst, α-Hop1, or G/R antibodies to detect Red1 (see below). (C) Southern blot of DSB analysis at the YCR048w hotspot (Wu and Lichten, 1994). p = 10 kb expected parental band; DSB fragments are indicated by the bracket.
A trivial explanation for the mek1-as1 inhibitor-resistant phenotype beginning at 3.5 h is that the cells have become impermeable to the inhibitor. This is not the case, however, as addition of inhibitor at 4 h is able to inactivate an analog-sensitive allele of CDC7 (Wan and Hollingsworth, unpublished data).
The earliest time at which MEK1 can act was determined by monitoring the appearance of the Mek1-as1 protein by using Western blots. Mek1-as1 was observed at 1 h, showing up 30 min after Red1 and Hop1 were first detected (Figure 1B). Together with the functional data, these experiments reveal a window of time for Mek1 kinase activity between 1 and 3.5 h after transfer to sporulation medium.
The timing of MEK1 function was correlated with a number of other meiotic events, such as DSB and prototroph formation, as well as meiotic progression (all occurring in the absence of inhibitor). Cells did not proceed through MI until after 4 h, by which time Mek1 kinase activity was dispensable. MEK1, therefore, acts solely in prophase (Figure 1A). Heteroallelic recombination at LEU2 increased dramatically between 1 and 2 h and reached a plateau by 3.5 h, correlating well with the onset of DSB formation at the YCR048w hotspot (Figure 1, A and C) (Wu and Lichten, 1994). There was a 30-min lag between the appearance of DSBs and resistance to inhibitor, suggesting that Mek1 functions during and/or immediately after DSBs are made (Figure 1A). At the 4-h time point, a significant number of DSBs are still present, yet inhibition of mek1-as1 has no phenotype. Therefore, either Mek1 kinase activity is not directly required for the repair of DSBs or Mek1 has already phosphorylated substrates that function in downstream repair events by 3.5 h.
Deletion of MEK1 reduces the steady state levels of meiotic DSBs (Leem and Ogawa, 1992; Xu et al., 1997). Inhibition of Mek1-as1 kinase activity also decreased steady-state levels of DSBs at the YCR048w meiotic hotspot (our unpublished data). When DSB processing is blocked, however, mek1 exhibits no reduction in DSBs at this hotspot, indicating that a wild-type number of breaks is formed (Pecina et al., 2002). These results suggest that the decrease in steady-state DSBs observed in mek1 diploids is due to increased resection or a difference in DSB repair compared with wild-type.
Mek1 Kinase Activity Is Required after the Formation of DSBs for the Prevention of DMC1-independent DSB Repair
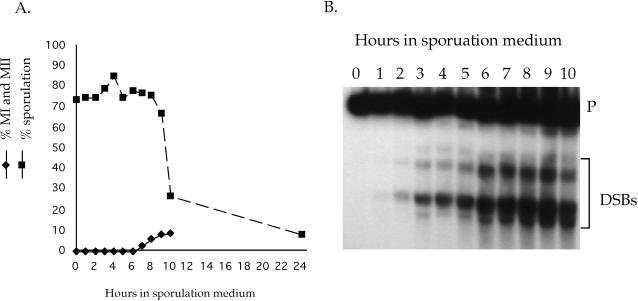
Deletion of MEK1 has previously been shown to alleviate the meiotic arrest triggered by the unrepaired DSBs present in a dmc1 mutant (Xu et al., 1997). To test whether Mek1 kinase activity is responsible, a time course was performed using a dmc1 mek1-as1 diploid. When sporulated in the absence of inhibitor, >90% of the dmc1 mek1-as1 cells were arrested in prophase after 10 h and no asci were observed after 48 h (Figure 2A). Addition of 1-Na-PP1 at 0 h completely abolished the sporulation defect, confirming the importance of Mek1 kinase activity for the dmc1 arrest (Figure 2A). Furthermore, the dmc1 arrest can be suppressed by addition of inhibitor well after DSBs have been made, demonstrating that Mek1 kinase activity is constitutively required to maintain the arrest (Figure 2). There was a steep drop in the proportion of cells that responded to inhibitor after 9 h, although DSBs were still present (Figure 2). One explanation is that cells become impermeable to the inhibitor after prolonged arrest at prophase. Alternatively, the chromosomes may become so badly fragmented that a second, MEK1-independent, checkpoint is activated. A third possibility is that the phosphatase needed to dephosphorylate Mek1's targets has become inactive.
Figure 2.
DSB formation, meiotic progression, and sporulation in a dmc1 mek1-as1 diploid. A dmc1 mek1-as1 diploid (NH520::pLW19) was transferred to sporulation medium at 30°C. Aliquots were taken at the indicated time points and assayed for various meiotic parameters. (A) Meiotic progression was followed by 4,6-diamidino-2-phenylindole staining cells at different time points in the absence of inhibitor. For sporulation, a final concentration of 1 μM 1-Na-PP1 was added at various time points indicated by squares, the cells were returned to 30°C until 48 h when the number of asci were counted by light microscopy. Sporulation is indicated by a dashed line. At least 200 cells were counted for each measurement. (B) DSB formation at the YCR048w hotspot measured in the absence of inhibitor as described in Figure 1.
The previous work also demonstrated that DSBs in mek1Δ dmc1 mutants are repaired (Xu et al., 1997). These experiments did not address, however, whether the observed repair is due to a lack of Mek1 kinase activity or the absence of the Mek1 protein; furthermore they did not determine whether the repair occurs because DSBs formed in the absence of MEK1 are somehow qualitatively different from those formed when MEK1 is present. The mek1-as1 dmc1 diploid was used to address these questions. Meiotic progression and DSBs were monitored in a dmc1 mek1-as1 diploid to which inhibitor was added at 4 h, a time at which the cells were completely arrested with unrepaired DSBs that were generated while Mek1 was active (Figure 3). After a 1-h lag, during which time there was a decrease in the number of DSBs, the cells began to progress through MI (Figure 3). By 10 h, >90% of the cells had completed both divisions, and the bulk of the DSBs were gone. After 48 h, 84% of the cells had sporulated, but only 3% of the spores were viable. Mek1 kinase activity is required, therefore, after DSB formation to prevent DMC1-independent repair.
Figure 3.
DSBs and meiotic progression in a dmc1 mek1-as1 diploid after addition of inhibitor at 4 h. (A) Meiotic progression and quantitation of DSB formation in a dmc1 mek1-as1 diploid (NH520::pLW19) after transfer to sporulation medium at 30°C. The arrow indicates the time of addition of a final concentration of 1 μM 1-Na-PP1 to the total culture. (B) Southern blot showing the DSBs at the YCR048w hotspot that are quantitated in A.
Phosphorylation of Red1 Is Independent of Mek1 Kinase Activity
Previous studies suggested that Red1 is a substrate of Mek1 (Bailis and Roeder, 1998; de los Santos and Hollingsworth, 1999). Our work used an epitope tagged allele of RED1 (RED1-3HA) that exhibits phosphorylated Red1 only when present in high copy number, suggesting that the tag is interfering with Red1 phosphorylation (de los Santos and Hollingsworth, 1999). Therefore, to test the idea that Red1 is a direct substrate of Mek1, a different way of detecting Red1 was needed. Toward this end, polyclonal antibodies were generated against a C-terminal fragment of Red1 fused to Gst that are effective both for IP experiments as well as for probing immunoblots (see MATERIALS AND METHODS). This serum (called the G/R antibodies) is specific for Red1 based on the following criteria: 1) the G/R antibodies IP and detect Red1 from RED1 but not red1 diploids (Figure 4, A and D); 2) they IP and detect Red1 from sporulating, but not vegetatively growing cells, as expected given that RED1 is a meiosis-specific gene (Thompson and Roeder 1989) (Figure 4B); and 3) they coimmunoprecipitate Hop1 in a MEK1-independent manner (our unpublished data). In addition to detecting Red1, the serum also contains antibodies against Gst. The G/R antibodies can also be used, therefore, to detect Gst-Mek1 (Figure 4, B and C).
Figure 4.
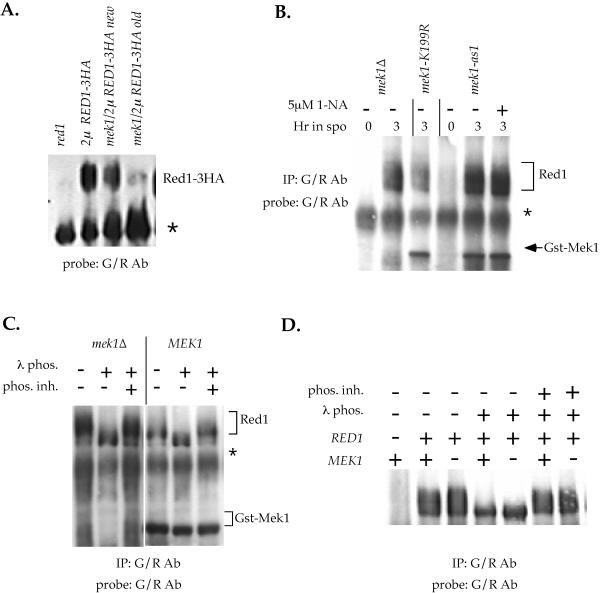
Red1 mobility in the absence of Mek1 kinase activity in the SK1 and BR strain backgrounds. (A) Mobility of Red1–3HA in the presence or absence of MEK1. Isogenic SK1 diploids YTS3::pRS306 (red1), YTS3/pNH212 (2μ RED1–3HA), NH177/pNH212 (red1 mek1/2μ RED1–3HA) new and NH177/pNH212 (red1 mek1/2μ RED1-3HA) old were sporulated for 3 h at 30°C. 50 μg soluble extract were fractionated on an 8% gel and probed with G/R antibodies to detect Red1. “New” indicates a recently transformed colony of NH177. “Old” indicates the transformant used for the experiments shown in de los Santos and Hollingsworth (1999). The asterisk indicates a cross-reacting band. (B) Mobility of endogenous Red1 in the absence of Mek1 kinase activity. Extracts from mek1 (YTS1ade2::pRS402), GST-mek1-K199R (YTS1ade2::pTS31) or GST-mek1-as1 (YTS1ade2::pTS30-Q247G) diploids were made at 0 and 3 h after transfer to sporulation at 30°C. G/R antibodies were used to simultaneously precipitate Red1 and Mek1 and the IPs were probed with G/R antibodies to detect Red1 and Gst-Mek1. In the mek1-as1 diploid, the culture was split and a final concentration of 5 μM 1-Na-PP1 was added to half the culture at 0 h as indicated. The asterisk indicates a cross-reacting band. (C) Phosphatase treatment of Red1 IPed in the presence or absence of MEK1. YTS1ade2::pRS402 (mek1) and YTS1ade2::pTS30 (GST-MEK1) diploids were sporulated in 20 ml of 2% potassium acetate for 4.5 h at 30°. Gst-Mek1 and Red1 were simultaneously immunoprecipitated with G/R antibodies. The IPs were treated with λ protein phosphatase as described in de los Santos and Hollingsworth (1999). The phosphatase inhibitors NaF and Na4P2O7 were used at 10 and 1 mM final concentrations, respectively. The Western blot was probed with G/R antibodies to detect Red1 and Gst-Mek1. The asterisk indicates a cross-reacting band. (D) Mobility of endogenous Red1 in the BR strain background. BR strains NH572 (red1), NH575::pLP37 (MEK1) and NH575::pRS306 (mek1) were sporulated in 750 ml of 2% potassium acetate for 17 h at 30°C. G/R antibodies (30 μl) were used to immunoprecipitate Red1. The IPs were split into equal aliquots and exposed to λ protein phosphatase and phosphatase inhibitors as described in de los Santos and Hollingsworth (1999). The Western blot was probed with G/R antibodies to detect Red1.
The prior biochemical evidence suggesting that Red1 is a substrate of Mek1 was based primarily on an increase in protein mobility exhibited by Red1 in the absence of MEK1 (Bailis and Roeder 1998; de los Santos and Hollingsworth, 1999). Using the G/R antibodies, we discovered that our published results showing a MEK1-dependent Red1–3HA mobility shift were an artifact peculiar to a particular transformant. Although Red1–3HA is reproducibly underphosphorylated in this transformant (Figure 4A, RED1-3HA old), analysis of Red1–3HA from mek1 red1 colonies newly transformed with the 2 μ RED1–3HA plasmid found no difference in mobility compared with Red1–3HA from a MEK1 diploid (Figure 4A, RED1–3HA new; our unpublished data). Because the phosphorylated form of Red1-3HA is only observed when the protein is overexpressed, the most likely explanation for this discrepancy is that the transformant used in our initial studies carries a low number of copies of the 2μ RED1–3HA plasmid. Consistent with this conclusion, the old mek1 2μ RED1–3HA transformant reproducibly exhibits less Red1–3HA protein than the new transformant (Figure 4A; de los Santos and Hollingsworth, 1999).
To examine the relevance of MEK1 on the mobility of endogenous Red1 in our SK1 strains, Mek1 kinase activity was abolished in one of three different ways: 1) deletion of MEK1, 2) mutation of the catalytic site of Mek1 (mek1-K199R; de los Santos and Hollingsworth, 1999), or 3) addition of inhibitor to a mek1-as1 diploid. In all cases, phosphorylated Red was observed, demonstrating that MEK1 is not required for the Red1 mobility shift (Figure 4B).
To ensure that the G/R antibodies recognize the phosphorylated form of Red1 and that our gel conditions can discriminate between unphosphorylated and phosphorylated forms of Red1, Red1 precipitated by the G/R antibodies was treated with λ protein phosphatase. The increase in Red1 mobility in the presence of active phosphatase confirmed that the G/R antibodies are precipitating the Red1 phosphoprotein (Figure 4C). The same pattern was observed with Red1 precipitated from a mek1 diploid, demonstrating that the phosphorylation-induced mobility shift of Red1 is independent of MEK1 (Figure 4C). Similar to Red1, an increase in mobility was detected for Mek1 in the presence of active phosphatase, consistent with previous reports that Mek1 is a phosphoprotein (Figure 4C) (Bailis and Roeder, 2000).
A MEK1-dependent Red1 mobility shift has also been reported in the BR strain background (Bailis and Roeder, 1998). To test whether the discrepancy in our results and those of Bailis and Roeder is due to strain background differences, RED1 and MEK1 were individually deleted from the BR strain background and Red1 mobility was analyzed in IPs using the G/R antibodies. Similar to the results obtained in our SK1 strains, deletion of MEK1 had no effect on Red1 mobility (Figure 4D). Treatment of the Red1 IPs with λ protein phosphatase resulted in the loss of the slower migrating forms of Red1, confirming that Red1 is a MEK1-independent phosphoprotein in these strains.
The Mek1-as1 Protein Has High Specificity for ATP Analogs in In Vitro Kinase Assays
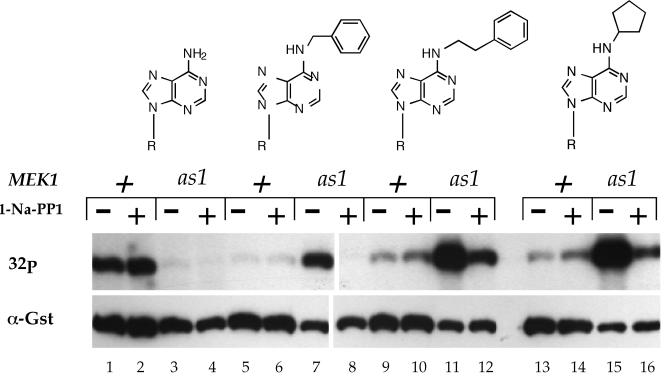
Not all phosphorylation events change the mobility of the target protein. The ability of Mek1-as1 to specifically use analogs of ATP provides an alternative approach for asking whether a kinase transfers a phosphate to another protein without depending upon mobility shifts as the assay. In vitro kinase assays performed with an as kinase and ATP analogs result in specific radioactive labeling of direct targets of the kinase (Shah and Shokat, 2002). Therefore, if Mek1 phosphorylates Red1, the Red1 protein might become radioactive when present in kinase assays by using Mek1-as1 and radioactively labeled ATP analogs. Before doing this experiment, we first confirmed that Mek1-as1 efficiently uses ATP analogs in vitro as predicted. Kinase assays were performed with Mek1 or Mek1-as1 partially purified from meiotic extracts using radioactive ATP or various analogs of ATP (see MATERIALS AND METHODS). Mek1 autophosphorylation was used as the assay for kinase activity (de los Santos and Hollingsworth, 1999). The relative amounts of Mek1 present in the assays were determined by probing the filters used for autoradiography with α-Gst antibodies. Mek1 exhibited efficient labeling with ATP, but not with the ATP analogs (Figure 5). The activity of the wild-type kinase was insensitive to the presence of the 1-Na-PP1 inhibitor (Figure 5, lanes 1 and 2, 5 and 6, 9 and 10, 13 and 14). The Mek1-as1 kinase, in contrast, exhibited very low activity with ATP. Given that mek1-as1 completely complements the spore inviability of a mek1Δ (Table 2), the in vitro result is most likely explained by the as1 mutation increasing the Km of the kinase for ATP. The increased Km is not a problem in vivo where the cellular ATP concentration is estimated to be 3 mM, but is a problem in vitro because the amount of cold ATP in the kinase assays cannot be raised sufficiently without titrating out the radioactively labeled ATP molecules (our unpublished data). In contrast to ATP, Mek1-as1 produced a strong signal with all three ATP analogs (Figure 5). Furthermore, autophosphorylation of Mek1-as1 was reduced in the presence of 5 μM 1-Na-PP1 (Figure 5). The addition of 10 μM inhibitor decreased kinase activity to background levels (Figure 6B). Combining the use of Gst-mek1-as1 and radioactive ATP analogs should, therefore, provide a way of specifically detecting direct substrates of Mek1.
Figure 5.
In vitro kinase assays with Mek1 and Mek1-as1 by using different analogs of ATP. The GST-MEK1 and GST-mek1-as1 diploids YTS1/pLW1 and YTS1/pLW3 were sporulated for 3 h at 30°C. Kinase assays were performed using glutathione-precipitated Mek1 as described in MATERIALS AND METHODS. The structures of the different analogs of ATP are drawn from left to right: ATP (lanes 1–4); N6-benzyl-ATP (lanes 5–8); N6-phenylethyl-ATP (lanes 9–12); and N6-cyclopentyl-ATP (lanes 13–16). 1-Na-PP1 (5 μM) was added to the reactions as indicated. 32P indicates the autoradiogram of the filter. α-Gst represents the same filter probed with α-Gst antibodies to detect Gst-Mek1. R, ribose, 5′-triphosphate.
Figure 6.
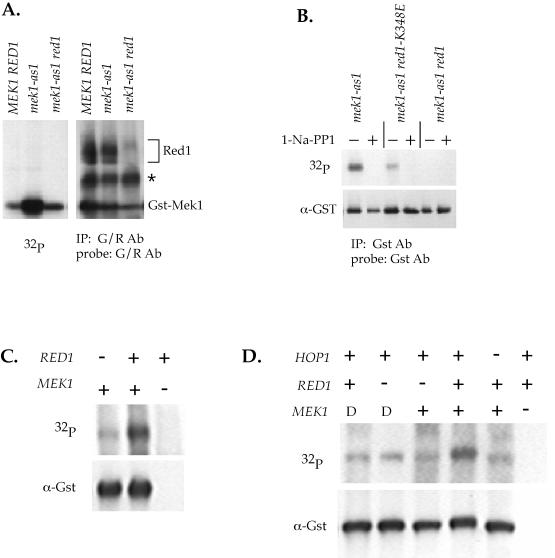
Mek1-as1/Mek1 kinase activity under different conditions. (A) Mek1-as1 activity in the absence of RED1. YTS1/pLW1 (2μ GST-MEK1), YTS1/pLW3 (2μ GST-mek1-as1) and NH423::pTS30-Q247G (GST-mek1-as1 red1) diploids were sporulated for 3 h at 30°C. Red1 and Mek1 were simultaneously immunoprecipitated using G/R antibodies. Kinase assays used N6-cyclopentyl-ATP. After autoradiography, the filter was probed with G/R antibodies. The asterisk indicates a cross-reacting band. (B) Mek1-as1 activity in a red1-K348E mutant. NH423::pTS30-Q247G::pSB3 (GST-mek1-as1 RED1), NH423::pTS30-Q247G::pSB3-K348E (GST-mek1-as1 red1-K348E), NH423::pTS30-Q247G::pRS306 (GST mek1-as1 red1) were sporulated for 3 h at 30°C. Mek1 was immunoprecipitated with α-Gst antibodies and kinase assays were performed using radioactive N6-benzyl-ATP in the presence or absence of 10 μM 1-NA-PP1. After exposure for autoradiography, the filter was probed with α-Gst antibodies to determine the relative amounts of Mek1 present. (C) Mek1 kinase activity in the absence of RED1. NH423::pTS30::pSB3 (GST-MEK1 RED1), NH423::pTS30::pRS306 (GST-MEK1 red1) and NH423::pRS402::pSB3 (mek1 RED1) diploids were sporulated for 3.5 h at 30°C. Gst-Mek1 was partially purified using glutathione Sepharose and used for kinase assays with radioactive ATP as described in de los Santos and Hollingsworth (1999). After exposure to film, the filter was probed with α-Gst antibodies to detect Gst-Mek1. (D) Mek1-T327D activity in the absence of RED1. NH423::pTS30::pSB3 (GST-MEK1 RED1), NH423::pTS30::pRS306 (GST-MEK1 red1), NH423::pTS33::pSB3 (GST-mek1-T327D RED1), NH423::pTS33::pRS306 (GST-mek1-T327D red1), NH423::pRS402::pSB3 (mek1 RED1), and NH550::pTS30 (GST-MEK1 hop1) diploids were sporulated for 3.5 h at 30°C and kinase assays and the Western blot performed as in A. –, a null allele; D, the GST-mek1-T327D allele.
To test whether Red1 is a direct target of Mek1-as1, in vitro kinase assays were performed using G/R IPs. The G/R antibodies simultaneously precipitate both Red1 and Mek1. Western blot analysis confirmed that both Red1 and Mek1 were present (Figure 6A). In the kinase assays, high levels of Mek1 autophosphorylation were observed in the mek1-as1 diploid, whereas no radioactive Red1 was detected (Figure 6A). Mek1-as1 is, therefore, not phosphorylating Red1 in vitro. A possible objection to this experiment is that Red1 may not get labeled in vitro because it has already been phosphorylated by Mek1 in vivo, before making the extracts. To test this idea, Red1 was immunoprecipitated from a mek1 diploid and mixed with Mek1-as1 that was precipitated using glutathione Sepharose. No labeling of Red1 was seen in this experiment either (our unpublished data). We conclude, therefore, that Red1 is not a substrate of Mek1.
A reduction in the level of Mek1-as1 kinase activity was seen in the absence of RED1 (Figure 6A). This result could simply be due to a difference in mek1-as1 copy number in the RED1 versus red1 diploids used in this experiment. The experiment was repeated with all of the diploids containing two integrated copies of mek1-as1 and similar results were obtained (Figure 6B). These data suggest that, rather than being phosphorylated by Mek1, RED1 is necessary to activate Mek1.
One caveat of these experiments is that the dependence on RED1 for wild-type levels of Mek1 kinase activity could be an artifact arising from the use of the Mek1-as1 protein. To determine whether this was the case, in vitro kinase reactions were performed using wild-type Mek1 and radioactive ATP. Similar to the experiments with the Mek1-as1 protein, Mek1 kinase was less active in the absence of RED1 (Figure 6C). The requirement for RED1 for maximum Mek1 kinase activity in vitro is therefore not specific to the Mek1-as1 protein.
Red1/Hop1 Complexes Are Required for Wild-Type Levels of Mek1-as1 Kinase Activity
Red1 interacts with both Hop1 and Mek1 (de los Santos and Hollingsworth, 1999; Bailis and Roeder, 1998). To test whether the interaction between Hop1 and Red1 is important for Mek1 kinase activity, kinase assays were performed in mek1-as1 diploids carrying the red1-K348E allele. This allele encodes a Red1 protein with a single amino acid change that specifically disrupts the formation of Red1/Hop1 complexes (Woltering et al., 2000). A reduction in Mek1-as1 kinase activity was observed in the red1-K348E diploid compared with wild type (Figure 6B). Mek1-as1 autophosphorylation from both the RED1 and red1-K348E strains was eliminated by addition of inhibitor, confirming that Mek1-as1 kinase activity is being measured (Figure 6B). As expected given these results, wild-type levels of Mek1 kinase activity also require HOP1 (Figure 6D).
A Functional Mek1 FHA Domain Is Required for Activation of the Kinase and for Interaction with Red1
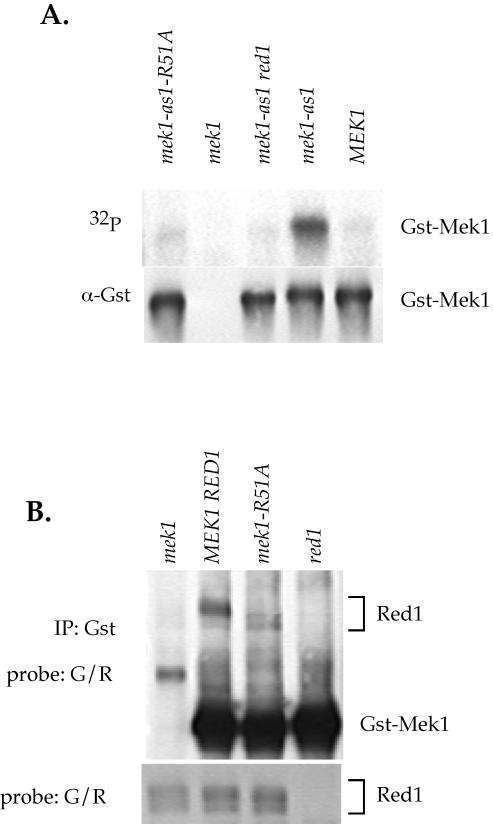
Mek1 contains an FHA domain, a highly conserved phospho-protein specific binding domain present is a variety of proteins (Durocher and Jackson, 2002). Mutation of a conserved arginine residue in the FHA1 domain of the DNA damage checkpoint kinase Rad53 disrupts its interaction with Rad9 without affecting the structure of the phosphopeptide binding site (Durocher et al., 1999, 2000). Mutation of the analogous arginine to alanine in the FHA domains of Mek1-as1 and Mek1 create null alleles (mek1-as1-R51A, <1% viable spores, 26 asci dissected; mek1-R51A, 2% viable spores, 74 asci dissected). The Mek1-as1-R51A and Mek1-R51A proteins also exhibit a reduction in Mek1 kinase activity in vitro (Figure 7A; our unpublished data).
Figure 7.
Effect of the mek1-R51A mutation on kinase activity and Red1 coimmunoprecipitation. (A) Mek1 kinase activity in a mek1-as1-R51A mutant. NH423::pTS30::pSB3 (GST-MEK1 RED1), NH423::pTS30-Q247G:: pSB3 (GST-mek1-as1 RED1), NH423::pTS30-Q247G::pRS306 (GST-mek1-as1 red1), NH423::pRS402::pSB3 (mek1 RED1), and NH423::pTS30-Q247G-R51A::pSB3 (GST-mek1-as1-R51A RED1) diploids were sporulated for 3.5 h at 30°C. The Gst-Mek1 proteins were precipitated with Gst antibodies and used in autophosphorylation assays with radioactively labeled N6-benzyl-ATP. After autoradiography, the filter was probed with Gst antibodies to detect Gst-mek1. (B) Red1 coimmunoprecipitation in a mek1-R51A mutant. NH423::pTS30::pSB3 (GST-MEK1 RED1), NH423::pTS30::pRS306 (GST-MEK1 red1), NH423::pRS402::pSB3 (mek1 RED1) and YTS1ade2::pTS30-R51A (GST-mek1-R51A) were sporulated for 3 h at 30°C. (All four strains are isogenic except that YTS1ade2 contains two copies of RED1, whereas the NH423 diploids have only one copy. All diploids contain two copies of the GST-mek1 allele). Gst-Mek1 was immunoprecipitated with 4 μl of Gst antibodies, and the IPs were probed with G/R antibodies to detect Red1 and Gst-mek1. The bottom panel shows a Western blot of the soluble extracts used for the IPs probed with G/R antibodies to detect Red1.
The mutant phenotype of the mek1-R51A allele suggests that a critical protein-protein interaction has been disrupted. Given the requirement of RED1 for Mek1 activation, and the fact that Red1 is phosphorylated in a MEK1-independent manner, a reasonable hypothesis is that the FHA domain of Mek1 is binding to phosphothreonines on Red1. Mek1 and Mek1-R51A were precipitated from meiotic extracts with α-Gst antibodies, and the IPs were probed with G/R antibodies. Both unphosphorylated and phosphorylated forms of Red1 were present in equal ratios in the MEK1- and mek1-R51A–soluble extracts (Figure 7B). Although Red1 was observed to coimmunoprecipitate with Mek1, no phosphorylated Red1 was present in the Mek1-R51A IP (Figure 7B). Some unphosphorylated Red1 is present, however, perhaps due to interaction with a different part of Mek1 or with another protein in the IP. We conclude that Mek1 binds to phosphorylated Red1 via its FHA domain.
A Conserved Threonine Residue in the Activation Segment of Mek1 Plays an Important Role in the Function of the Kinase
Many kinases are regulated by phosphorylation of a specific threonine in a part of the protein called the “activation segment,” which is flanked by the highly conserved DFG and PE residues (Johnson et al., 1996). The fact that Mek1 is phosphorylated during meiosis raised the possibility that Mek1 might be similarly regulated by phosphorylation of this threonine (located at position 327). A variety of amino acid substitutions were introduced at position 327, and the resulting alleles were assayed for their ability to complement the mek1 spore inviability phenotype. Changing T327 to alanine significantly decreased spore viability, regardless of the copy number of the allele (Table 3). Changing T327 to aspartic acid, a negatively charged amino acid that may mimic the phosphorylated state, complemented nearly as well as wild-type when overexpressed and was significantly improved over mek1-T327A, even with just two copies (Table 3). This result supports the idea that, analogous to other kinases, Mek1 may be activated by phosphorylation of T327. When T327 was substituted for serine, an intermediate phenotype was observed, indicating that the kinase responsible for phosphorylating Mek1 on this residue is not restricted to threonine (Table 3).
Table 3.
Sporulation and spore viability in various mek1 mutants
| Strain/plasmid | Relevant genotype | % spore viability (no. asci) | % sporulationa |
|---|---|---|---|
| YTS4/YEplac181 | mek1 | 2.4 (52) | 83.5 |
| YTS4/pDW15 | 2μ MEK1 | 78.4 (104) | 36.6 |
| YTS4/pDW16 | 2μ mek1-K199R | 5.7 (128) | 78.9 |
| YTS4/pDW17 | 2μ mek1-T327A | 13.9 (182) | 83.3 |
| YTS4/pDW18 | 2μ mek1-T327D | 68.7 (130) | 88.3 |
| YTS4/pDW19 | 2μ mek1-T327S | 54.8 (146) | 90.1 |
| NH423::pTS30::pSB3 | MEK1 RED1 | 94.2 (26) | 84.5 |
| NH423::pRS402::pSB3 | mek1 RED1 | <1.0 (26) | 83.1 |
| NH423::pTS32::pSB3 | mek1-T327A RED1 | 14.4 (26) | 87.0 |
| NH423::pTS33::pSB3 | mek1-T327D RED1 | 51.9 (26) | 92.5 |
| NH423::pTS33::pRS306 | mek1-T327D red1 | 4.0 (25) | 75.0 |
Transformants were patched onto selective medium and then transferred to spo plates. Spore viability was monitored by tetrad dissection and sporulation by light microscopy. At least 200 cells were counted for each strain.
Mutation of Mek1 T327 to glutamic acid has been reported to cause a prophase arrest when overexpressed (Bailis and Roeder, 2000). This allele also contains a mutation of T331 to glutamic acid. Overexpression of mek1-T327D in our SK1 strains had no effect on sporulation, although overexpression of wild-type GST-MEK1 did reduce the number of asci (Table 3). The discrepancy between our results and those in literature may be due to differences in strain background, different levels of overexpression as a result of using different vectors, the difference between glutamic acid and aspartic acid or to the fact that we mutated only T327. It is clear from our results, however, that the presence of a negative charge at T327 is sufficient to allow MEK1 to function in the formation of viable spores.
If RED1 is required to activate Mek1 by facilitating phosphorylation of T327, this requirement should be bypassed by the T327D mutation. Mek1-T327D kinase partially purified from isogenic RED1 and red1 diploids was assayed for autophosphorylation by using radioactive ATP. Although red1 reduced the activity of the wild-type kinase, red1 had no effect on the activity of Mek1-T327D (Figure 6D). mek1-T327D does not rescue the spore inviability of red1 (Table 3), however, consistent with the idea that RED1 has additional roles in meiosis beyond activating Mek1.
DISCUSSION
The Role of Mek1 in DMC1-independent DSB Repair
There are two RecA-like strand transfer proteins used for meiotic recombination in budding yeast, Dmc1 and Rad51 (Yves-Masson and West, 2001). Dmc1 is meiosis specific, and functions predominantly in recombination between homologs (Bishop et al., 1992). The other, Rad51, mediates recombination in vegetative and meiotic cells and participates in both intersister and interhomolog events (Schwacha and Kleckner, 1997). When DMC1 is deleted, DSBs become hyper-resected and accumulate because of the failure to form single end invasion intermediates with either the homolog or the sister chromatid (Bishop et al., 1992; Hunter and Kleckner, 2001). As a result, in the SK1 background, the meiotic recombination checkpoint is triggered and the cells arrest in prophase (Xu et al., 1997). dmc1 DSBs are repaired if arrested cells are returned to growth by transfer to rich, glucose-containing medium (Arbel et al., 1999; Bishop et al., 1999). Although the disappearance of DSBs could be due to number of repair processes such as synthesis-dependent strand annealing or break-induced replication, the most likely mechanism is sister chromatid repair (SCR). When joint molecules were examined in dmc1 diploids under return to growth (RTG) conditions, mostly intersister JMs were observed (Schwacha and Kleckner, 1997). Furthermore, repair of dmc1 DSBs and cell viability under RTG conditions is dependent upon RAD54, which is used predominantly for mediating SCR in meiosis (Arbel et al., 1999; Bishop et al., 1999). Deletion of RED1 allows repair of dmc1 DSBs and the formation of joint molecules, but these JMs are between sister chromatids (Schwacha and Kleckner, 1997; Bishop et al., 1999). These observations have led to the idea that there are two ways the cell ensures that crossovers occur between homologous chromosomes during meiosis: first, by using the meiosis-specific Dmc1 protein to promote interhomolog strand invasion, and second, by creating a barrier to SCR that is mediated by RED1.
Our work supports the hypothesis of a RED1-dependent barrier to SCR and provides a molecular explanation for how this barrier might be achieved. Similar to red1, mek1Δ diploids also exhibit repair of dmc1 DSBs (Xu et al., 1997). In addition, both red1 and mek1 show increased levels of meiotic ectopic sister chromatid exchange by a genetic assay (Thompson and Stahl, 1999). Therefore Mek1 also likely plays a role in the barrier to SCR. We have used an inhibitor-sensitive allele of Mek1 to show that Mek1 kinase activity is necessary to maintain the dmc1 arrest after DSB formation. Subsequent inhibition of Mek1 kinase activity is sufficient to allow disappearance of the breaks and sporulation. That the breaks are being repaired, and not just highly resected, is evidenced by high spore viability in mek1-as1 dmc1 spo13 diploids in the presence of inhibitor (Hollingsworth, unpublished data). The key questions then are what substrate(s) is Mek1 phosphorylating and how does this phosphorylation prevent SCR?
Potential Targets of Mek1
SCR is presumably mediated by Rad51, so any type of phosphorylation event that interferes with Rad51 function should have the desired result of preventing recombination between sisters. In vitro, Rad51 function is facilitated by the presence of the single-stranded binding protein complex RPA, as well as mediator proteins such as Rad52, Rad55, and Rad57, which load Rad51 onto RPA-bound ssDNA (Symington, 2002). RPA exhibits meiosis-specific phosphorylation and therefore could be a target of Mek1 (Brush et al., 2001). However, given that Rad51 foci are phenotypically wild type in a dmc1 mutant, it would seem that Rad51 is being efficiently loaded onto DNA, which makes proteins that promote Rad51 loading less likely targets (Bishop, 1994). Rad51 itself could be a substrate, but given that Rad51 also plays a role in interhomolog recombination, the Mek1-derived phosphate could not generally affect Rad51 activity but would have to do so in a way specific to sister chromatid invasion. A potential target that we favor is the Rad54 protein. In vitro, Rad54 stimulates the ability of Rad51 to form D-loops, similar to the SEI intermediates formed in vivo (Petukhova et al., 1999). RAD54 is required for repair of meiotically induced DSBs in haploid, but not diploid, cells under RTG conditions, consistent with a role in mediating sister chromatid recombination (Arbel et al., 1999). As mentioned previously, the repair of dmc1 DSBs after RTG requires RAD54. The interpretation is that RTG removes the barrier to SCR, allowing Rad54/Rad51-mediated strand invasion to occur. Finally, overexpression of RAD54 during meiosis suppresses the dmc1 sporulation defect by repair of the DSBs (Bishop et al., 1999). These authors proposed that RAD54 overexpression “has the ability to overcome a constraint that normally makes conversion of DSBs to recombination products depend on DMC1” (Bishop et al., 1999). We propose that phosphorylation of Rad54 by Mek1 interferes with its ability to promote Rad51-mediated SCR. Mek1 may be unable to phosphorylate all of the Rad54 protein when it is overexpressed, resulting in a pool of unphosphorylated Rad54 that can now work with Rad51 to allow SCR.
The Role of Red1 in Mek1 Kinase Activation
red1 and mek1 share many similar phenotypes such as defects in DSB formation, sister chromatid cohesion, and partner choice, recombination, spore viability, and the meiotic recombination checkpoint (Xu et al., 1997; Bailis and Roeder, 1998; Thompson and Stahl, 1999). These similarities have been interpreted in light of a model for Mek1/Red1 function that is based on the premise that Red1 is phosphorylated by Mek1 (de los Santos and Hollingsworth, 1999; Bailis and Roeder, 2000). This model was based primarily on experiments showing a loss of the Red1 phosphoprotein in mek1 diploids. Our current work strongly suggests that this model is wrong. Our published result showing a change in Red1–3HA mobility in the absence of MEK1 was an artifact created by using a particular transformant that did not contain enough copies of the partially functional RED1–3HA allele to generate phosphorylated protein. Furthermore, we failed to see an effect of mek1Δ on Red1 mobility in the BR strain background, in contradiction to published results (Bailis and Roeder, 1998, 2000). Additional evidence that Red1 is not a Mek1 target is that Mek1-as1 failed to label Red1 in vitro. The finding that RED1 is required for maximal levels of Mek1 kinase activity is consistent with the idea that, rather than being a Mek1 substrate, RED1 functions upstream of Mek1. The RED1 requirement for activation of Mek1 is not an artifact of using the Mek1-as1 protein, as the same result was obtained using Mek1 and radioactive ATP. Furthermore, the need for RED1 to activate Mek1 can be bypassed by using a mutation that constitutively activates Mek1. Therefore, rather than being a substrate of Mek1, Red1 acts upstream to enhance Mek1 kinase activity.
A Model for Hop1/Red1/Mek1 Function in the Regulation of Meiotic DSB Repair
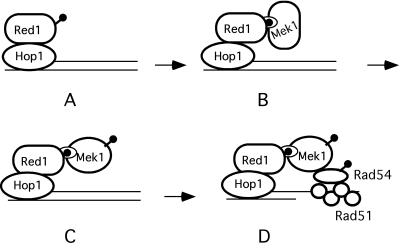
HOP1 is required to activate Mek1, as are Red1/Hop1 complexes. Hop1 is a DNA binding protein with a strong preference for binding to GC-rich sequences (Muniyappa et al., 2000). In addition, Hop1 protects against the degradation of linear DNA fragments in vitro, suggesting a role near the ends of breaks (Kironmai et al., 1998). We propose that Hop1 binds to sites where DSBs will form, thereby recruiting Red1 to these sites as well (de los Santos and Hollingsworth, 1999) (Figure 8A). In this way, the regional preference of Red1 for GC-rich isochores could be explained (Schwacha and Kleckner, 1997). Hop1 localization to chromosomes is independent of DSB formation, as expected if Hop1 is bound to DNA before the breaks are initiated (Smith and Roeder, 1997). Although this idea contradicts cytological data that indicate the Hop1 localization to chromosomes depends upon RED1 (Smith and Roeder, 1997), it is consistent with the fact that hop1 mutants exhibit more severe reductions in recombination and DSBs than red1 and that DSBs formed in a red1 mutant are dependent upon HOP1 (Rockmill and Roeder, 1990; Baumgartner and Hollingsworth, unpublished data). One explanation for this conundrum is that the amount of Hop1 present at DSB sites was not sufficient to be detected by the cytological methods that were used.
Figure 8.
Model for Hop1/Red1/Mek1 function during meiosis. (A) Hop1/Red1 complexes bind to DNA at sites where DSBs will form. (B) Phosphorylated Red1 forms a complex with the FHA domain of Mek1. (C) Mek1 is activated by phosphorylation of T327. (D) Mek1 phosphorylates substrates involved in the barrier to SCR. Lollipops indicate phosphates.
Once Hop1/Red1 complexes are localized to DNA, phosphothreonines on Red1 may provide a recognition sequence for the FHA domain of Mek1 (located between amino acids 47 and 102) (Figure 8B). This idea is analogous to the situation in vegetative cells that have sustained DNA damage. In this case, Rad9 becomes phosphorylated and phosphothreonines on Rad9 act to recruit Rad53, an Mek1 paralog, through its FHA domains to sites of DNA damage (Durocher et al., 1999). Substitution of a conserved arginine with alanine within the Rad53 FHA1 domain (R70A) abolishes the interaction with Rad9 (Durocher et al., 1999). The analogous mutation in the Mek1 FHA domain (R51A) creates a null allele with regard to spore viability, as well as reducing Mek1 kinase activity and disrupting the interaction between Mek1 and Red1. According to our model, binding of Mek1 to phosphorylated Red1 then allows activation of the kinase by phosphorylation of threonine 327 by an as yet unidentified kinase (Figure 8C). Consistent with our model, Mek1 localization to chromosomes requires both HOP1 and RED1 and Mek1 is not phosphorylated in the absence of RED1 (Bailis and Roeder, 1998). The FHA domain, however, is not the sole means by which Mek1 may be localized to chromosomes. The first 63 amino acids of Mek1 are sufficient to bring Mek1 to chromosomes (Bailis and Roeder, 1998). It is possible that Mek1 may also interact via a different domain with Hop1, given that overexpression of MEK1 suppresses specific point mutations in the C-terminal tail of the Hop1 protein (Niu and Hollingsworth, unpublished data). Once activated, Mek1 phosphorylates protein(s) that inhibit DSB repair by SCR, for example, Rad54 (see above) (Figure 8D).
In summary, this work provides a molecular model for how three meiosis-specific chromosomal core components, Hop1, Red1, and Mek1, interact with recombination proteins to ensure that crossovers occur between nonsister chromatids of homologous chromosomes. A chemical genetic approach has been used to show that Mek1 functions after DSB formation to prevent DMC1-independent DSB repair, perhaps by creating a barrier to SCR. Current models in the literature in which Red1 is a substrate of Mek1 must be reevaluated given biochemical and genetic evidence indicating that Red1 is necessary to activate Mek1 kinase activity. The ability to use ATP analogs in combination with the Mek1-as protein should facilitate the identification of protein substrates required for this barrier.
Acknowledgments
We thank Neta Dean, Janet Leatherwood, Aaron Neiman, Rolf Sternglanz, and Jeff Ubersax for helpful discussions. JoAnne Engebrecht, Jette Foss, Alastair Goldman, Aaron Neiman, and two anonymous reviewers from a previous submission gave useful comments on the manuscript. Doug Bishop and Doug Kellogg provided useful reagents. Cellular Genomics generously provided the labeled analogs. We thank Brittany Larkin, Dana Woltering, and Dana Turney for excellent technical support. This work was supported by National Institutes of Health grant GM-50717 to N.M.H and AI44009 to K.M.S.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–07–0499. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0499.
References
- Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. [DOI] [PubMed] [Google Scholar]
- Arbel, A., Zenvirth, D., and Simchen, G. (1999). Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or. TID1. EMBO J. 18, 2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis, J.M., and Roeder, G.S. (1998). Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 12, 3551–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis, J.M., and Roeder, G.S. (2000). Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell 101, 211–221. [DOI] [PubMed] [Google Scholar]
- Bailis, J.M., Smith, A.V., and Roeder, G.S. (2000). Bypass of a meiotic check-point by overproduction of meiotic chromosomal proteins. Mol. Cell. Biol. 20, 4838–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom-Slack, C.A., Ross, L.O., and Dawson, D.S. (1997). Chiasmata, crossovers and meiotic chromosome segregation. Adv. Genetics 35, 253–284. [DOI] [PubMed] [Google Scholar]
- Bishop, A.C., Buzko, O., and Shokat, K.M. (2001). Magic bullets for protein kinases. Trends Cell Biol. 11, 167–172. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K. (1994). RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell 79, 1081–1092. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., Nikolski, Y., Oshiro, J., Chon, J., Shinohara, M., and Chen, X. (1999). High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells 4, 425–443. [DOI] [PubMed] [Google Scholar]
- Bishop, D.K., Park, D., Xu, L., and Kleckner, N. (1992). DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation and cell cycle progression. Cell 69, 439–456. [DOI] [PubMed] [Google Scholar]
- Brush, G.S., Clifford, D.M., Marinco, S.M., and Bartrand, A.J. (2001). Replication protein A is sequentially phosphorylated during meiosis. Nucleic Acids Res. 29, 4808–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos, T., and Hollingsworth, N.M. (1999). Red1p: a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J. Biol. Chem. 274, 1783–1790. [DOI] [PubMed] [Google Scholar]
- Durocher, D., and Jackson, S.P. (2002). The FHA domain. FEBS Letters 513, 58–66. [DOI] [PubMed] [Google Scholar]
- Durocher, D., Henckel, J., Fersht, A.R., and Jackson, S.P. (1999). The FHA domain is a modular phosphopeptide recognition motif. Mol Cell 4, 387–394. [DOI] [PubMed] [Google Scholar]
- Durocher, D., Taylor, I.A., Sarbassova, D., Haire, L.F., Westcott, S.L., Jackson, S.P., Smerdon, S.J., and Yaffe, M.B. (2000). The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6, 1169–1182. [DOI] [PubMed] [Google Scholar]
- Engebrecht, J., Hirsch, J., and Roeder, G.S. (1990). Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell 62, 927–937. [DOI] [PubMed] [Google Scholar]
- Hollingsworth, N.M., and Johnson, A.D. (1993). A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by over-expression of two other meiosis-specific genes, RED1, and REC104. Genetics 133, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N.M., and Byers, B. (1989). HOP1: a yeast meiotic pairing gene. Genetics 121, 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N.M., and Ponte, L. (1997). Genetic interactions between HOP1, RED1 and MEK1 suggest that MEK1 regulates assembly of axial element components during meiosis in the yeast, Saccharomyces cerevisiae. Genetics 147, 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth, N.M., Ponte, L., and Halsey, C. (1995). MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 9, 1728–1739. [DOI] [PubMed] [Google Scholar]
- Hunter, N., and Kleckner, N. (2001). The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 106, 59–70. [DOI] [PubMed] [Google Scholar]
- Johnson, L.N., Noble, M.E.M., and Owen, D.J. (1996). Active and inactive protein kinases: structural basis for regulation. Cell 19, 149–158. [DOI] [PubMed] [Google Scholar]
- Keeney, S. (2001). Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52, 1–53. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., Kikuchi, A., Fujii-Nakata, T., Turck, C.W., and Murray, A.W. (1995). Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol. 130, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai, K.M., Muniyappa, K., Friedman, D.B., Hollingsworth, N.M., and Byers, B. (1998). DNA-binding properties of Hop1 protein, a synaptonemal complex component from Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leem, S.-H., and Ogawa, H. (1992). The MRE4 gene encodes a novel protein kinase homologue required for meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 20, 449–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Meuwissen, R.L.J., Offenberg, H.H., Dietrich, A.J.J., Riesewijk, A., van Iersel, M., and Heyting, C. (1992). A coiled-coil related protein specific for synapsed regions of meiotic prophase chromosomes. EMBO J. 11, 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniyappa, K., Anuradha, S., and Byers, B. (2000). Yeast meiosis-specific protein Hop1 binds to G4 DNA and promotes its formation. Mol. Cell. Biol. 20, 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecina, A., Smith, K.N., Mezard, C., Murakami, H., Otha, K., and Nicolas, A. (2002). Target stimulation of meiotic recombination. Cell 111, 173–184. [DOI] [PubMed] [Google Scholar]
- Petukhova, G., Van Komen, S., Vergano, S., Klein, H., and Sung, P. (1999). Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274, 29453–29462. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., and Roeder, G.S. (1990). Meiosis in asynaptic yeast. Genetics 126, 563–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill, B., and Roeder, G.S. (1991). A meiosis-specific protein kinase homologue required for chromosome synapsis and recombination. Genes Dev. 5, 2392–2404. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S., and Bailis, J.M. (2000). The pachytene checkpoint. Trends Genet. 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Schwacha, A., and Kleckner, N. (1997). Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90, 1123–1135. [DOI] [PubMed] [Google Scholar]
- Shah, K., and Shokat, K. (2002). A chemical genetic screen for direct v-Src substrates reveals ordered assembly of a retrograde signaling pathway. Chem. Biol. 9, 35–47. [DOI] [PubMed] [Google Scholar]
- Smith, A.V., and Roeder, G.S. (1997). The yeast Red1 protein localizes to the cores of meiotic chromosomes. J. Cell Biol. 136, 957–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym, M., Engebrecht, J., and Roeder, G.S. (1993). ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72, 365–378. [DOI] [PubMed] [Google Scholar]
- Symington, L.S. (2002). Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66, 630–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, D.A., and Stahl, F.W. (1999). Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics 153, 621–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, E.A., and Roeder, G.S. (1989). Expression and DNA sequence of RED1, a gene required for meiosis I chromosome segregation in yeast. Mol. Gen. Genet. 218, 293–301. [DOI] [PubMed] [Google Scholar]
- Vershon, A.K., Hollingsworth, N.M., and Johnson, A.D. (1992). Meiotic induction of the yeast HOP1 gene is controlled by positive and negative regulatory elements. Mol. Cell. Biol. 12, 3706–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witucki, L.A., Huang, X., Shah, K., Liu, Y., Kyin, S., Eck, M.J., and Shokat, K. (2002). Mutant tyrosine kinases with unnatural nucleotide specificity retain the structure and phospho-acceptor specificity of the wild-type enzyme. Chem. Biol. 9, 25–33. [DOI] [PubMed] [Google Scholar]
- Woltering, D., Baumgartner, B., Bagchi, S., Larkin, B., Loidl, J., de los Santos, T., and Hollingsworth, N.M. (2000). Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell. Biol. 20, 6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T.-C., and Lichten, M. (1994). Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science 263, 515–518. [DOI] [PubMed] [Google Scholar]
- Xu, L., Weiner, B.M., and Kleckner, N. (1997). Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 11, 106–118. [DOI] [PubMed] [Google Scholar]
- Yves-Masson, J., and West, S.C. (2001). The Rad51 and Dmc1 recombinases: a non-identical twin relationship. Trends Biochem. Sci. 26, 131–136. [DOI] [PubMed] [Google Scholar]