Abstract
T cells transformed by Herpesvirus saimiri express seven viral U-rich noncoding RNAs of unknown function called HSURs. We noted that conserved sequences in HSURs 1 and 2 constitute potential binding sites for three host-cell microRNAs (miRNAs). Coimmunoprecipitation experiments confirmed that HSURs 1 and 2 interact with the predicted miRNAs in virally transformed T cells. The abundance of one of these miRNAs, miR-27, is dramatically lowered in transformed cells, with consequent effects on the expression of miR-27 target genes. Transient knockdown and ectopic expression of HSUR 1 demonstrate that it directs degradation of mature miR-27 in a sequence-specific and binding-dependent manner. This viral strategy illustrates use of a ncRNA to manipulate host-cell gene expression via the miRNA pathway.
Herpesvirus saimiri (HVS) infects T cells and causes aggressive leukemias and lymphomas in New World primates (1). In transformed marmoset T cells, the most abundant HVS transcripts are seven small noncoding RNAs (ncRNAs) called HSURs (H. saimiri U-rich RNAs) (2–4). HSURs exhibit structural but little sequence similarity to cellular small nuclear RNAs (snRNAs). HSURs are encoded by all HVS subgroups; HSURs 1 and 2 (Fig.1A) are the most highly conserved and the only snRNAs expressed by the closely related Herpesvirus ateles (5). Because HSURs are dispensable for transformation in vitro (6, 7), their strong conservation suggests an in vivo role in infected monkeys. HSURs 1 and 2 contain highly conserved AU-rich 5′-end sequences (Fig. 1A and figs. S1 and S2) that are similar to AU-rich elements (AREs) found in the 3′ untranslated regions (3′UTRs) of short-lived mRNAs (8–10). HSURs 1 and 2 are responsible for the up-regulation of a handful of host proteins that are hallmarks of T-cell activation (11) and may contribute to an enhanced growth rate (7) of transformed cells.
Fig. 1.
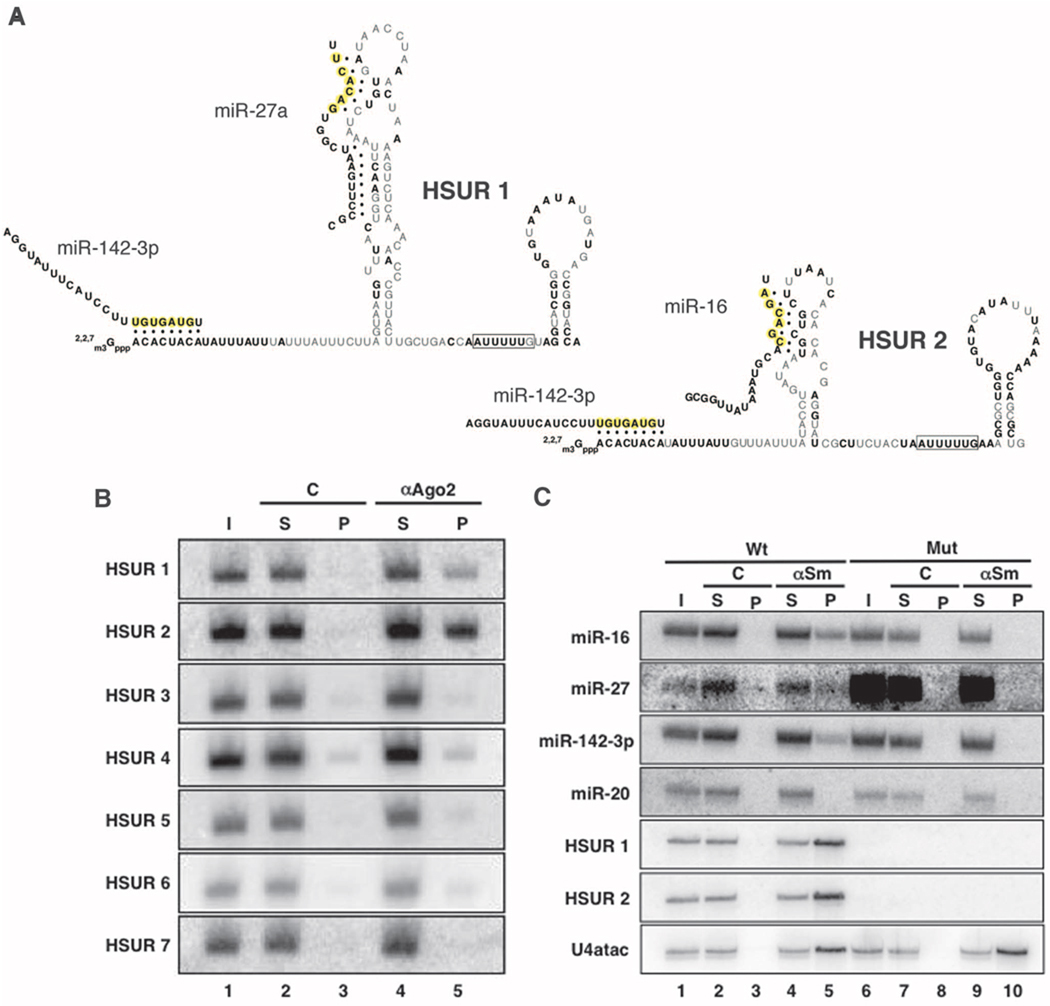
HSURs 1 and 2 bind host miRNAs in virally transformed T cells. (A) Sequences and predicted secondary structures of HSURs 1 and 2. Bold nucleotides are perfectly conserved in all available genome sequences from independent isolates of HVS A, B, and C strains and also in H. ateles (figs. S1 and S2). Complementarity between HSURs and miRNAs is represented by dots; miRNA seed regions are in yellow. (B) Coimmunoprecipitation of HSURs from extracts of virally transformed marmoset T cells with antibody to Flag (lane 3) or antibody to Ago2 (lane 5). I, input (5%); S, supernatant (5%); P, pellet (100%). (C) Coimmunoprecipitation of miRNAs from extracts of virally transformed marmoset T cell lines expressing (Wt, lanes 1 to 5) or lacking HSURs 1 and 2 (Mut, lanes 6 to 10) with Y12 antibody (αSm, lanes 4, 5 and 9, 10) or nonimmune serum (C, lanes 2, 3 and 7, 8). I, input (2%); S, supernatant (2%); P, pellet (100%). Northern blots in (B) and (C) were probed for HSURs, miRNAs, or U4atac, as an αSm immunoprecipitation control.
Comparisons of HSUR 1 (fig. S1) and HSUR 2 (fig. S2) between HVS strains identified stretches of perfectly or highly conserved sequences (Fig. 1A, bold nucleotides). Bioinformatic searches then revealed complementarity between these HSUR sequences and three microRNAs (miRNAs) expressed in T cells: miR-142-3p, miR-27, and miR-16 (Fig. 1A) (12).
Coimmunoprecipitation experiments on extracts of virally transformed marmoset T cells using antibodies to Ago2 showed that HSURs 1 and 2 were selectively present as compared with control immunoprecipitates, whereas all other HSURs (Fig. 1B, lanes 3 and 5) and cellular small nuclear ribonucleoproteins (snRNPs) (fig. S3) did not detectably associate with micro-ribonucleoproteins (miRNPs). Immunoprecipitation was then performed on extracts from T-cell lines transformed with either the wild-type HVS strain A11 or a mutant deleted for HSURs 1 and 2 (7) by using antibodies to Sm proteins, which recognize both cellular and viral snRNPs (3). Figure 1C (lanes 5 and 10) reveals the association of miR-16, miR-27, and miR-142-3p, and not of the control miR-20, but only when HSURs 1 and 2 are present. Psoralen [aminomethyltrioxsalen (AMT)] crosslinking experiments (fig. S4) (13) confirmed the existence of in vivo interactions between miR-27 and HSURs.
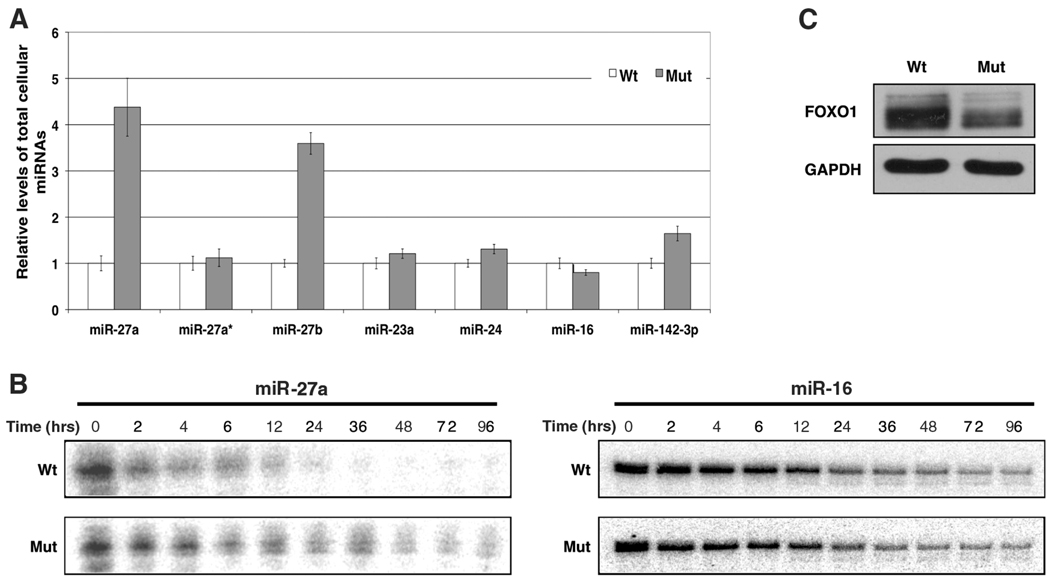
We noticed a distinct difference in the overall level of miR-27 in the marmoset T-cell line transformed by wild-type HVS as compared with that of the mutant lacking HSURs 1 and 2 (Fig. 1C and fig. S5). The miR-27 family includes miR-27a and miR-27b, which are transcribed from different chromosomes and differ by only one nucleotide near the 3′ end. Quantitative real-time polymerase chain reaction (PCR) confirmed the higher abundance of both miR-27a and miR-27b in transformed T cells lacking HSURs 1 and 2 (Fig. 2A). Levels of miR-23a and miR-24, two miRNAs contained in the same primary transcript as miR-27a (14), were unchanged (Fig. 2A and fig. S5), which suggested posttranscriptional differences in the expression of miR-27 between these two cell lines. The abundance of the precursor miRNAs (pre-miRNAs) for miR-27a and miR-27b (fig. S6) did not differ between the two cell lines, nor did that of the passenger strand of the miR-27a duplex (Fig. 2A), which suggests that miRNA processing by Drosha and Dicer (15) is not altered.
Fig. 2.
The presence of HSURs 1 and 2 affects miR-27a abundance, decay, and target expression. (A) Relative levels of different mature miRNAs in virally transformed marmoset T cells expressing (Wt) or lacking (Mut) HSURs 1 and 2 were determined by means of quantitative real-time PCR. (B) Pulse-chase assay assessing the decay of radioactively labeled synthetic miR-27a and miR-16. (C) Western blot analysis of FOXO1 in marmoset T cells transformed by HVS expressing (Wt) or lacking (Mut) HSURs 1 and 2.
To determine whether the difference in the abundance of miR-27 is due to a change in the stability of the mature miRNA, we designed a pulse-chase strategy using synthetic miRNA duplexes (16) in which only the guide strand was radio-actively labeled. After nucleofection (the “pulse”), we monitored the miRNA remaining over time (the “chase”; Fig. 2B) in marmoset T cell lines transformed by either wild-type HVS or mutant HVS lacking HSURs 1 and 2. miR-27a was degraded more rapidly in the wild-type transformed cells, whereas no difference was observed for either miR-16, which is predicted to bind HSUR 2 (Fig. 1A), or for the control miR-20a (fig. S7).
HSURs 1 and 2 do not affect the steady-state levels of host mRNAs in virally transformed marmoset T cells, except for eight genes (8, 11) that are not predicted targets of HSUR-bound miRNAs (17). We analyzed the levels of fork-head box 1 (FOXO1) protein, whose mRNA is a validated target of miR-27 (18). The difference in miR-27 abundance correlates with up-regulation of the FOXO1 protein (Fig. 2C) in the presence of HSURs 1 and 2, which suggests that these HVS ncRNAs perturb host gene expression via the miRNA pathway.
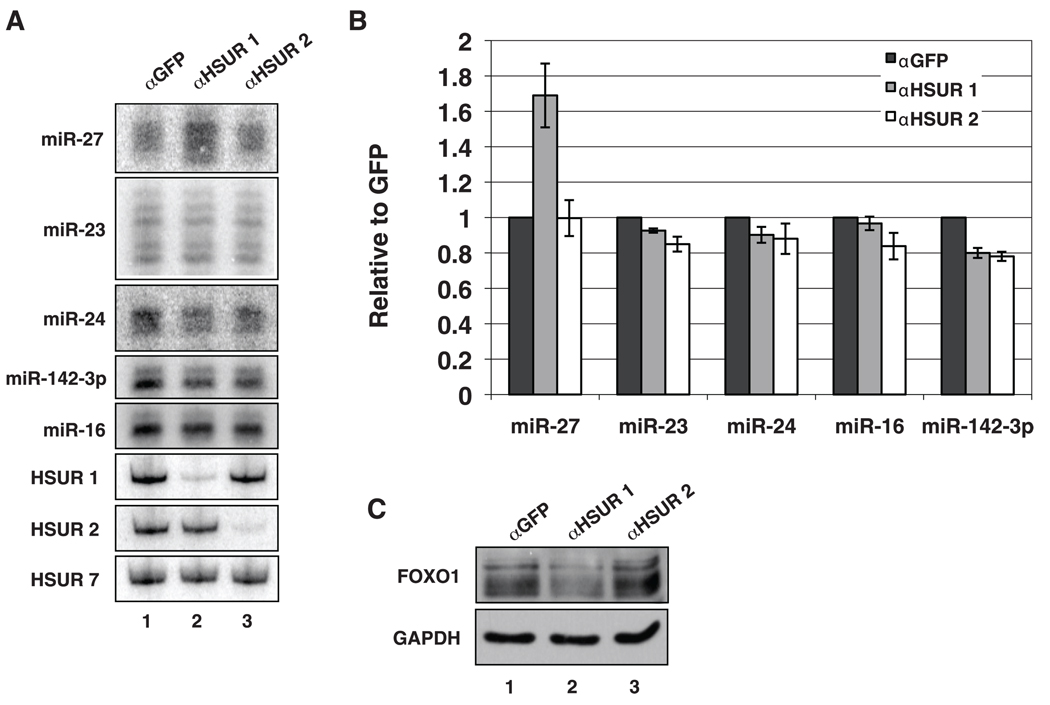
To confirm that the difference in miR-27 levels does not result from accumulated mutations in the two HVS-transformed T cell lines, we treated cells that contained wild-type HVS with chimeric oligonucleotides that effectively induce degradation of complementary nuclear RNAs (19). Knockdown of HSUR 1 but not of HSUR 2 correlated with higher levels of miR-27 (Fig. 3, A and B) and with lower levels of the miR-27 target protein, FOXO1 (Fig. 3C), which suggests that HSUR 1 is specifically involved in regulating miR-27.
Fig. 3.
HSUR 1 regulates the abundance of miR-27 in virally transformed T cells. (A) Northern blot analyses of miRNAs and HSURs after nucleofection with chimeric oligonucleotides anti-sense to GFP (lane 1), HSUR1 (lane 2), or HSUR 2 (lane 3). (B) Quantification of miRNAs from three independent experiments performed as in (A). (C) Western blot of FOXO1 in HVS-transformed marmoset T cells nucleofected as described in (A).
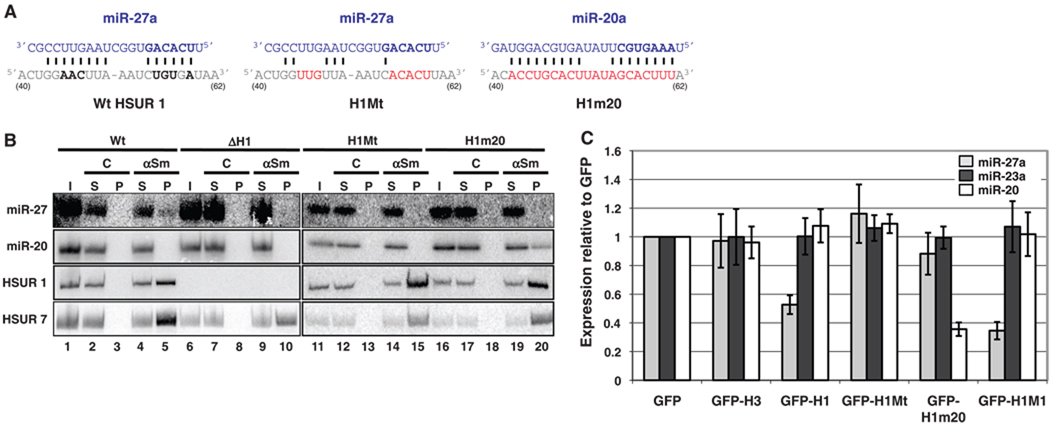
Direct base-pairing between HSUR 1 and miR-27 is required to control miRNA abundance. Human Jurkat T cells were stably transfected with a plasmid containing HVS DNA that encodes all seven HSURs, including their endogenous transcription and processing signals (7). Precipitation of HSUR snRNPs with antibodies to Sm proteins confirmed their association with miR-27 in extracts of this cell line (Fig. 4B, lanes 1 to 5). In contrast, antibodies to Sm proteins did not coimmunoprecipitate miR-27 from extracts of Jurkat T cells stably transfected with a plasmid deleted for the HSUR 1 gene (Fig. 4B, lanes 6 to 10). Likewise, mutation of the conserved miR-27 binding site in HSUR 1 [Fig. 4A, HSUR 1 mutant (H1Mt)] abolished the immunoprecipitation of miR-27 (Fig. 4B, lanes 11 to 15). Furthermore, mutations in HSUR 1 that were designed to produce complementarity to miR-20a (Fig. 4A, H1m20) enabled a previously unknown interaction with miR-20 (Fig. 4B, lanes 16 to 20).
Fig. 4.
HSUR 1 down-regulates mature miRNAs in a sequence-specific and binding-dependent manner. (A) Partial sequences of HSUR 1 (positions 40 to 62) and its mutants (in red) H1Mt and H1m20. Bold nucleotides are perfectly conserved (Fig. 1A). (B) Coimmunoprecipitation of miRNAs with αSm, as in Fig. 1C, from extracts of Jurkat T cells stably expressing HSURs 2 to 7 and either wild-type HSUR 1 (Wt, lanes 1 to 5), no HSUR 1 (ΔH1, lanes 6 to 10), mutant HSUR 1 H1Mt (lanes 11 to 15), or mutant HSUR 1 H1m20 (lanes 16 to 20). (C) miRNA levels in Jurkat T cells fluorescence-activated cell sorted for GFP after transient transfection with empty vector (GFP) or with plasmids expressing GFP and the following: HSUR 3 (GFP-H3), Wt HSUR 1 (GFP-H1), H1Mt (GFP-H1Mt), H1m20 (GFP-H1m20), or H1M1 (GFP-H1M1).
Expression of wild-type HSUR 1 alone (fig. S8) in Jurkat T cells (fig. S9) is sufficient to down-regulate the level of miR-27a as compared with transfection with the empty vector [Fig. 4C; green fluorescent protein (GFP)]. Direct interaction between HSUR 1 and miR-27 is required because cells transfected with a H1Mt that is unable to bind miR-27 (Fig. 4B) have levels of miR-27 comparable with those of cells transfected with the empty vector. Moreover, the miR-20a level was substantially lower after transfection of the HSUR 1mutant (H1m20) that binds this miRNA (Fig. 4B). Together, these results indicate that base-pairing to an internal site in HSUR 1 is both necessary and sufficient to direct a mature miRNA into a cellular degradation pathway.
The ARE-like sequence in HSUR 1 is known to induce in vivo decay of HSUR 1 itself (9), suggesting that the ARE could be involved in the HSUR 1–dependent decay of miR-27. We transfected Jurkat T cells with a mutant HSUR 1 containing two U→G substitutions in the ARE (H1M1) that were previously shown to stabilize and raise cellular levels of HSUR 1 (9). This mutation resulted in higher levels of HSUR 1 (fig. S9) and did not alleviate but produced a more pronounced down-regulation of the abundance of miR-27 as compared with wild-type HSUR 1 (Fig. 4C), indicating that HSUR 1 directs the degradation of miRNAs by an ARE-independent mechanism.
We have demonstrated that HSUR 1 and 2 snRNPs directly bind specific host miRNPs in virally transformed T cells. Whereas the interaction of miR-27 with an internal site in HSUR 1 results in the degradation of this miRNA, the binding of miR-142-3p and miR-16 to HSURs 1 and 2 does not result in their lowered levels (Fig. 2, A and B, and fig. S5). Nonetheless, mutational alteration of its binding site in HSUR 2 indicates that the interaction with miR-16 also occurs via base-pairing (fig. S10), and it is conceivable that if this base-pairing were stronger, decay would be induced. Because HSURs are comparable in abundance with the bound miRNAs in virally transformed T cells (table S1), it seems unlikely that they could effectively compete with mRNA targets and act as miRNA sponges (20) even though down-regulating the activity of these miRNAs might be advantageous for the virus. For instance, miR-16 is reported to target cell-cycle and apoptosis regulators such as Bcl-2 and cyclins D1 and E1 (21, 22), but we do not observe differences in levels of miR-16 target proteins in the presence versus absence of HSURs 1 and 2 (fig. S11). The functional importance of the interaction between HSURs 1 and 2 and miR-16 and miR-142-3p requires further investigation.
It is not yet clear how down-regulation of miR-27 benefits HVS. Down-regulation of the same host miRNA has been reported for another herpesvirus, murine cytomegalovirus, upon infection of cell lines and primary macrophages apparently also at the posttranscriptional level (23). Only a few targets of miRNA-27, including the transcription factors FOXO1, RUNX1 and PAX3, have been validated (18, 24, 25). Thus, identification of additional targets of miR-27 in T cells transformed with HVS is needed, as well as elucidation of the molecular mechanism by which association with HSUR 1 leads to miR-27 decay.
Supplementary Material
Footnotes
Supporting Online Material
www.sciencemag.org/cgi/content/full/328/5985/1563/DC1
Materials and Methods
Figs. S1 to S11
Table S1
References
References and Notes
- 1.Ensser A, Fleckenstein B. Adv. Cancer Res. 2005;93:91. doi: 10.1016/S0065-230X(05)93003-0. [DOI] [PubMed] [Google Scholar]
- 2.Biesinger B, Trimble JJ, Desrosiers RC, Fleckenstein B. Virology. 1990;176:505. doi: 10.1016/0042-6822(90)90020-r. [DOI] [PubMed] [Google Scholar]
- 3.Lee SI, Murthy SC, Trimble JJ, Desrosiers RC, Steitz JA. Cell. 1988;54:599. doi: 10.1016/s0092-8674(88)80004-7. [DOI] [PubMed] [Google Scholar]
- 4.Wassarman DA, Lee SI, Steitz JA. Nucleic Acids Res. 1989;17:1258. doi: 10.1093/nar/17.3.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albrecht JC. J. Virol. 2000;74:1033. doi: 10.1128/jvi.74.2.1033-1037.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ensser A, Pfinder A, Müller-Fleckenstein I, Fleckenstein B. J. Virol. 1999;73:10551. doi: 10.1128/jvi.73.12.10551-10555.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murthy SC, Trimble JJ, Desrosiers RC. J. Virol. 1989;63:3307. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cook HL, Mischo HE, Steitz JA. Mol. Cell. Biol. 2004;24:4522. doi: 10.1128/MCB.24.10.4522-4533.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan XC, Myer VE, Steitz JA. Genes Dev. 1997;11:2557. doi: 10.1101/gad.11.19.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myer VE, Lee SI, Steitz JA. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1296. doi: 10.1073/pnas.89.4.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook HL, et al. Curr. Biol. 2005;15:974. doi: 10.1016/j.cub.2005.04.034. [DOI] [PubMed] [Google Scholar]
- 12.Landgraf P, et al. Cell. 2007;129:1401. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Annu. Rev. Biochem. 1985;54:1151. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Science. 2001;294:853. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 15.Kim VN, Han J, Siomi MC. Nat. Rev. Mol. Cell Biol. 2009;10:126. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 16.Hwang HW, Wentzel EA, Mendell JT. Science. 2007;315:97. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 17.Materials and methods are available as supporting material on Science Online.
- 18.Guttilla IK, White BA. J. Biol. Chem. 2009;284:23204. doi: 10.1074/jbc.M109.031427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ideue T, Hino K, Kitao S, Yokoi T, Hirose T. RNA. 2009;15:1578. doi: 10.1261/rna.1657609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebert MS, Neilson JR, Sharp PA. Nat. Methods. 2007;4:721. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimmino A, et al. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13944. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Q, et al. Nucleic Acids Res. 2008;36:5391. doi: 10.1093/nar/gkn522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buck AH, et al. RNA. 2010;16:307. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ben-Ami O, Pencovich N, Lotem J, Levanon D, Groner Y. Proc. Natl. Acad. Sci. U.S.A. 2009;106:238. doi: 10.1073/pnas.0811466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crist CG, et al. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13383. doi: 10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.We thank A. Giraldez for initial bioinformatic searches, R. C. Desrosiers for plasmids and cell lines, R. Jobava for technical assistance, K. Tycowski and K. Riley for critical commentary, and A. Miccinello for editorial assistance. This work was supported by grant CA16038 from the NIH. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. J.S. is an investigator at the Howard Hughes Medical Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.