Abstract
In the budding yeast Saccharomyces cerevisiae, phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2) is synthesized by a single phosphatidylinositol 3-phosphate 5-kinase, Fab1. Cells deficient in PtdIns(3,5)P2 synthesis exhibit a grossly enlarged vacuole morphology, whereas increased levels of PtdIns(3,5)P2 provokes the formation of multiple small vacuoles, suggesting a specific role for PtdIns(3,5)P2 in vacuole size control. Genetic studies have indicated that Fab1 kinase is positively regulated by Vac7 and Vac14; deletion of either gene results in ablation of PtdIns(3,5)P2 synthesis and the formation of a grossly enlarged vacuole. More recently, a suppressor of vac7Δ mutants was identified and shown to encode a putative phosphoinositide phosphatase, Fig4. We demonstrate that Fig4 is a magnesium-activated PtdIns(3,5)P2-selective phosphoinositide phosphatase in vitro. Analysis of a Fig4-GFP fusion protein revealed that the Fig4 phosphatase is localized to the limiting membrane of the vacuole. Surprisingly, in the absence of Vac14, Fig4-GFP no longer localizes to the vacuole. However, Fig4-GFP remains localized to the grossly enlarged vacuoles of vac7 deletion mutants. Consistent with these observations, we found that Fig4 physically associates with Vac14 in a common membrane–associated complex. Our studies indicate that Vac14 both positively regulates Fab1 kinase activity and directs the localization/activation of the Fig4 PtdIns(3,5)P2 phosphatase.
INTRODUCTION
PtdIns(3,5)P2 is a recently identified phosphoinositide (Whiteford et al., 1997; Dove et al., 1997) that was originally only detectable in the budding yeast, Saccharomyces cerevisiae, after exposure to hyperosmotic stress for short periods of time (Dove et al., 1997). Increases in PtdIns(3,5)P2 synthesis during hyper-osmotic shock has been proposed to regulate vacuole size during adaptation to water loss (Bonangelino et al., 2002). However, PtdIns(3,5)P2 is now known to be present in yeast cells that have not been osmotically shocked (Bonangelino et al., 2002; Gary et al., 2002). PtdIns(3,5)P2 is synthesized by Fab1, a phosphatidylinositol(3)-phosphate 5-kinase (Cooke et al., 1998; Gary et al., 1998). Deletion of FAB1 results in undetectable levels of PtdIns(3,5)P2, slow growth, a grossly enlarged and poorly acidified vacuole, an inability to grow at 38°C and defects in cargo selection for protein sorting within the multivesicular (MVB) body sorting pathway (Yamamoto et al., 1995; Cooke et al., 1998; Gary et al., 1998; Odorizzi et al., 1998). Maximal PtdIns(3,5)P2 synthesis in yeast also requires the products of two other genes, VAC7 (Gary et al., 1998) and VAC14 (Bonangelino et al., 2002; Dove et al., 2002). Consequently, vac7 and vac14 deletion mutants display phenotypes in common with fab1 mutants i.e., grossly enlarged and poorly acidified vacuoles (Bonangelino et al., 1997, 2002; Gary et al., 1998; Dove et al., 2002). Moreover, both vac7 and vac14 are each required for maximal Fab1-mediated PtdIns(3,5)P2 synthesis during hyper-osmotic shock (Gary et al., 1998; Bonangelino et al., 2002; Dove et al., 2002). Interestingly, from a genetic screen designed to identify additional regulators of PtdIns(3,5)P2 levels, we have previously reported that fig4 mutations suppress the vacuole size defect and temperature sensitivity of vac7 mutants by restoring PtdIns(3,5)P2 levels (Gary et al., 2002).
FIG4 was originally identified as a pheromone-induced gene (Erdman et al., 1998) and is one of four genes in S. cerevisiae (SAC1, SJL2/INP52, SJL3/INP53, and FIG4), whose gene products each contain a domain first identified in Sac1, namely the sac domain (reviewed in Hughes et al., 2000a). The sac domain is composed of seven conserved motifs that define a phosphoinositide phosphatase enzymatic activity (Guo et al., 1999; Hughes et al., 2000b). The sixth motif contains the sequence CX5R(T/S) that is thought to represent the catalytic residues of the sac domain (Guo et al., 1999; Hughes et al., 2000b). In vitro, the sac domains of Sac1, Sjl2 and Sjl3, dephosphorylate PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 (Guo et al., 1999; Hughes et al., 2000b). PtdIns(4,5)P2 is not a substrate for yeast sac domain-containing proteins (Guo et al., 1999; Hughes et al., 2000b). However, a human sac domain-containing protein does exist that selectively dephosphorylates PtdIns(4,5)P2 and PtdIns(3,4,5)P3 (hSac2, Minagawa et al., 2001). In addition to containing sac domains, Sjl2 and Sjl3 contain a C-terminal type II PtdIns(4,5)P2 5-phosphatase domain (Stolz et al., 1998; Srinivasan et al., 1997). Consequently, Sjl2 and Sjl3 have the capacity to dephosphorylate all four phosphoinositides synthesized in yeast (Guo et al., 1999; Hughes et al., 2000b; Stefan et al., 2002). Because no phosphoinositide phosphatase activity was demonstrated for Fig4 in vitro, its substrate preference was unknown.
Deletion of FIG4 by itself does not result in any significant change in phosphoinositide levels (Guo et al., 1999; Gary et al., 2002). This result suggests that either the pool of lipid that Fig4 metabolizes is small and tightly regulated (Guo et al., 1999; Gary et al., 2002) or that in the absence of Fig4, other sac domain-containing proteins maintain phosphoinositide levels (Gary et al., 2002; Stefan et al., 2002). The demonstration that deletion of FIG4 suppresses vac7Δ phenotypes (Gary et al., 2002) strongly indicates that Fig4 is a PtdIns(3,5)P2 phosphatase in vivo and that the pool of PtdIns(3,5)P2 is tightly regulated (Gary et al., 2002). Collectively the genetic evidence suggests that in yeast, PtdIns(3,5)P2 levels are determined by the balance of cocommitted pathways of Vac14- and Vac7-activated Fab1 kinase–mediated PtdIns(3,5)P2 synthesis (Gary et al., 1998, 2002), and Fig4–mediated PtdIns(3,5)P2 dephosphorylation (Gary et al., 2002).
The repertoire of yeast sac domain-containing proteins is known (reviewed in Hughes et al., 2000a), and the consequences of their inactivation, either alone or in combination, on the phosphoinositide levels in vivo has been determined (Foti et al., 2001; Gary et al., 2002; Stefan et al., 2002). However, the regulatory mechanisms and proteins that govern the cellular activity and localization of these proteins are not known. In this article we demonstrate that Fig4 is a PtdIns(3,5)P2-specific phosphoinositide phosphatase that localizes to the limiting membrane of the yeast vacuole. Further, we identify Vac14 as a Fig4 interacting protein that both regulates PtdIns(3,5)P2 levels through the control of Fig4 localization and the positive regulation of Fab1 kinase.
MATERIALS AND METHODS
Strains and Media
The genotypes of S. cerevisiae strains used in this study are listed in Table 1. All yeast strains were grown in YPD or SD minimal media containing the necessary amino acid supplements. Routine growth and manipulation of yeast strains were performed using the methods described in Rose et al. (1990).
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3, 112 ura3-52 his3-Δ 200 trp1-Δ 901 lys2-801 suc2-Δ 9 | Robinson et al., 1988 |
| JGY134 | SEY6210; vac7Δ::HIS3 | Gary et al., 2002 |
| JGY137 | SEY6210; fig4Δ::LEU2 vac7Δ::HIS3 | Gary et al., 2002 |
| JGY138 | SEY6210; fig4Δ::LEU2 | Gary et al., 2002 |
| JGY145 | SEY6210; vac14Δ::TRP1 | This study |
| JGY146 | SEY6210; fig4Δ::LEU2 vac14Δ::TRP1 | This study |
| SRY1 | SEY6210; FIG4-GFP:HIS3MX6 | This study |
| SRY2 | SEY6210; FIG4-GFP:HIS3MX6 vac14Δ::HIS3 | This study |
| SRY3 | SEY6210; FIG4-GFP:HIS3MX6 vac7Δ::HIS3 | This study |
| SRY4 | SEY6210; VAC14-GFP:HIS3MX6 | This study |
| SRY5 | SEY6210; VAC14-GFP:HIS3MX6 vac7Δ::HIS3 | This study |
| DAY1 | SEY6210; fig4Δ::LEU2 vac7Δ::HIS3 vac14Δ::TRP1 | This study |
| SRY6 | SEY6210; FIG4Δ761-879-GFP:HIS3MX6 | This study |
| SRY7 | SEY6210; FIG4-GFP:HIS3MX6 vps4Δ::TRP1 | This study |
| SRY8 | SEY6210; VAC14-GFP:HIS3MX6 vps4Δ::TRP1 | This study |
| SRY9 | SEY6210; VAC14-GFP:HIS3MX6 fig4Δ::LEU2 | This study |
| SRY10 | SEY6210; VAC14-GFP:HIS3MX6 fab1Δ::HIS3 | This study |
| SRY11 | SEY6210; FIG4-GFP:HIS3MX6 fab1Δ::HIS3 | This study |
| SRY12 | SEY6210; VAC14-GFP:HIS3MX6 vac7Δ::HIS3 | This study |
Genetic and DNA Manipulations
Restriction and DNA-modifying enzymes were purchased from Roche (Indianapolis, IN) and Life Technologies (Gaithersburg, MD). PCR reactions were performed with PfuTurbo (Stratagene, La Jolla, CA) and AmpliTaq (Applied Biosystems, Foster City, CA) DNA polymerases for ORF and diagnostic reactions, respectively. Standard molecular biology techniques were used for DNA manipulations (Maniatis et al., 1992) and PCR reactions were conducted as directed by manufacturer's instructions. Yeast transformations were performed as described by Ito et al. (1983) and yeast genomic DNA isolated by the method of Hoffman and Winston (1987).
Wild-type strains expressing Fig4-GFP, Fig4Δ761–879-GFP, and Vac14-GFP were engineered by transformation of PCR-amplified genomic integration constructs using GFP-His3MX6 plasmids as templates (Longtine et al., 1998) and primers specific for FIG4 and VAC14. Chromosomal integration was verified by PCR analysis, and expression of Fig4-GFP, Fig4Δ761–879-GFP and Vac14-GFP fusion proteins was confirmed by Western blot analysis.
The FIG4 ORF was amplified from pRS416 FIG4 (Gary et al., 2002) using oligonucleotide primers that incorporated unique BamHI and XhoI restriction enzyme sites at the 5′ and 3′ of FIG4, respectively. The PCR product was digested with BamHI and XhoI and subcloned into BamHI-XhoI–digested pET-15b (Novagen, Madison, WI), generating an N-terminally His-tagged FIG4 allele for expression in Escherichia coli. N-terminally His-tagged fig4-1 (G519R) was subsequently generated by site-directed mutagenesis of the pET-15b FIG4 plasmid, using the Quik-change kit (Stratagene), and confirmed by DNA sequencing.
Determination of Fig4 Phosphoinositide Phosphatase Activity
Fig4 and fig4-1 were expressed in BL21-Star E. coli (DE3) (Invitrogen Life Technologies, Carlsbad, CA) as His-tagged full-length fusion proteins. His-tagged Sac1p(1–507) (Maehama et al., 2000) was a generous gift from Jack Dixon (University of California, San Diego). Competent cells were then transformed according to the manufacturers instructions (Invitrogen Life Technologies).
Protein expression was induced overnight at 30°C by the addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (Roche). His-tagged Fig4 and fig4-1 and His-tagged Sac1p(1–507) (Maehama et al., 2000) were purified from bacterial lysates using Ni2+-agarose affinity resin (Qiagen Inc., Valencia, CA) as essentially described by Maehama et al. (2000) and the manufacturer's instructions. Briefly, bacteria from a 1-liter culture were disrupted with sonication in 30 ml of 50 mM Tris-HCl (pH 8), 300 mM NaCl, 20 mM imidazole-HCl (pH 8) containing one dissolved Complete EDTA-free protease inhibitor cocktail tablet (Roche). Triton X-100 was added (0.5% vol/vol), and the lysate centrifuged at 18, 000 × g for 20 min to remove insoluble material. His-tagged proteins were then isolated from the soluble extract by the addition of 2 ml of equilibrated Ni2+-agarose affinity resin and incubated for 2 h at 4°C. Resins were then washed four times with lysis buffer and twice with lysis buffer without detergent. Phosphoinositide phosphatase activity was then assayed for using His-tagged proteins bound to Ni2+-agarose affinity resin.
Phosphoinositide phosphatase activity assays were conducted as described by Taylor and Dixon (2001) using fluorescent substrates of C6-C6-BODIPY-FLPtdIns(3)P, C6-C6-BODIPY-FLPtdIns(4)P, C6-C6-BODIPY-FLPtdIns(3,5)P2, and C6-C6-BODIPY-FLPtdIns(4,5)P2 (Echelon Biosciences Inc., Salt Lake City, UT). Assays were conducted with ∼50 μl of resin bound with His-tagged phosphatase, in a final volume of 100 μl of 50 mM Tris-HCl (pH 7.5) containing a bath-sonicated liposome mixture of 4 μg phosphatidylethanolamine (Avanti Polar Lipids, Alabaster, AL) and 1 μg of one of the four fluorescent phosphoinositides. After 30 min at 30°C in the presence or absence of 1 mM MgCl, the phosphatase reactions were terminated, and the reaction products were analyzed by TLC using K6 silica gel A 20 × 20-cm glass-backed TLC plates (Whatman Inc., Clifton, NJ) as described by Taylor and Dixon (2001). The fluorescent phosphoinositide spots were visualized under ultraviolet light and quantitated by fluorimetry. Images of TLC plates exposed to ultraviolet light were captured using a Nikon “COOLPIX” digital camera (Nikon Corporation, Tokyo, Japan).
FM4-64 Labeling of Yeast Vacuoles
Vacuole membranes were visualized in vivo using cells labeled with the fluorescent dye FM4-64 (Molecular Probes, Eugene, OR). Labeling was performed as previously described by Vida and Emr (1995), and the cells were observed by fluorescent microscopy.
Fluorescence Microscopy
Fluorescent images of FM4-64–labeled cells, and Fig4-GFP and Vac14-GFP–expressing cells were obtained using a Zeiss Axiovert S1002TV (Thornwood, NY) inverted fluorescent microscope equipped with rhodamine and FITC filters. Images collected were then processed using a Delta Vision deconvolution system (Applied Precision, Seattle, WA) and Adobe Photoshop 6.0 (Adobe Systems Inc., San Jose, CA).
Fractionation, Immunoprecipitation, and Western Blot Analysis
For subcellular fractionation, 10 A600 U of VAC14-GFP cells grown at 26°C to midlogarithmic phase were converted to spheroplasts (Darsow et al., 1997) and then harvested by centrifugation at 500 × g. Spheroplasts were resuspended gently in 1 ml ice-cold lysis buffer (200 mM sorbitol, 50 mM potassium acetate, 20 mM HEPES, pH 7.2, 2 mM EDTA), supplemented with one Complete EDTA-free protease inhibitor cocktail tablet (Roche) and then subjected to 14 strokes in an ice-cold Dounce tissue homogenizer. Lysates were divided into two 0.5-ml aliquots and centrifuged at 4°C for 20 min at 13,000 × g to generate P13 pellets. The 13,000 × g supernatant fractions were centrifuged at 100,000 × g for 1 h at 4°C in a Beckman TLA100.3 rotor, resulting in the production of S100 supernatant fractions and P100 pellet fractions. Triton X-100 was then added to each fraction to a final concentration of 1% (vol/vol), and the fractions were incubated on ice for 30 min with frequent agitation. Detergent-insoluble material was then removed by centrifugation at 18,000 × g for 20 min. Of the two duplicate detergent-soluble supernatants collected for each cell fraction generated, one was TCA-precipitated and acetone-washed for the determination of the total amount of Vac14-GFP and Fig4 present in the supernatant, whereas the second was subject to immunoprecipitation of Vac14-GFP, using Mouse mAb against GFP (Santa Cruz Biotechnology Inc., Santa Cruz, CA) and Protein A-sepharose beads (Amersham Biosciences, Piscataway, NJ). Protein A-sepharose bound immunocomplexes were collected by brief centrifugation and washed three times with ice-cold lysis buffer containing 1% Triton X-100 and three times with detergent-free lysis buffer.
Western blot analysis of SDS-PAGE resolved proteins were performed as previously described by Gary et al. (1998). GFP and HA fusion proteins were detected using mouse monoclonal antibodies against GFP (Santa Cruz Biotechnology Inc.) and HA (12CA5; Roche), respectively. Native Fig4 and Vam3 (Darsow et al., 1997) were detected using rabbit polyclonal antibodies generated by the Emr laboratory. Horseradish peroxidase–conjugated goat anti-mouse and anti-rabbit antibodies (Zymed Laboratories Inc., South San Francisco, CA) and SuperSignal pico chemiluminescent substrate (Pierce Biotechnology Inc., Rockford, IL) were used in concert with x-ray film (Eastman Kodak Company, Rochester, NY) to visualize immunoreactive proteins.
In Vivo Analysis of Phosphoinositides
Analysis of phosphoinositide levels was carried out as previously described by Gary et al. (2002). Cells were labeled with 60 μCi of myo-[2-3H]inositol (Amersham Biosciences) in SD-inositol media for 45 min. Phosphoinositides were then extracted by precipitation with the addition of ice-cold perchloric acid to a final concentration of 4.5% (Whiteford et al., 1996). The phospholipids contained within each cell precipitate were deacylated by treatment with methylamine (Hawkins et al., 1986). Briefly, 0.5 ml of methylamine reagent (10.7% methylamine, 45.7% methanol, 11.4% 1-butanol) was added to each cell precipitate and incubated in a 53°C heat block for 50 min. Unreacted methylamine was then removed in vacuo, and the dried pellet was resuspended in 300 μl sterile water. After a second sequence of drying in vacuo and resuspension in 300 μl sterile water, an equal volume of 1-butanol/ethylether/formic acid ethyl ester (20:4:1) was added. The samples were vortexed for 5 min and centrifuged at 14, 000 × g for 2 min. The aqueous phase containing the [3H]glycero-phosphoinositides were transferred to new tubes and the extraction repeated once more with 1-butanol/ethylether/formic acid ethyl ester (20:4:1). Finally, the aqueous phase was collected and dried in vacuo.
Samples were resuspended in sterile water and 18 × 106 cpm quantities of [3H]glycero-phosphoinositides were analyzed using an anion-exchange Partisphere SAX column (Whatman Inc.) coupled to a gold high-performance liquid chromatography (HPLC) system (Beckman Coulter Inc., Fullerton, CA) and an on-line radiomatic detector (Packard Instrument Company, Meriden, CT) utilizing Ultima Flo scintillation fluid (Packard).
RESULTS
Fig4 is a PtdIns(3,5)P2-specific Phosphoinositide Phosphatase
Fig4 is predicted to be a phosphoinositide phosphatase not only on the basis of genetic evidence (Gary et al., 2002), but also on the basis of sequence homology to the known phosphoinositide phosphatase sac domain (Erdman et al., 1998; Hughes et al., 2000a). However, attempts to demonstrate Fig4 phosphatase activity have been unsuccessful on the basis of in vivo phosphoinositide analysis of fig4 deletion mutants (Guo et al., 1999; Gary et al., 2002). Therefore we elected to test directly whether Fig4 is a phosphoinositide phosphatase using phosphatase assays conducted with recombinant Fig4 protein.
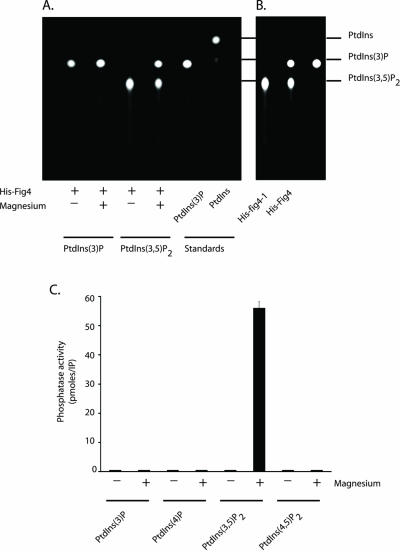
His-tagged full-length Fig4 was expressed in E. coli and purified using Ni2+-agarose affinity resin (MATERIALS AND METHODS). Phosphoinositide phosphatase activity was then assayed using fluorescent derivatives of all four phosphoinositides synthesized in yeast (PtdIns(3)P, PtdIns(4)P, PtdIns(3,5)P2, and PtdIns(4,5)P2). Fluorescent derivatives of phosphoinositides have been successfully used to determine substrate preferences of recombinant phosphoinositide phosphatases, including PTEN and myotubularin (Taylor and Dixon, 2001), and myotubularin-related proteins 1 and 2 (Buj-Bello et al., 2002; Nelis et al., 2002). His-tagged Fig4 phosphatase activity was observed only against PtdIns(3,5)P2 and only when 1 mM magnesium was included in the assay mixture (Figure 1A). The reaction product of Fig4–mediated dephosphorylation migrated with fluorescent PtdIns(3)P and not PtdIns, indicating that only a single phosphate was removed from the inositol head group of PtdIns(3,5)P2. Surprisingly PtdIns(3)P was not a substrate for Fig4 phosphatase (Figure 1A). We were concerned that fluorescent PtdIns(3)P might be a poor substrate for sac domain phosphoinositide phosphatases. However, His-tagged Sac1p(1–507) (Taylor and Dixon, 2001) readily dephosphorylated fluorescent PtdIns(3)P (our unpublished results).
Figure 1.
Fig4 is a magnesium-activated PtdIns(3,5)P2-specific phosphoinositide phosphatase. (A) Recombinant His-tagged full-length Fig4 was assayed in the absence and presence of 1 mM magnesium chloride using fluorescent PtdIns(3)P and PtdIns(3,5)P2 as described in MATERIALS AND METHODS. Reaction products were analyzed by TLC, and the positions of the fluorescent phosphoinositides were visualized with ultraviolet light. (B) Recombinant His-tagged fig4-1 and Fig4 were assayed for phosphatase activity in the presence of 1 mM magnesium using fluorescent PtdIns(3,5)P2 as substrate. (C) Quantitation of His-tagged Fig4 phosphatase activity against fluorescent derivatives of all four known phosphoinositides synthesized in S. cerevisiae.
A mutant allele of FIG4, fig4-1, was originally identified from a genetic screen as a suppressor of vac7Δ mutant phenotypes. fig4-1 contains an arginine instead of glycine at amino acid 519, a position that is located within the seventh conserved motif of the sac domain (Gary et al., 2002). Consequently, the mutation harbored in fig4-1 is predicted to ablate catalytic activity of the Fig4 phosphatase (Gary et al., 2002). We next determined the in vitro phosphatase activity of His-tagged fig4-1 using PtdIns(3,5)P2 as substrate. In the presence of 1 mM magnesium, fig4-1 was unable to catalyze the dephosphorylation of PtdIns(3,5)P2 (Figure 1B). This result not only confirms the prediction that fig4-1 is an inactive phosphatase (Gary et al., 2002), but also eliminates the trivial possibility that magnesium in combination with an unidentified E. coli protein is responsible for catalyzing the dephosphorylation of PtdIns(3,5)P2 in our in vitro phosphatase assays.
Having determined the conditions required to measure Fig4 phosphatase activity in vitro, we next determined whether Fig4 dephosphorylated PtdIns(4)P and PtdIns(4,5)P2. Neither phosphoinositides were Fig4 phosphatase substrates (Figure 1C); therefore we conclude that Fig4 is a magnesium-activated PtdIns(3,5)P2-specific phosphatase.
Fig4 Localizes to the Vacuole Membrane
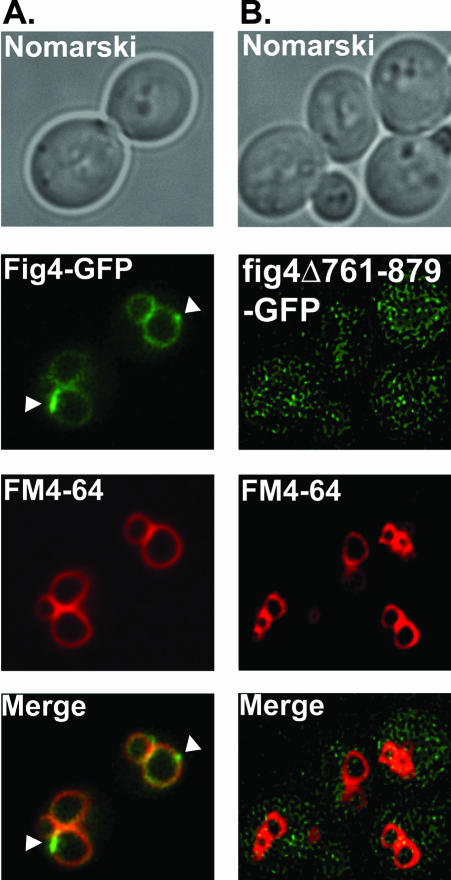
Having demonstrated that Fig4 is a PtdIns(3,5)P2 phosphatase in vitro, we next sought to understand the regulatory mechanisms that govern the cellular localization of Fig4. To begin to address this question, we wanted to observe the cellular localization of Fig4 in vivo. To achieve this, the chromosomal copy of the FIG4 locus was tagged with GFP at the 3′ end. The fusion protein was expressed as a full-length protein under the control of the native FIG4 promoter and is therefore expressed at physiological levels. Fig4-GFP localized predominantly to the vacuole membrane (Figure 2A). Faint Fig4-GFP fluorescence was also detected in the cytoplasm (Figure 2A). To visualize vacuolar membranes directly, FIG4-GFP cells were incubated with FM4-64, a lipophilic fluorescent dye that is transported into cells via the endocytic pathway and ultimately accumulates at vacuolar membranes (Vida and Emr, 1995). Colocalization of Fig4-GFP and FM4-64–stained vacuoles was observed (Figure 2A), demonstrating that Fig4-GFP localizes to the limiting membrane of the vacuole.
Figure 2.
Fig4 localizes to the limiting membrane of the vacuole. Wild-type cells expressing (A) Fig4-GFP or (B) fig4Δ761–879-GFP were labeled with the fluorescent dye FM4-64 for 10 min at 26°C. The dye was washed away and the cells were chased for 60 min at 26°C. The cells were concentrated and chilled on ice, and the localization of FM4-64 and Fig4-GFP was compared by fluorescent microscopy.
Interestingly, we noted that there appeared to be areas of concentrated Fig4-GFP fluorescence on the vacuole membrane (highlighted by white arrow heads, Figure 2A). These patches of Fig4-GFP fluorescence were not coincident with areas where vacuole membranes were closely apposed (Figure 2A), indicating that these patches likely define areas enriched in Fig4 phosphatase.
This result is consistent with Fig4 being a peripheral membrane-associated protein that regulates the levels of PtdIns(3,5)P2 at the vacuole membrane necessary for vacuolar size control. We did not observe GFP fluorescence within intracellular punctate structures distinct from the vacuolar membrane (Figure 2A), indicating that little or no Fig4 is present on other structures, such as Golgi, endosomes, or endoplasmic reticulum.
Because Fig4 lacks an obvious transmembrane domain (Erdman et al., 1998; Gary et al., 2002), we next sought to determine the region of Fig4 that was required for vacuole localization. Fig4 orthologues contain sequences C-terminal to the sac domain devoid of any recognizable protein motif (catalytic or otherwise); we therefore reasoned that the C-terminus of Fig4 might be responsible for targeting the phosphatase to the vacuole. To address this possibility, the chromosomal copy of the FIG4 locus was tagged with GFP, such that a truncated Fig4-GFP fusion protein was generated that was deleted for 119 amino acids at the C-terminus. Analysis of the localization of Fig4Δ761–879-GFP revealed that the fusion protein was no longer localized to the limiting membrane of the vacuole, but instead was observed exclusively within the cytoplasm of the cell (Figure 2B). This result indicates that the C-terminus of Fig4 is required for proper protein folding and/or contains amino-acid sequences necessary to target the phosphatase domain to the vacuole.
Vac14 Recruits Fig4 to the Vacuole
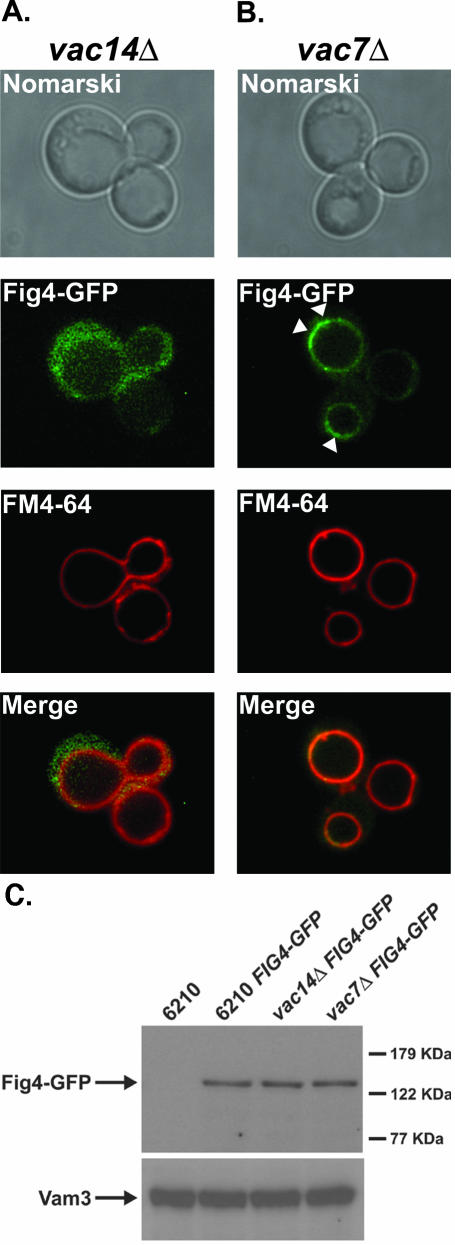
We next wanted to understand how Fig4 associates with the vacuole membrane. We reasoned that one of the known regulators of the Fab1 PtdIns(3)P 5-kinase, Vac7 or Vac14, may recruit the Fig4 phosphatase to the vacuole. Consequently, the cellular localization of Fig4-GFP in FM4-64–stained vac14Δ and vac7Δ mutants was compared with that of wild-type cells (Figure 3, A and B). In vac14Δ mutants, Fig4-GFP fluorescence was observed only within the cytoplasm (Figure 3A). No phosphatase signal was detected at the vacuole membrane (Figure 3A). In contrast, in vac7Δ mutants, Fig4-GFP fluorescence was observed at the limiting membrane of the grossly enlarged vacuoles, with faint fluorescence detectable within the cytoplasm (Figure 3B). This pattern of fluorescence was essentially identical to that of Fig4-GFP in wild-type cells (Figure 2A). Moreover, more intense patches of Fig4-GFP fluorescence were again observed on the vacuole; however, this time they were more pronounced because of the grossly enlarged vacuole morphology of vac7Δ mutants (highlighted by white arrow heads, Figure 3B).
Figure 3.
Vac14 recruits Fig4 phosphatase to the limiting membrane of the vacuole. The vacuoles of (A) vac14Δ and (B) vac7Δ mutants expressing Fig4-GFP were visualized by FM4-64 staining as described in MATERIALS AND METHODS. The localization of FM4-64 and Fig4-GFP were compared by fluorescent microscopy. (C) The total cellular protein content of wild-type, vac7Δ, and vac14Δ mutants were precipitated with TCA and resolved by SDS-PAGE. Fig4-GFP was detected by Western blotting with anti-GFP antibody. Western blotting with anti-Vam3 served as a loading control.
Genetic inactivation of FIG4 stabilizes PtdIns(3,5)P2 levels in vac7Δ mutants and suppresses the large vacuole phenotype exhibited by vac7Δ mutants (Gary et al., 2002). Consequently preservation of the large vacuole phenotype of vac7Δ mutants in vac7Δ FIG4-GFP mutants (compare the FM4-64 staining of wild-type cells FIG4-GFP in Figure 2A with that of vac7Δ FIG4-GFP mutants in Figure 3B) demonstrates that the Fig4-GFP fusion protein is functional in vivo.
Because the vacuoles of vac14Δ and vac7Δ mutants are both grossly enlarged, they would be expected to share the same altered physical properties. Therefore, the fact that Fig4-GFP localizes to the grossly enlarged vacuoles of vac7Δ mutants eliminates a trivial possibility that the altered vacuole properties of vac14Δ mutants, and not the direct loss of Vac14 protein, are prohibiting the association of Fig4 with the vacuole membrane.
The failure of Fig4-GFP to localize correctly in the absence of Vac14 could possibly be due to protein instability in vac14Δ mutants, i.e., the GFP fluorescence observed in vac14Δ mutants could be generated by proteolysis of the Fig4-GFP fusion. Therefore, we determined by Western blot analysis, the steady state levels of Fig4-GFP in wild-type, vac14Δ, and vac7Δ mutants (Figure 3C). Inspection of Western blots obtained from total cell extracts of equal loading from wild-type, vac14Δ, and vac7Δ mutants revealed that intact Fig4-GFP was expressed at comparable levels in all strain backgrounds (Figure 3C). Therefore, we can conclude that the absence of Vac14 does not influence Fig4-GFP stability, but rather directly influences the capacity of Fig4 phosphatase to localize to the vacuole (Figure 3A).
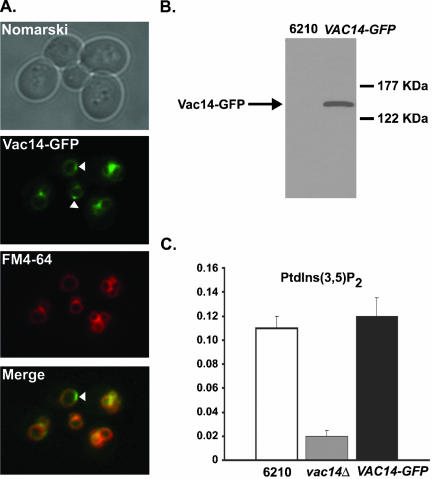
Vac14 Localizes to the Vacuole Membrane
If Vac14 determines Fig4 localization to the vacuole, then Vac14 itself also should localize to the limiting membrane of the vacuole. To observe Vac14 localization in vivo, the chromosomal copy of the VAC14 locus was tagged in frame with GFP at the 3′ end. Vac14-GFP localized almost exclusively to the vacuole membrane, with faint fluorescence detectable in the cytoplasm (Figure 4A). As with Fig4-GFP, we observed patches of more intense Vac14-GFP fluorescence on the vacuole membrane (highlighted with white arrows, Figure 4A) that could not be accounted for by close apposition of vacuole membranes and were not detected as bright patches with FM4-64 (Figure 4A). This indicates that there could exist on the vacuole membrane domains enriched for Fig4 and Vac14. Colocalization of FM4-64 and Vac14-GFP fluorescence confirmed that Vac14 localized to the limiting membrane of the vacuole (Figure 4A). In common with Fig4-GFP fluorescence, we did not detect Vac14-GFP fluorescence within punctate structures distinct from the vacuolar membrane, indicating that Vac14 is not localized to Golgi or endosomal membranes.
Figure 4.
Vac14 localizes to the limiting membrane of the vacuole. (A) Vac14-GFP–expressing wild-type cells were labeled with the fluorescent dye FM4-64 to visualize the vacuole membranes. The localization of FM4-64 and Vac14-GFP were compared by fluorescent microscopy. (B) The total cellular protein content of wild-type and wild-type expressing Vac14-GFP were TCA-precipitated and resolved by SDS-PAGE. Vac14-GFP was detected by Western blotting using anti-GFP antibody. (C) Quantitation of PtdIns(3,5)P2 in wild-type, vac14Δ, and VAC14-GFP strains. 3H-labeled phosphoinositides were isolated, resolved and measured as described in MATERIALS AND METHODS. The levels of PtdIns(3,5)P2 are expressed as a percentage of the total 3H-labeled phosphoinositides analyzed by HPLC.
FM4-64 fluorescence also revealed that the vacuole morphology of VAC14-GFP cells were equivalent to wild-type cells (compare the FM4-64 staining of FIG4-GFP cells in Figure 2A with that of VAC14-GFP cells in Figure 4A). This demonstrates that the Vac14-GFP fusion protein is functional. Further, Vac14-GFP is expressed as a full-length protein (Figure 4B), and VAC14-GFP strains synthesize wild-type quantities of PtdIns(3,5)P2 (Figure 4C).
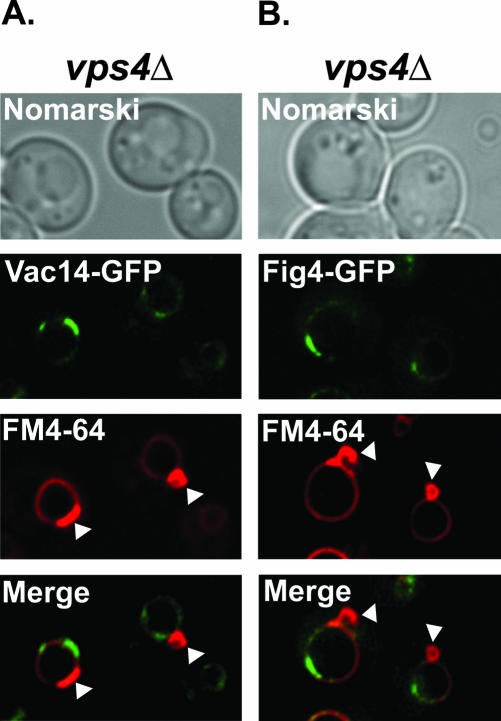
To directly address the possibility that the patches of Vac14-GFP and Fig4-GFP fluorescence represent late endosomes, either fusing with, or closely opposed to the vacuole membrane, we took advantage of the phenotype exhibited by class E VPS mutants. Class E proteins are essential for formation of MVBs (reviewed in Katzmann et al., 2002). Deletion of any of the class E VPS genes, such as VPS4 (Babst et al., 1997) results in the accumulation of endosomal proteins in large aberrant endosome structures, commonly referred to as the class E compartment (Piper et al., 1995; Rieder et al., 1996). The class E compartment can be visualized directly by FM4-64 staining and is recognizable as an intense patch or dot of fluorescence adjacent to the vacuole (Rieder et al., 1996). Analysis of Vac14-GFP (Figure 5A) and Fig4-GFP (Figure 5B) localization in vps4Δ mutants revealed that the localization pattern of both fusion proteins was unaffected in a class E VPS mutant. GFP fluorescence remained visible on the vacuole membrane and within intense patches on the membrane (Figure 5, A and B). Further, these patches of Vac14-GFP (Figure 5A) and Fig4-GFP (Figure 5B) fluorescence remained distinct from the FM4-64–positive class E compartment (highlighted by white arrow heads) in vps4Δ mutants. This result demonstrates that Vac14-GFP and Fig4-GFP are not localized to endosomes.
Figure 5.
Vac14 and Fig4 do not localize to the E compartment of vps4Δ mutants. The vacuoles and the E compartments of vps4Δ mutants expressing either Vac14-GFP (A) or Fig4-GFP (B) were visualized by FM4-64 staining. The localization of the GFP fusion proteins relative to the FM4-64–positive E compartments (highlighted by white arrow heads) were compared by fluorescence microscopy.
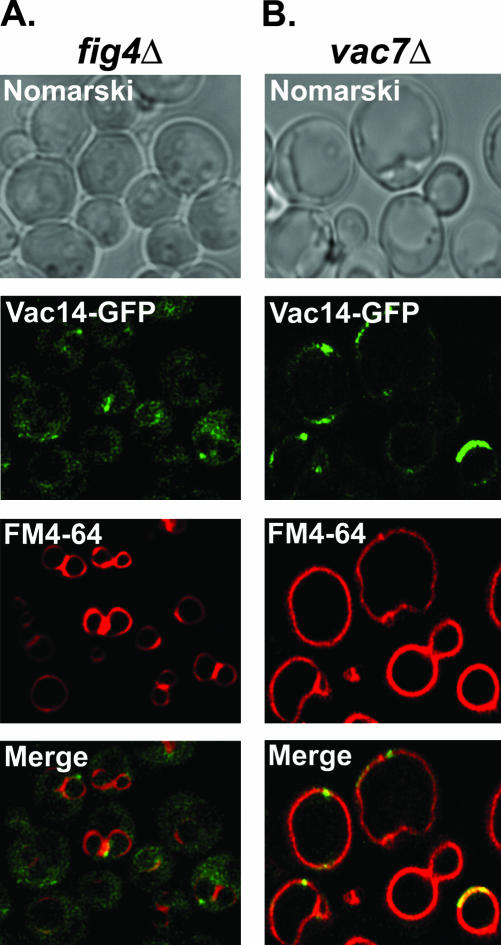
We next determined the localization of Vac14-GFP in fig4Δ mutants and vac7Δ mutants. Compared with wild-type cells, Vac14-GFP fluorescence was more readily observed in the cytoplasm of fig4Δ cells (Figure 6A). However, Vac14-GFP remained at the limiting membrane of the vacuole, and the distinctive patches of vacuole Vac14-GFP fluorescence remained visible in fig4Δ mutants (Figure 6A). This result indicates that Vac14 localization to the vacuole membrane is not absolutely dependent on the presence of the Fig4 phosphatase. Consistent with this interpretation, fig4Δ mutants do not exhibit phenotypes exhibited by Δvac14 mutants (Gary et al., 2002). Therefore, Vac14 is functional in fig4Δ mutants. However, the result does indicate that the localization of Vac14 to the limiting membrane of the vacuole is optimized by the presence of Fig4. In contrast, Vac14-GFP fluorescence was observed almost exclusively at the limiting membrane of the grossly enlarged vacuoles of vac7Δ mutants (Figure 6B). Furthermore, intense patches of Vac14-GFP fluorescence were again observed on the vacuole (Figure 6B). This pattern of fluorescence was essentially identical to that of Vac14-GFP in wild-type cells (Figure 4A). This result indicates that Vac7 does not control the vacuole localization of either Vac14 or Fig4.
Figure 6.
Vac14 localizes to the vacuole more efficiently when associated with Fig4. Vac14-GFP localization was determined in FM4-64–labeled fig4Δ (A) or vac7Δ (B) mutants by fluorescence microscopy.
Fab1 Kinase Is Required for Vacuole Localization of both Vac14 and Fig4
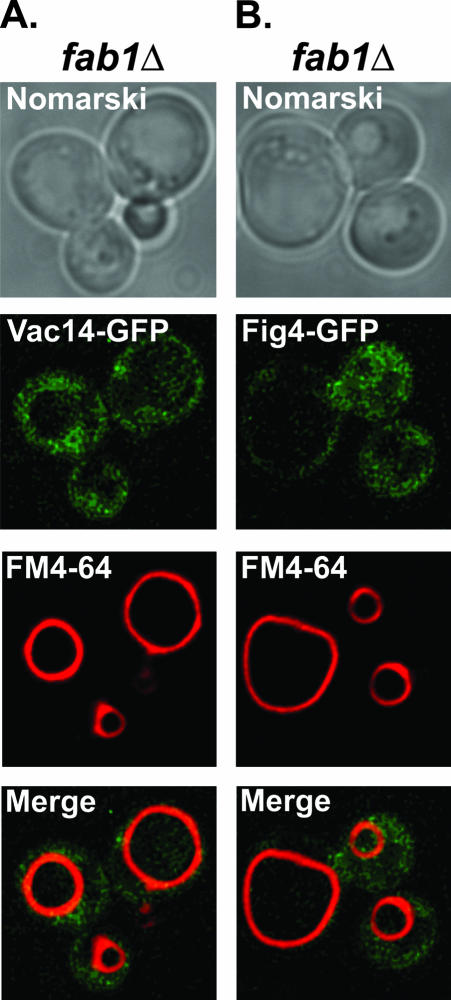
Bonangelino et al. (2002) reported that significantly reduced amounts of Vac14 fractioned with the vacuoles of fab1Δ mutants. Consequently, we determined the localization of Vac14-GFP and Fig4-GFP in fab1Δ mutants. Both Vac14-GFP (Figure 7A) and Fig4-GFP (Figure 7B) failed to localize to the grossly enlarged vacuoles of fab1Δ mutants. Taken together, our results indicate that Fab1 kinase, but not Vac7, regulates the vacuole localization of Vac14 and Fig4. However, because we and Bonangelino et al. (2002) have been unsuccessful in detecting a direct protein-protein interaction between Fab1 and Vac14, additional proteins are most likely required to mediate Vac14 localization to the vacuole.
Figure 7.
Fab1 kinase is required for vacuole localization of Vac14 and Fig4. (A) Vac14-GFP and (B) Fig4-GFP localization were determined in FM4-64–labeled fab1Δ mutants by fluorescence microscopy.
Fig4 Interacts with Vac14 at the Vacuole Membrane
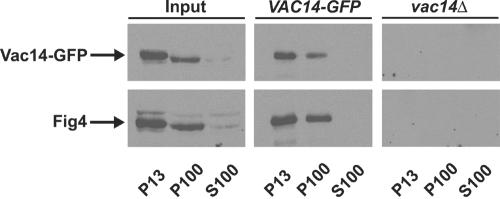
To date, no physical interactions have been detected between any one of the proteins that constitute the known machinery for regulation of PtdIns(3,5)P2 levels, namely the Fab1 kinase, Vac7, Vac14, and Fig4 phosphatase. However, the Vac14-dependent association of Fig4 with vacuole membranes (Figure 3A) and the vacuolar localization of Vac14 (Figure 4A) indicates that Vac14 may physically interact with Fig4. To test this directly, we determined whether both proteins could be coimmunoprecipitated in a common protein complex. Lysates from wild-type Vac14-GFP–expressing cells were fractionated into P13, P100, and S100 fractions. Vac14-GFP was immunoprecipitated from the detergent-solubilized P13 and P100 fractions, but not from the S100 fraction (Figure 8). Western blot analysis was then undertaken to determine whether Fig4 coimmunoprecipitated with Vac14-GFP. Fig4 was detected in immunoprecipitates of Vac14-GFP from both the P13 and P100 fractions (Figure 8). Fig4 did not fractionate to the S100 fraction (Figure 8). These results suggest that Vac14 and Fig4 physically associate within a common membrane associated protein complex.
Figure 8.
Vac14 interacts with Fig4. Coimmunoprecipitation of Vac14-GFP and Fig4 from P13 and P100 fractions. Lysates from wild-type cells expressing Vac14-GFP were fractionated into P13, S100, and P100 fractions, and Vac14-GFP immunoprecipitated from detergent-solubilized fractions, as described in MATERIALS AND METHODS. Vac14-GFP was resolved by SDS-PAGE and visualized by Western blotting using anti-GFP antibody. Fig4 was detected by Western blotting with anti–Fig4 antisera.
Fig4 Mislocalization in vac14Δ Mutants Does Not Contribute to vac14Δ Phenotypes
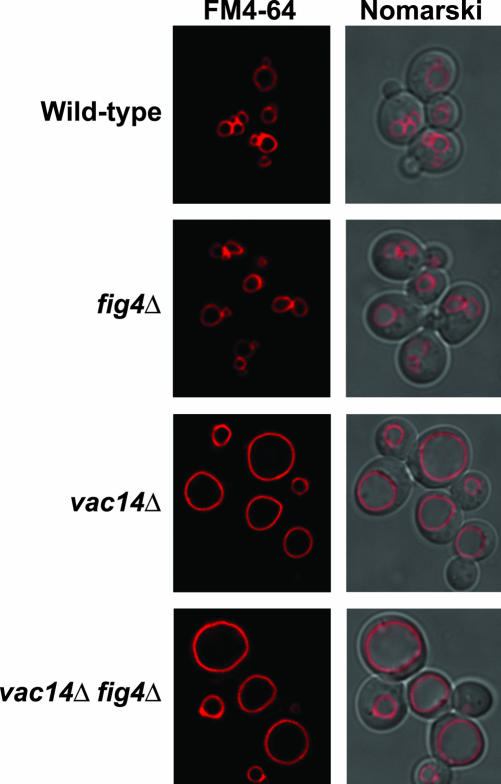
Because vac14Δ mutants are unable to synthesize wild-type levels of PtdIns(3,5)P2 (Bonangelino et al., 2002; Dove et al., 2002) and Fab1 kinase is localized to both endosomes and vacuole membranes (Gary et al., 1998; Bonangelino et al., 2002; Dove et al., 2002), we wanted to ascertain whether or not the mislocalization of the PtdIns(3,5)P2-specific Fig4 phosphatase contributed to the phenotypes exhibited by vac14Δ mutants. We reasoned that an increased level of nonvacuolar Fig4 could in theory gain access to endosomal pools of PtdIns(3,5)P2. Therefore, we deleted FIG4 from vac14Δ strains and examined both the vacuole morphology and phosphoinositide levels of vac14Δ fig4Δ double mutants. FM4-64 staining of the double mutants revealed that vac14Δ fig4Δ double mutants exhibited single grossly enlarged vacuoles (Figure 9). In vivo phosphoinositide analysis of vac14Δ fig4Δ double mutants revealed that PtdIns(3,5)P2 levels were identical to those of vac14Δ single mutants (Table 2). Therefore, this result indicates that mislocalized Fig4 does not contribute to the phenotype exhibited by vac14Δ mutants.
Figure 9.
Mislocalized Fig4 phosphatase does not contribute to vac14Δ phenotypes. Vacuoles of wild-type, fig4Δ, vac14Δ, and vac14Δ fig4Δ strains were visualized by FM4-64 staining and fluorescence microscopy.
Table 2.
3′-Phosphoinositide levels in wild-type, fig4Δ, vac14Δ, and vac14Δ fig4Δ mutants
| Strain | PtdIns(3)P | PtdIns(3,5)P2 |
|---|---|---|
| Wild-type | 2.60 ± 0.12 | 0.10 ± 0.01 |
| fig4Δ | 1.74 ± 0.02 | 0.07 ± 0.01 |
| vac14Δ | 3.07 ± 0.04 | 0.02 ± 0.01 |
| vac14Δ fig4Δ | 1.70 ± 0.01 | 0.02 ± 0.01 |
PtdIns(3)P and PtdIns(3,5)P2 levels in wild-type, fig4Δ, vac14Δ, and vac14Δ fig4Δ mutants, recorded after HPLC analysis of glycerophosphoinositides derived from the extracted phosphoinositides of [3H]-labeled cells. The values reported are expressed as a percentage of the total [3H]cpm glycerophosphoinositides eluted and are representative of at least three independent experiments.
Genetic inactivation of FIG4 suppresses the phenotypes associated with vac7Δ mutants (Gary et al., 2002); however, vac14Δ fig4Δ double mutants exhibit identical phenotypes to that of vac14Δ mutants (Figure 9). Consistent with this result, there was no increased PtdIns(3,5)P2 synthesis detected in vac14Δ fig4Δ double mutants compared with vac14Δ mutants (Table 2). Therefore, fig4Δ mutations neither contribute to, nor suppress vac14Δ phenotypes.
The failure of fig4Δ to suppress vac14Δ might be accounted for by the continued turnover of PtdIns(3,5)P2 by Sjl2, Sjl3, and/or SacI. However, deletion of these sac domain-containing proteins also failed to suppress vac14Δ (our unpublished results). Taken together, these results indicate that in the absence of Vac14, Vac7-dependent activation of Fab1 kinase is attenuated. Therefore, Vac14 is required for maximal Vac7-dependent activation of the Fab1 kinase.
Vac14 Is Required for fig4Δ Suppression of vac7Δ
The data presented demonstrate that Vac14 functions to recruit Fig4 to the vacuole membrane. However, if this were the sole role of Vac14, then one would predict that vac14Δ mutants would synthesize increased amounts of PtdIns(3,5)P2. Instead vac14Δ mutants are unable to synthesize wild-type levels of PtdIns(3,5)P2 (Bonangelino et al., 2002; Dove et al., 2002) and vac7Δ vac14Δ double mutants exhibit the same phenotypes as both the single vac7Δ and vac14Δ deletion mutants (Bonangelino et al., 1997). Moreover, from our analysis of vac14Δ fig4Δ double mutants in this article, we have data that indicate that Vac14 is required for maximal Vac7-dependent activation of the Fab1-kinase. Taken together, these results indicate that Vac14 also performs a second cellular function of positively regulating Fab1 kinase activity, in addition to recruiting Fig4 phosphatase to the vacuole.
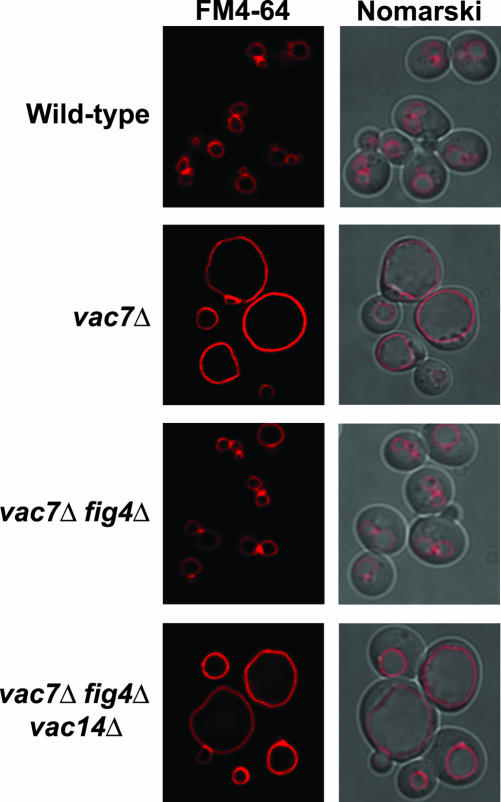
To test whether Vac14 regulates Fab1 kinase activity, we reasoned that deletion of VAC14 from vac7Δ fig4Δ double mutants would eliminate Fab1 kinase activity. Analysis of vac7Δ fig4Δ vac14Δ triple mutants revealed that these mutants exhibited vac7Δ phenotypes; enlarged vacuoles (Figure 10) and in vivo phosphoinositide analysis determined that these mutants synthesized undetectable levels of PtdIns(3,5)P2 (Table 3). Consequently, Vac14 is required for PtdIns(3,5)P2 synthesis in the absence of Vac7 and Fig4. Therefore, Vac14 positively regulates Fab1 kinase. This additional function of Vac14 explains why vac14Δ mutants fail to suppress vac7Δ mutants.
Figure 10.
Vac14 is required for fig4Δ suppression of vac7Δ. The vacuole morphologies of wild-type, vac7Δ, vac7Δ fig4Δ, and vac7Δ fig4Δ vac14Δ strains were visualized by fluorescent staining with FM4-64 and fluorescence microscopy.
Table 3.
3-Phosphoinositide levels in wild-type, vac7Δ, vac7Δ fig4Δ, and vac7Δ fig4Δ vac14Δ mutants
| Strain | PtdIns(3)P | PtdIns(3,5)P2 |
|---|---|---|
| Wild-type | 2.25 ± 0.12 | 0.11 ± 0.01 |
| vac7Δ | 1.41 ± 0.02 | 0 |
| vac7Δ fig4Δ | 1.81 ± 0.04 | 0.07 ± 0.01 |
| vac7Δ fig4Δ vac14Δ | 1.48 ± 0.04 | 0 |
PtdIns(3)P and PtdIns(3,5)P2 levels in wild-type, vac7Δ, vac7Δ, fig4Δ, and vac7Δ fig4Δ vac14Δ mutants recorded after HPLC analysis of glycerophosphoinositides derived from the extracted phosphoinositides of [3H]-labeled cells. The values reported are expressed as a percentage of the total [3H]cpm glycerophosphoinositides eluted and are representative of at least three independent experiments.
DISCUSSION
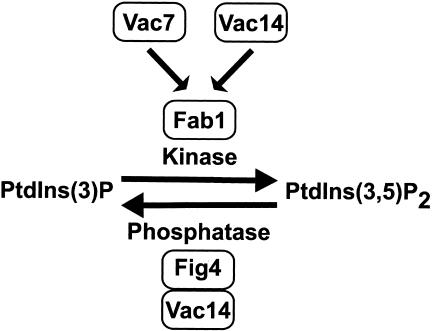
From a genetic screen designed to identify regulators of PtdIns(3,5)P2 levels, a mutant allele of FIG4 was isolated as a suppressor of the vac7Δ phenotypes (Gary et al., 2002). In this article we show for the first time that the sac domain-containing protein Fig4 is a PtdIns(3,5)P2-selective phosphatase in vitro. Further, we have demonstrated that Vac14 regulates membrane association of the Fig4 phosphatase with the vacuole in vivo and have determined that Vac14 and Fig4 exist in a common membrane-associated protein complex. Consistent with Vac14 determining Fig4 phosphatase localization to the vacuole, we have shown that Vac14 localizes to the limiting membrane of the vacuole in vivo. Together, our findings suggest a complex mechanism for Vac14 regulation of PtdIns(3,5)P2 levels necessary for vacuole size control; Vac14 functions both as an activator of Fab1 kinase and as a regulator of the Fig4 sac domain-containing phosphoinositide phosphatase (Figure 11). Our data suggest that these two antagonistic functions of Vac14 in regulating PtdIns(3,5)P2 levels might occur through the existence of domains enriched for either Vac14 or Fig4 on the limiting membrane of the vacuole.
Figure 11.
Model. Vac14 regulates PtdIns(3,5)P2 phosphatase activity at the vacuole membrane by both positively regulating Fab1 kinase and controlling Fig4 phosphatase localization.
Fig4 Is a Magnesium-activated PtdIns(3,5)P2-specific Phosphatase
Previous work by Guo et al. (1999) and Hughes et al. (2000b) identified the sac domains of Sac1, Sjl2, and Sjl3 as phosphoinositide phosphatases that have the capacity in vitro to dephosphorylate PtdIns(4)P, PtdIns(3)P, and PtdIns(3,5)P2. Our biochemical analysis of Fig4 has determined that Fig4 is a phosphoinositide phosphatase that selectively dephosphorylates PtdIns(3,5)P2. This difference in phosphoinositide substrate preferences for each of the sac domains of Fig4, SacI, Sjl2, and Sjl3, is perhaps surprising, but recent work has identified a sac domain containing protein in humans (hSac2) that can only dephosphorylate PtdIns(4,5)P2 and PtdIns(3,4,5)P3 (Minagawa et al., 2001). Therefore as noted by Minagawa et al. (2001), different sac domain-containing proteins could each have their own phosphoinositide preferences. Biochemically uncharacterized orthologues of Fig4 exist in humans (KIAA0274/hFig4/hSac3; Erdman et al., 1998; Hughes et al., 2000a; Minagawa et al., 2001) and mouse (AI326867). Like Fig4, these mammalian orthologues differ from other sac domain-containing proteins by virtue of an extended C-terminus that is devoid of any known phosphoinositide phosphatase motif but that contains a unique 120-amino acid region of homology immediately adjacent to the sac domain (Erdman et al., 1998; Hughes et al., 2000a). Consequently Fig4, hFig4, and mouse Fig4 may represent a subclass of sac domain-containing proteins that selectively dephosphorylate PtdIns(3,5)P2.
A second notable biochemical difference between the phosphatase activities of Fig4 compared with Sac1, Sjl2 and Sjl3 was the magnesium activation of Fig4 phosphatase activity in vitro. The significance of this apparent dependence on or activation by magnesium is not clear. Magnesium has been noted to inhibit the sac domain phosphatase activities of Sjl2 and Sjl3 (Guo et al., 1999), but to activate hSac2 (Minagawa et al., 2001). However, the concentration of magnesium that elicits detectable Fig4 phosphatase activity in vitro, i.e., 1 mM, is well in the range of physiological intra-cellular concentrations of magnesium measured in S. cerevisiae (Beeler et al., 1997). And in yeast the vacuole is a major site of magnesium deposition (Beeler et al., 1997). Consequently, Fig4 phosphatase localized to the vacuole membrane could well be exposed to high local concentrations of magnesium, as these ions flux across the vacuole membrane.
Role of Fig4 Phosphatase in Regulating PtdIns(3,5)P2 Levels at the Vacuole
The Fab1 kinase has been shown previously to localize to both endosomes and vacuole membranes (Bonangelino et al., 1997, 2002; Gary et al., 1998; Dove et al., 2002), consistent with the roles of PtdIns(3,5)P2 in MVB protein sorting and vacuole size control (Odorizzi et al., 1998; Gary et al., 1998, 2002; Bonangelino et al., 2002; Dove et al., 2002). Our analysis of the cellular localization of endogenous levels of Fig4-GFP in living cells has revealed that Fig4 phosphatase is primarily localized to the limiting membrane of the vacuole. Because we did not observe any punctate Fig4-GFP fluorescence signals distinct from the vacuole membrane and the signal did not colocalize to the E compartment in vps4Δ mutants, Fig4 phosphatase is unlikely to be associated with endosomes. Therefore, we assume that Fig4 phosphatase activity is restricted to the vacuole membrane, where it could regulate the levels of PtdIns(3,5)P2 produced by the Fab1 kinase either at the vacuole or at prevacuolar compartments that ultimately dock and fuse with the vacuole. Fig4 phosphatase may function to terminate the MVB-associated PtdIns(3,5)P2 signal when MVBs fuse with the vacuole. Fig4 could also function to regulate the levels of PtdIns(3,5)P2 synthesized at the vacuole membrane necessary for organelle size control (Gary et al., 2002). The existence of a vacuole localized PtdIns(3,5)P2-specific phosphatase enzyme also explains why fig4 mutants were identified as suppressors of vac7Δ (Gary et al., 2002). Vac7 is localized to the vacuole and regulates the activity of Fab1 kinase (Gary et al., 1998). In the absence of Vac7, the low levels of PtdIns(3,5)P2 produced are stabilized in cells missing Fig4 function.
Vac14 Recruits Fig4 Phosphatase to the Vacuole
Fig4, Sjl2, and Sjl3 are peripheral membrane-associated proteins (Stolz et al., 1998; Gary et al., 2002), consequently their ability to dephosphorylate phosphoinositides depends entirely on their relative abilities to localize directly to intra-cellular membranes and/or structures closely apposed to their substrates. In contrast, Sac1 is an integral membrane protein of the endoplasmic reticulum (Foti et al., 2001). Interestingly, Sjl2 and Sjl3 have both been shown to relocalize from a diffuse cytosolic localization to actin patches during osmotic shock (Ooms et al., 2000). However, no interacting proteins have been identified that were responsible for determining the localization of Sjl2 and Sjl3. Although one possible candidate does exist, the Sjl2/Sjl3-interacting protein Bsp1 (Wicky et al., 2003). Our studies of Fig4 localization have uncovered an unanticipated function of Vac14 in determining the vacuole localization of Fig4 and in doing so identified for the first time a mechanism by which a sac domain phosphatase is recruited to access its phosphoinositide substrate. Moreover, we have demonstrated that Vac14 and Fig4 physically associate in a common membrane-associated protein complex. Consistent with this observation is a recent report identifying Fig4 as an interacting protein of Vac14 using the yeast two-hybrid system (Dove et al., 2002).
Endogenous levels of Vac14-GFP in living cells are localized principally to the limiting membrane of the vacuole. Although a faint diffuse fluorescence signal was observed in the cytoplasm, we did not detect any pools of Vac14-GFP on punctate structures. Further, Vac14-GFP fluorescence did not coalesce into the E compartment in vps4Δ mutants. Therefore, like Fig4, we conclude that Vac14 is localized primarily to the limiting membrane of the vacuole and is not associated with endosomes. This result further supports a model in which Vac14 recruits the Fig4 phosphatase to the vacuole and indicates that the Vac14-Fig4 phosphatase complex regulates the vacuolar pool of PtdIns(3,5)P2.
Interestingly, we find that Vac14 localizes to the vacuole membrane more efficiently when associated with Fig4. Consequently, fig4-1 may stabilize Vac14 localization at the vacuole membrane, such that the fig4-1 mutant suppresses vac7Δ phenotypes better than loss of Fig4, i.e., fig4Δ mutants (Gary et al., 2002).
Our studies have shown that Fab1 kinase regulates the localization of the Vac14-Fig4 phosphatase complex to the limiting membrane of the vacuole. This effect is specific to the loss of Fab1 kinase, as opposed to a nonspecific effect precipitated by the formation of a grossly enlarged vacuole, because Vac7 is not required for the vacuole localization of the Vac14-Fig4 phosphatase complex. However, future endeavors will focus on investigating whether there exists a direct interaction between Fab1 kinase and the Vac14-Fig4 phosphatase complex, or instead, the effect on localization is mediated indirectly by unidentified proteins. Presently we favor the second idea, because we and Bonangelino et al. (2002), who first reported that Fab1 kinase influences Vac14 vacuole association, have been unable to demonstrate a direct protein-protein interaction between Fab1 and Vac14.
A common mechanism for the regulation of the Fig4 phosphatase is likely to exist in mammalian cells, as orthologues of both Fig4 (Erdman et al., 1998; Hughes et al., 2000b; Minagawa et al., 2001) and Vac14 (Bonangelino et al., 2002; Dove et al., 2002; Davy and Robinson, 2003) are present in mammals.
Role of Vac14 in Fab1 Kinase–mediated PtdIns(3,5)P2 Synthesis
Having demonstrated that Vac14 determines the vacuole localization of a PtdIns(3,5)P2-specific phosphatase, we entertained the idea that the reduced synthesis of detectable PtdIns(3,5)P2 levels in vac14Δ mutants might be attributable to mislocalized Fig4 phosphatase gaining access to endosomal pools of PtdIns(3,5)P2. However, we found that vac14Δ fig4Δ double mutants exhibited grossly enlarged vacuoles and were unable to synthesize elevated levels of PtdIns(3,5)P2. Instead the failure of fig4Δ to suppress vac14Δ indicated that Vac7-dependent activation of Fab1 kinase was compromised in the absence of Vac14.
Our demonstration that Vac14 is required for fig4Δ suppression of vac7Δ mutants directly indicates that Vac14 positively regulates Fab1 kinase activity. This result is consistent with the model in which Vac14 functions upstream of Fab1 kinase and PtdIns(3,5)P2 synthesis (Bonangelino et al., 2002; Dove et al., 2002). In contrast to Vac14-dependent recruitment of Fig4 to the limiting membrane of the vacuole, Fab1 localization to the vacuole is not dependent on Vac14 (Bonangelino et al., 2002; Dove et al., 2002). Further, no physical association has been detected between Vac14 and Fab1 (Bonangelino et al., 2002). Consequently, the mechanism of Vac14-dependent activation of Fab1 kinase remains unclear.
The data presented in this article demonstrates that Vac14 functions in at least two discrete mechanisms of regulating PtdIns(3,5)P2 levels (Figure 11): Fab1 kinase activation and recruitment of Fig4 phosphatase to the sites of PtdIns(3,5)P2 synthesis at the vacuole membrane. Future work will focus on how Vac14 performs these two seemingly antagonistic functions. One plausible model that explains how Vac14 functions in this dual capacity is the existence of domains on the vacuole membrane that are enriched for either Vac14 or Fig4. This would allow the stoichiometry of Vac14 and Fig4 to fluctuate across the vacuole membrane to favor either PtdIns(3,5)P2 synthesis (at regions of high Vac14, but lower or no Fig4 content) or PtdIns(3,5)P2 dephosphorylation (at regions of low or no Vac14, but higher Fig4 content). In support of this model, we did note the existence of patches of intense Fig4-GFP and Vac14-GFP fluorescence on the vacuole membrane. Future studies will attempt to determine the colocalization of Fig4 and Vac14, such that the relative positions of these intense patches of fluorescence can be determined.
The existence of domains on the vacuole membrane that serve to direct discrete function has been proposed previously with the discovery that proteins that regulate homotypic vacuole fusion are enriched in areas of vacuole contact, known as the vertex ring domain (Wang et al., 2003). The domains on the vacuole membrane that are enriched for either Vac14 or Fig4 are distinct from that of the vertex ring domain, because they do not coincide with areas of vacuole contact and instead might function as sites of membrane recycling necessary to maintain vacuolar size.
Acknowledgments
We acknowledge the invaluable contributions of Drs. Jonathan Gary and Trey Sato to the early stages of this project. We thank Drs. Vicki Sciorra, Anjon Audhya, Christopher Stefan, and Bill Parrish for critical reading of the manuscript and helpful suggestions. This work was supported by a grant from the National Institutes of Health (CA-58689 to S.D.E.). S.D.E. is an investigator of the Howard Hughes Medical Institute.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–05–0297. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0297.
References
- Babst, M., Sato, T.K., Banta, L.M., and Emr, S.D. (1997). Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Katzmann, D.J., and Emr, S.D. (2002). ESCRT III: an endosomal-associated heteroologomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- Beeler, T., Bruce, K., and Dunn, T. (1997). Regulation of cellular Mg2+ by Saccharomyces cerevisiae. Biochim. Biophys. Acta 1323, 310–318. [DOI] [PubMed] [Google Scholar]
- Bonangelino, C.J., Catlett, N.L., and Weisman, L.S. (1997). Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol. Cell Biol. 17, 6847–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino, C.J., Nau, J.J., Duex, J.E., Brinkman, M., Wurmser, A.E., Gary, J.D., Emr, S.D., and Weisman, L.S. (2002). Osmotic-stress induced increase in phosphatidylinositol 3, 5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156, 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buj-Bello, A., Furling, D., Tronchere, H., Laporte, J., Lerouge, T., Butler-Browne, G.S., and Mandel, J.L. (2002). Muscle-specific alternative splicing of mytubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 11, 2297–2307. [DOI] [PubMed] [Google Scholar]
- Cooke, F.T., Dove, S.K., McEwen, R.K., Painter, G., Holmes, A.B., Hall, M.N., Michell, R.H., and Parker, P.J. (1998). The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr. Biol. 8, 1219–1222. [DOI] [PubMed] [Google Scholar]
- Darsow, T., Rieder, S.E., and Emr, S.D. (1997). A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol. 138, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, B.E., and Robinson, M.L. (2003). Congenital hydrocephalus in hy3 mice is caused by a frameshift mutation in Hydin, a large novel gene. Hum. Mol. Genet. 12, 163–170. [DOI] [PubMed] [Google Scholar]
- Dove, S.K., Cooke, F.T., Dougllas, M.R., Sayers, L.G., Parker, P.J., and Michell, R.H. (1997). Osmotic stress activates phosphatidylinositol 3, 5-bisphosphate synthesis. Nature 390, 187–192. [DOI] [PubMed] [Google Scholar]
- Dove, S.K., McEwen, R.K., Mayes, A., Hughes, D.C., Beggs, J.D., and Michell, R.H. (2002). Vac14 controls PtdIns(3, 5)P2 synthesis and Fab1-dependent protein trafficking to the multivesicular body. Curr. Biol. 12, 885–893. [DOI] [PubMed] [Google Scholar]
- Erdman, S., Lin, L., Malczynski, M., and Snyder, M. (1998). Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140, 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foti, M., Audhya, A., and Emr, S.D. (2001). Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol. Biol. Cell 12, 2396–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, J.D., Wurmser, A.E., Bonangelino, C.J., Weisman, L.S., and Emr, S.D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, J.D., Sato, T.K., Stefan, C.J., Bonangelino, C.J., Weisman, L.S., and Emr, S.D. (2002). Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. 13, 1238–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S., Stolz, L.E., Lemrow, S.M., and York, J.D. (1999). SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 274, 12990–12995. [DOI] [PubMed] [Google Scholar]
- Hawkins, P.T., Stephens, L., and Downes, C.P. (1986). Rapid formation of inositol 1, 3, 4, 5-tetrakisphosphate and inositol 1, 3, 4-trisphosphate in rat parotid glands may both result indirectly from receptor-stimulated release of inositol 1, 4, 5-trisphosphate from phosphatidylinositol 4, 5-bisphosphate. Biochem. J. 238, 507–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, C.S., and Winston, F. (1987). A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57, 267–272. [DOI] [PubMed] [Google Scholar]
- Hughes, W.E., Cooke, F.T., and Parker, P.J. (2000a). Sac phosphatase domain proteins. Biochem. J. 350, 337–352. [PMC free article] [PubMed] [Google Scholar]
- Hughes, W.E., Woscholski, R., Cooke, F.T., Patrick, R.S., Dove, S.K., McDonald, N.Q., and Parker, P.J. (2000b). SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologues Inp52p and Inp53p phosphatases. J. Biol. Chem. 275, 801–808. [DOI] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformations of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Odorizzi, G., and Emr, S.D. (2002). Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 12, 898–905. [DOI] [PubMed] [Google Scholar]
- Maehama, T., Taylor, G.S., Slama, J.T., and Dixon, J.E. (2000). A sensitive assay for phosphoinositide phosphatases. Anal. Biochem. 279. 248–250. [DOI] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. (1992). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Minagawa, T., Ijuin, T., Mochizuki, Y., and Takenawa, T. (2001). Identification and characterization of a sac domain-containing phosphoinositide 5-phosphatase. J. Biol. Chem. 276, 22011–22015. [DOI] [PubMed] [Google Scholar]
- Nelis, E., Erdem, S., Tan, E., Lofgren, A., Ceuterick, C., De Jonghe, P., Van Broeckhoven, C., Timmerman, V., and Topaloglu, H. (2002). A novel homozygous missense mutation in the mytotubularin-related protein s gene associated with recessive Charcot-Marie-Tooth disease with irregularly folded myelin sheaths. Neuromuscul. Disord. 12, 869–873. [DOI] [PubMed] [Google Scholar]
- Ooms, L.M., McColl, B., Wiradjaja, F., Wijayaratnam, A.P., Gleeson, P., Gething, M.J., Sambook, J., and Mitchell, C.A. (2000). The yeast inositol polyphosphate 5-phosphatases Inp52p and Inp53p translocate to actin patches following hyperosmotic stress: mechanism for regulating phosphatidylinositol 4, 5-bisphosphate at plasma membrane invaginations. Mol. Cell Biol. 20, 9376–9390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, G., Babst, M., and Emr, S.D. (1998). Fab1p PtdIns(3) 5-kinase function is essential for protein sorting in the multivesicular body. Cell 95, 847–858. [DOI] [PubMed] [Google Scholar]
- Piper, R.C., Cooper, A.A., Yang, H., and Stevens, T.H. (1995). VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.S., Klionsky, D.J., Banta, L.M., and Emr, S.D. (1988). Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in delivery and processing of multiple vacuolar hydrolases. Mol. Cell Biol. 8, 4936–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, S.E., Banta, L.M., Kohrer, K., McCaffery, J.M., and Emr, S.D. (1996). Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant, Mol. Biol. Cell 6, 985–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., and Hieter, P. (1990). Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Srinivasan, S., Seaman, M., Nemoto, Y., Daniell, L., Suchy, S.F., Emr, S.D., De Camilli, P., and Nussbaum, R. (1997). Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur. J. Cell Biol. 74, 350–360. [PubMed] [Google Scholar]
- Stefan, C.J., Audhya, A., and Emr, S.D. (2002). The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4, 5)-bisphosphate. Mol. Biol. Cell 13, 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz, L.E., Huynth, C.V., Thorner, J., and York, J.D. (1998). Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics 148, 1715–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, G.S., and Dixon, J.E. (2001). An assay for phosphoinositide phosphatases utilizing fluorescent substrates. Anal. Biochem. 295, 122–126. [DOI] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Merz, A.J., Collins, K.M., and Wickner, W. (2003). Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 160, 365–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford, C.C., Best, C., Kazlauskas, A., and Ulug, E.T. (1996). D-3 phosphoinositide metabolism in cells treated with platelet-derived growth factor. Biochem. J. 31, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford, C.C., Brearley, C.A., and Ulug, E.T. (1997). Phosphatidylinositol 3, 5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem. J. 323, 597–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky, S., Frischmuth, S., and Singer-Kruger, B. (2003). Bsp1p/Ypr171p is an adapter that directly links some synaptojanin family members to the cortical actin cytoskeleton in yeast. FEBS Lett. 537, 35–41. [DOI] [PubMed] [Google Scholar]
- Yamamoto, A., DeWald, D.B., Boronenkov, I.V., Andrson, R.A., Emr, S.D., and Koshland, D. (1995). Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol. Biol. Cell 5, 525–539. [DOI] [PMC free article] [PubMed] [Google Scholar]