Abstract
Background
Environmental variables such as moisture availability are often important in determining species prevalence and intraspecific diversity. The population genetic structure of dominant plant species in response to a cline of these variables has rarely been addressed. We evaluated the spatial genetic structure and diversity of Andropogon gerardii populations across the U.S. Great Plains precipitation gradient, ranging from approximately 48 cm/year to 105 cm/year.
Methodology/Principal Findings
Genomic diversity was evaluated with AFLP markers and diversity of a disease resistance gene homolog was evaluated by PCR-amplification and digestion with restriction enzymes. We determined the degree of spatial genetic structure using Mantel tests. Genomic and resistance gene homolog diversity were evaluated across prairies using Shannon's index and by averaging haplotype dissimilarity. Trends in diversity across prairies were determined using linear regression of diversity on average precipitation for each prairie. We identified significant spatial genetic structure, with genomic similarity decreasing as a function of distance between samples. However, our data indicated that genome-wide diversity did not vary consistently across the precipitation gradient. In contrast, we found that disease resistance gene homolog diversity was positively correlated with precipitation.
Significance
Prairie remnants differ in the genetic resources they maintain. Selection and evolution in this disease resistance homolog is environmentally dependent. Overall, we found that, though this environmental gradient may not predict genomic diversity, individual traits such as disease resistance genes may vary significantly.
Introduction
Intraspecific variation has been proposed to vary across space according to peak or gradient models [1]. In the peak model, species traits such as diversity vary from the core of the species range to the periphery. For example, Green et al. [2] found allozyme diversity of Rana pretiosa was greatest at the center of the species range, corresponding to the peak model. In the gradient model, intraspecific variation changes from one end of a species range to the other, often corresponding to environmental variation. The gradient model has been demonstrated to apply to variation of both molecular markers and heat and cold tolerance phenotypes in Drosophila melanogaster [3], [4]. Few studies have addressed whether the gradient or peak model applies to plant intraspecific diversity. Richards et al. [5] hypothesized that intraspecific diversity would decrease in progressively more stressful environments for plant species able to reproduce clonally. However, they did not find any trend between diversity and environment. Genetic isolation by distance [6] may influence spatial genetic structure due to clonal growth, limited dispersal, genetic drift, selection, and density.
Andropogon gerardii (Vitman) is the dominant plant species in the North American tallgrass prairie ecoregion. Because of its importance in conservation and restoration ecology, and more recently in potential biofuel systems, the spatial genetic structure of A. gerardii has been an ongoing subject of interest [7], [8], [9]. Gustafson et al. [7] did not find a statistically significant relationship between random amplified polymorphic DNA (RAPD) similarity and geographic distance overall, though some prairies exhibited this trend. Most of the genetic variation (89%) was found within populations and 11% among populations. Gustafson et al. [8] also found that local prairie remnants and restored populations were genetically different than non-local remnants and cultivars. This suggests that the geographic origin of seed selected for restoration purposes may be important if the restoration goal is to produce a prairie plant community similar to the historical prairie at a particular site. Other restoration and conservation goals may include development of plant communities resilient to climate and land use change [10], which will require a much more complete understanding of the ecological genomics of species such as A. gerardii [11], [12],[13], including the role of disease resistance genes.
Plant disease resistance may vary across environmental gradients [14], [15], [16], contributing to the environmental gradient experienced by pathogens, but the population genetic structure of disease resistance genes across these gradients has rarely been addressed in natural plant populations [17], [18]. To our knowledge, no reports have been made of resistance genes or homologs varying in diversity across an environmental gradient distinct from overall genomic diversity across that gradient [17]. Roughly one percent of the protein-coding genes in plant genomes are disease resistance genes, with 207 predicted resistance genes in Arabidopsis, approximately 400 in Populus, and more than 500 in Oryza [19]. The importance of this class of genes in plant genomes is also demonstrated by the unusually high level of selection found at disease resistance loci [20], [21]. Burdon and Thrall [16] have made impressive progress in understanding the spatial dynamics of phenotypic resistance and pathogenicity in the natural flax-flax rust pathosystem. Within-population dynamics for disease resistance have also been addressed through modeling [22], [23], [24], [25], [26], [27]. However, typical coevolution model assumptions about costs and benefits of resistance for plants and virulence for pathogens, are often not appropriate for actual populations [26], [27]. Field studies of resistance gene diversity in natural populations have been helpful in identifying the various types of selection occurring at these loci [28] and indicating that different regions of resistance gene sequences are under different types of selection [29].
This study examines the diversity of a resistance gene homolog in A. gerardii. This species presents a unique opportunity to address questions about the distribution of resistance gene variation. It has a relatively continuous spatial distribution across a wide range of environments in the Great Plains and has common pathogens with its more-studied relative Zea mays. Understanding the ecology and evolution of disease resistance genes in such widespread and abundant native plant species could have direct applications to agriculture, where many crop species are grown in monocultures over large areas. The spatial structure of resistance genes influences epidemic intensity in many systems [30], and the effect of this spatial structure, itself, has the potential to vary across gradients [31]. Higher interspecific plant diversity has been observed to decrease disease in tallgrass prairie plants [32] and intraspecific resistance gene diversity has the potential to provide similar ecosystem services for disease regulation in A. gerardii populations [33]. Patterns of disease resistance variability in A. gerardii may also be important in the development of biofuel systems based on prairie plant species, including ‘low-input high-diversity’ systems [34].
No disease resistance genes have been cloned from A. gerardii, however we report the presence of a homolog of the maize gene Rxo1 in A. gerardii. Zhao et al. [35] demonstrated that Rxo1 confers resistance to Burkholderia andropogonis and a resistance reaction to the rice bacterial streak pathogen Xanthomonas oryzae pv. oryzicola after transfer to rice, suggesting conserved functionality across different genera of grasses. It is known that Rxo1 recognizes avirulent B. andropogonis in maize, and that the same maize isolates of B. andropogonis infect A. gerardii [S. H. Hulbert, unpublished]. In addition, Rxo1 recognizes not only avirulent B. andropogonis, but also avrRxo1 in Xanthomonas oryzae pv. oryzicola [35], suggesting that Rxo1 recognizes conserved components of a bacterial effector or group of effectors [36] or recognizes modifications to a cellular component that is a common effector target [37]. These results suggest that the A. gerardii Rxo1 homolog may recognize avirulent B. andropogonis strains.
Burkholderia andropogonis is pathogenic to at least 51 plant species in 15 families of monocots and dicots [38]. It has been observed to cause low levels of bacterial stripe disease on A. gerardii at Konza Prairie Biological Station in Kansas under wet conditions in spring [39]. B. andropogonis infects sorghum and maize in warm and humid areas and is disseminated by splashing water [40]. Leaf surface moisture typically supports higher infection rates for bacterial foliar pathogens [38], [41], and bacterial stripe and leaf spot of maize caused by B. andropogonis tends to be more severe during periods of wet and warm weather [42]. Likewise, severity of bacterial brown leaf spot of citrus caused by B. andropogonis is associated with rainstorms in Florida [43].
The widespread distribution of A. gerardii across the North American Great Plains allows for the examination of resistance gene homolog diversity across an east to west gradient in precipitation. Net primary productivity increases by a factor of 5.5 from the Shortgrass Steppe Long Term Ecological Research Site in northeastern Colorado to the Konza Prairie Biological Station in Kansas [44]. Alexander et al. [45] found decreased disease (smut and rust) on Carex blanda field and herbarium specimens from western Kansas compared to eastern Kansas. This trend was attributed to the drier conditions in western Kansas providing an environment less suitable for the disease, the fact that western Kansas was at the edge of the range for C. blanda, and that western populations were smaller and more isolated, decreasing the potential for successful dispersal. Disease in A. gerardii caused by B. andropogonis likely also changes across this environmental gradient. The scale of selection for resistance can be relatively small. For example, differences in selection for herbivore resistance were found within a 4 ha stand of oak seedlings [46]. Variation in selection at the Rxo1 homolog may result in different levels of diversity among populations of A. gerardii.
In this study, our first objective was to determine genome-wide diversity of A. gerardii across the U.S. Great Plains precipitation gradient using AFLP markers. We found novel evidence for spatial genetic structuring in A. gerardii, but not evidence for a change in genomic diversity across the gradient. Second, we assessed diversity in Rxo1 homologs in the same natural populations of A. gerardii. We found that diversity in the resistance gene homologs decreased with decreasing precipitation across the gradient, in a pattern distinct from that of genomic diversity. The patterns of AFLP and Rxo1 homolog diversity within and between prairies also provide perspectives on the spatial scale at which intraspecific variation occurs for both the entire genome and for a presumably adaptive gene family, as context for planning tallgrass prairie conservation and restoration.
Materials and Methods
Tissue Collection
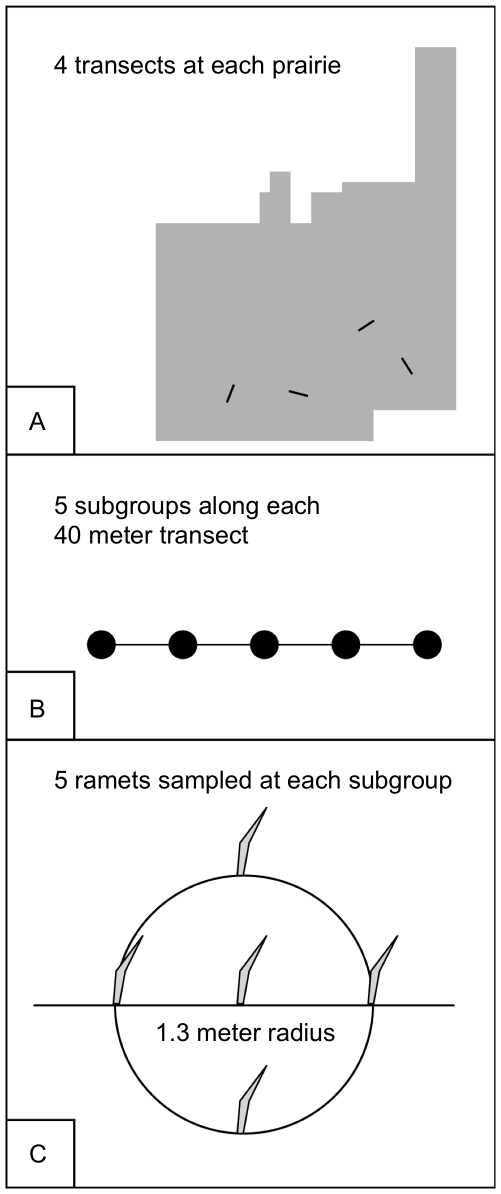
Tallgrass prairie remnants can be found in much of the eastern US, but are most common in the Great Plains where a sharp east-west precipitation gradient occurs (Table 1; Fig. 1). We selected native tallgrass prairie locations at approximately the same latitude across this gradient (Table 1; Fig. 2). Within each prairie, four 40 m transects were established with five sampling points at 10 m intervals, for a total of 5 samples per point and 25 samples per transect (Fig. 3). Each ramet sample consisted of a 15 cm basal cutting made from two leaves placed in a sealed plastic bag with 15 ml of silica gel (Demis Products, Lithonia, GA and Miracle Coatings, Anaheim, CA) for storage up to one year at room temperature. One hundred tissue samples were collected from each prairie. Tissues were also collected from 65 clones in Boulder, Colorado, that had been mapped and characterized to ploidy level by Keeler [9] in plots 28, 52, 61, and 102 established by Jane and Carl Bock in the City of Boulder Open Space and Mountain Parks. DNA was extracted according to Doyle & Doyle [47] with several modifications (Appendix S1).
Table 1. Tallgrass prairies sampled, with state, UTM coordinates, AFLP sample size, Rxo1-RFLP sample size and average annual precipitation from the nearest weather station (1971–2000).
| Prairie | State | UTM coordinates | AFLP sample size | Rxo1-RFLP sample size | Average annual precip. (cm/year) |
| Tucker Prairie | Missouri | 15 N 587416 E, 4311575 N | 71 | 100 | 104.7 |
| KPBS1 | Kansas | 14 N 709536 E, 4327791 N | 84 | 100 | 88.39 |
| TLI2 | Kansas | 14 N 624616 E, 4292829 N | 85 | 100 | 81.76 |
| Wilson Lake | Kansas | 14 N 527770 E, 4310686 N | 70 | 100 | 65.18 |
| Smoky Valley Ranch | Kansas | 14 N 328103 E, 4306146 N | 97 | 100 | 47.63 |
| Boulder3 | Colorado | 13 N 475769 E, 4427696 N | 63 | 63 | 50.62 |
Konza Prairie Biological Station.
The Land Institute.
Tissues from Boulder were used only in ploidy analyses.
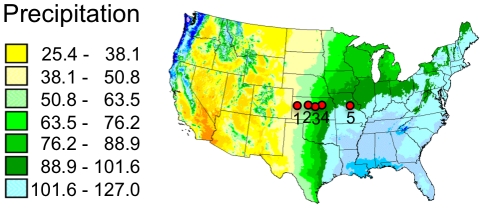
Figure 1. Location of prairies sampled across a significant precipitation gradient in the Great Plains.
Prairie locations are (1) Smoky Valley Ranch, (2) Wilson Lake, (3) The Land Institute, (4) Konza Prairie Biological Station, and (5) Tucker Prairie. The background shading indicates an interpolation of annual average precipitation (cm) from 1961–1990 (where the precipitation map is copyright PRISM Climate Group, Oregon State University, http://prism.oregonstate.edu).
Figure 2. Collection sites for Andropogon gerardii.
Figure 3. Sampling methodology at different scales.
Each prairie (A) contained four transects. Each 40 m transect (B) contained a series of five subgroups. Five ramets were sampled at each subgroup (C).
AFLP Genotyping
AFLP fingerprinting was performed according to Vos et al. [48] with many modifications (Appendix S1). Sixty-three repeatable AFLP polymorphic fragments were scored for the 6FAM-labeled EcoRI-AAA/MseI-CTG primer-pair and 32 polymorphic fragments were scored for the HEX-labeled EcoRI-ACC/MseI-CTG primer-pair. Capillary electrophoresis was used to identify fragments and GeneMarker version 1.6 software (SoftGenetics, State College, PA) was used to score AFLP markers. Statistical analyses described below were conducted using R software [49].
Rxo1 Homolog Identification
The sequences of the maize Rxo1 gene and its four other tightly linked family members from the maize inbred B73 (GenBank accessions AY935244-AY935248) were compared to identify conserved sequences near the predicted start codon and nucleotide binding site (NBS) regions. Two conserved primers were designed from these sequences: ATG forward (5′-GAAGGGATGGCAGAGATTGCTGT-3′) and MHD reverse (5′-CAAGTTCACGCACAAGATCATGCAT-3′). These primers amplified fragments of approximately 1500 nt from genomic DNA of a single A. gerardii plant (clone B10, ploidy unknown) obtained from Konza Prairie Biological Station. Cloning and partial sequence analysis of these fragments identified three different sequences highly similar to the maize Rxo1 gene which were then used to design RACE 5′ and 3′ PCR primers for 5′-TCTTTTTCAGGACAAGAACA-3′ and 5′-CGACGGAGCATGCTTCATGTTGCAGAGAG-3′ for 3′, respectively. Transcripts corresponding to the full coding region were then amplified with primers that were conserved among the RACE PCR amplicons; the primers 5′BRxo forward (5′-GGTATTGGGGCTAAAGAGTGAGCTTTGAC-3′) corresponded to sequences in the 5′UTR and 3′BRxo including the predicted stop codon (5′-GAGTTCTGCTATTTCATCTTAACCGGGCC-3′). RACE PCRs and cDNA amplifications were performed using the BD smart RACE cDNA amplification Clontech kit (Mountain View, CA). One cDNA corresponding to the full coding region had a 2793 nt open reading frame that was approximately 91% identical to the maize Rxo1 coding region (Genbank sequence # pending).
Rxo1 Homolog Amplification and Restriction Enzyme Digestion
We amplified sequences from the leucine-rich repeat (LRR) encoding region from A. gerardii clones originating from diverse locations (Appendix S1). A primer pair was designed to amplify an 810 bp fragment (5′-AGATTCTCGACGAGTTGCTGTGCT-3′ and 5′-AGCCTAAGAAGCCCATTTCCGTGA-3′). The primers did not require pre-screening for polymorphism, and amplified fragments from plant samples collected throughout the gradient and beyond in Colorado, Illinois, and Indiana. Four restriction enzymes were used to perform RFLP assays: BsaMI, RsaI, TaqI (Promega), and MseI (New England Biolabs). The PCR amplicons were digested individually. Digestion products were electrophoresed on 2% agarose gels and scored for the presence or absence of fragments. Five fragments were scored for BsaMI, nine for MseI, nine for RsaI, and 14 for TaqI. We found the RFLP markers to be greater than 99% repeatable.
Relatedness of Individuals of Different Ploidy
Like many perennial plant species, A. gerardii exists in populations with multiple ploidy levels [50]. Populations of mixed ploidy may complicate estimates of genetic diversity if the ploidy levels of individuals are unknown and genetic diversity varies with ploidy. A. gerardii populations consist predominantly of hexaploids and enneaploids with some individuals of intermediate ploidy levels [50], [51]. In general, hexaploids are more common in eastern prairies whereas western prairies (west of the Missouri River) have populations with mixed ploidy [50]. The origin and mechanism for maintenance of enneaploids is not known since they have not been found to have a reproductive fitness advantage in the field [52], at least in the short term, and crosses within or among any ploidy level very rarely yield enneaploid individuals [52], [39], [53]. In order to examine the distribution of resistance gene homologs in A. gerardii, it is necessary to account for the potential effects of spatial trends in ploidy. Population analyses of A. gerardii may be influenced by two factors. First, enneaploid and hexaploid individuals may be acting as separate populations. This could influence our results if enneaploids and hexaploids do not contribute equally to our diversity measures and if the proportion of enneaploids varies among prairies. Second, enneaploids may exhibit more AFLP amplicons compared to hexaploids because enneaploids contain 50% more chromosomes. In order to determine if enneaploids and hexaploids were acting as distinct populations, we measured AFLP dissimilarity among and within individuals of the two ploidy groups from the Boulder population. In order to test if enneaploid individuals were more diverse than hexaploid individuals we calculated the average number of AFLP fragments amplified in enneaploids and compared this to the hexaploid average. Assuming the polymorphic fragments analyzed in this study are distributed equally across chromosomes, we expected to find more fragments in enneaploids than in hexaploids. These ploidy analyses were conducted with data from the mapped Boulder population to determine if there is an inherent relationship between ploidy and diversity in A. gerardii.
Dissimilarity and Geographic Distance
Scale of sampling is important in determining spatial autocorrelation in clonal species [54]. To test for spatial structure at different scales using the AFLP and Rxo1-RFLP datasets, a similarity matrix with an entry for each pairwise comparison of ramets from all prairies outside Colorado was calculated for each of the two datasets. To estimate pairwise genetic similarity for this study, we used the simple match coefficient (s), which Kosman & Leonard [55] argue is the most appropriate index for polyploid species, to calculate genetic similarity (the number of matches, in terms of shared absence or presence of a DNA fragment, divided by the total number of possible matches). Dissimilarity was calculated as one minus similarity (1-s). The mean dissimilarities of all ramet pairs at a particular distance from each other were used to calculate the overall mean dissimilarity for that distance. For example, since there were five plants in the first transect subgroup of a transect and five in the second transect subgroup, the mean of all of the comparisons among these two transect subgroups was used as one datum in calculating the overall 10 meter mean dissimilarity. Mean dissimilarity was calculated for approximately 1.8 m (average within-subgroup distance), 10 m, 20 m, 30 m, 40 m, 2139 m (average distance between transects), and 473,025 m (average distance between prairies). In order to test for spatial genetic structure, a regression was performed for mean dissimilarity versus distance between sampled locations. This test is similar to a Mantel test [56]. The test was performed with the AFLP marker data. We evaluated whether the slope parameter estimate was significantly greater than would be expected by chance by comparing it to parameter estimates from 1000 random permutations of the data (details in Appendix S1).
Shannon's information measure was used to determine the proportion of diversity among populations: H′ = −Σ pi log pi, where pi is the frequency of the amplified fragment. H′ was calculated for each population and used to determine the average population diversity: Ha. The total diversity for all individuals used in this study was calculated and denoted as Hs.
Diversity and Precipitation
To determine the proportion of diversity among populations, the average population diversity was subtracted from the species diversity and this value was divided by the species diversity. This analysis was done with both AFLP and RFLP data. We chose two statistics to measure within-prairie diversity. First we calculated the average Rxo1 and AFLP dissimilarity for each prairie. This measure of diversity includes haplotype diversity in addition to diversity of individual alleles since individual haplotypes were used in calculating dissimilarity. Shannon's information measure was also calculated for each population for both AFLP and Rxo1 data. This measure of diversity considers allelic diversity alone. We used R to perform linear regression analyses of both Rxo1 and AFLP diversity measures, with annual average precipitation (Table 1) as the predictor in separate analyses. Precipitation data for each prairie were estimated as the average annual precipitation from the nearest weather station between 1971 and 2000 [57].
Results
AFLP Dissimilarity and Ploidy
Samples collected in Boulder, Colorado, were from mapped clones of known ploidy. AFLP dissimilarity was calculated among hexaploid clones (n = 38), among eaneaploid clones (n = 21), and between hexaploid and enneaploid clones. The mean dissimilarity for each of these comparisons was found to be 0.10 indicating equal dissimilarity both within and among ploidy groups. There was no difference in average fragment number among ploidy levels. Average number of fragments for hexaploids was 59.37 (n = 38, 58.76–59.97 95% confidence interval) and 59.33 (n = 21, 58.25–60.42 95% confidence interval) for enneaploids. Similarly, the average number of Rxo1-RFLP fragments for hexaploids was 10.71 (n = 38, 10.59–10.83 95% confidence interval and 10.62 (n = 21, 10.41–10.83 95% confidence interval) for enneaploids.
Dissimilarity and Geographic Distance
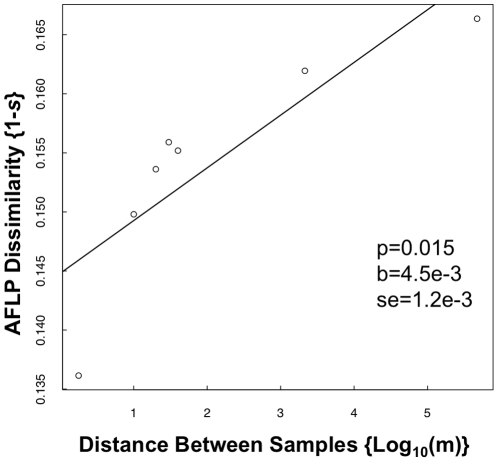
Mean pairwise dissimilarity based on AFLP markers increased with increasing distance between individuals (Fig. 4). These data indicate the presence of spatial genetic structure in the A. gerardii populations (which may ultimately be described best by an asymptotic function). The trend of increasing dissimilarity with increasing distance was also significant using the Rxo1-RFLP data (data not shown). We found 6.35% of the AFLP variation to be present among populations based on Shannon's information measure. For Rxo1-RFLP data, 13.00% of the variation was distributed among populations.
Figure 4. AFLP dissimilarity by geographic distance.
Points from left to right are plotted at 1.8 m (within subgroup), 10 m (between adjacent subgroups), 20 m, 30 m, 40 m, 2139 m (between transects), and 473,025 m (between prairies). The slope parameter (‘b’), and 95% confidence interval from a randomization test are indicated.
Rxo1 Dissimilarity Relative to AFLP Dissimilarity
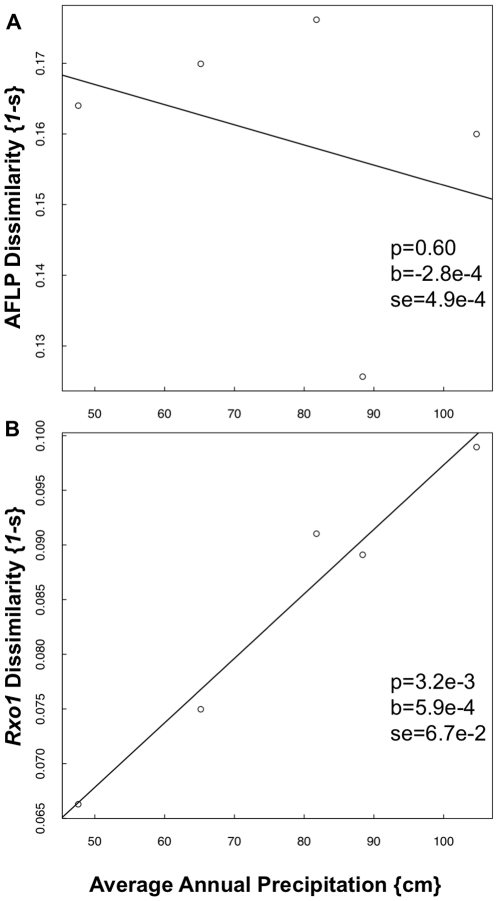
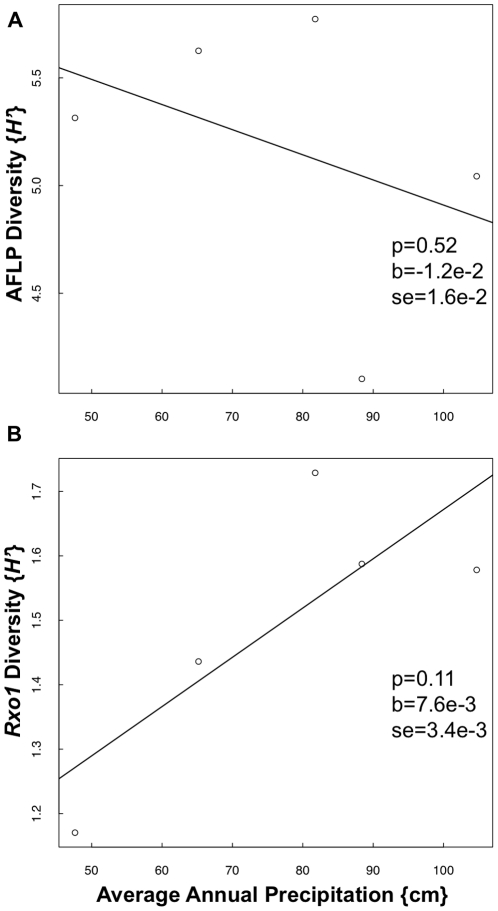
Mean Rxo1 homolog dissimilarity must be examined in the context of mean AFLP dissimilarity to determine whether the pattern in dissimilarity across longitude for Rxo1 is different from the pattern for the genome as a whole. We found a positive relationship between average annual precipitation and Rxo1 homolog dissimilarity (Fig. 5). We did not find a significant relationship between AFLP dissimilarity and precipitation (Fig. 5). For Shannon's information measure, we found a similar trend with marginal significance between Rxo1 diversity and precipitation, and no relationship between AFLP diversity and precipitation (Fig. 6). These data indicate that there is a positive relationship between Rxo1 diversity and precipitation, and that this relationship is not due to a positive relationship between precipitation and genome-wide diversity, as measured by AFLP diversity.
Figure 5. AFLP (a) and Rxo1 (b) dissimilarity across precipitation.
The mean dissimilarity for each prairie was calculated and plotted according to average annual precipitation. The p-value (‘p’), slope parameter (‘b’) and standard error (‘se’) from a linear regression model are indicated.
Figure 6. Shannon's information measure (H′) calculated for AFLP (a) and Rxo1 (b) results for each prairie as a function of average annual precipitation.
The p-value (‘p’), slope parameter (‘b’) and standard error (‘se’) from a linear regression model are indicated.
Discussion
A previous study of A. gerardii identified decreasing RAPD similarity with increasing distance within a subset of populations, though this trend was not consistent among prairies [7]. Our results demonstrate spatial genetic structuring among prairies. In our study, 6.3% and 13% of the AFLP and Rxo1-RFLP variation, respectively, were found among populations. Gustafson et al. [7] reported 11% of the variation was among prairies in Arkansas populations. The observed spatial genetic structure and estimated levels of population differentiation are significant for (1) validating the quality of the AFLP data and (2) providing preliminary data for decisions about minimum tallgrass prairie reserve acreage and strategies for selection of A. gerardii seed sources for prairie restoration. For the latter purpose, it will also be important to consider variation for genes under a range of types of relevant selection [11], [12].
There was a positive correlation between Rxo1 homolog diversity and precipitation. This correlation was not simply the result of genome-wide influences on A. gerardii, because there was no trend in AFLP dissimilarity across the same gradient. In contrast, a geographic cline in Cf-2 homolog diversity was consistent with a cline in diversity of neutral variation in natural populations of Solanum pimpinellifolium [17]. Information about which pathogen species and strains interact with this resistance gene locus in A. gerardii will be needed to fully assess the potential role of disease pressure in producing the gradient of Rxo1 homolog diversity.
A compelling explanation for the Rxo1 diversity cline is the presence of a gradient of disease pressure associated with the precipitation gradient. If a pathogen is found to interact with the Rxo1 homologous locus (loci) in a coevolutionary manner, measurements of disease pressure in a common garden experiment of clones with known resistance levels across the precipitation gradient could be used to test the hypothesis that the diversity cline is associated with a gradient of disease pressure.
Rxo1 homolog diversity was significantly higher in the eastern prairies. This may be due to selection for multiple alleles or gene family members in the disease-conducive environments, which would be consistent with the Red Queen hypothesis [58] applied to a plant-pathogen coevolutionary arms race [59], [60]. Diversifying selection characteristic of such an arms race has been observed in disease resistance loci that interact directly with pathogen effector proteins [61]. Alternatively, selection at loci with only single functional alleles can also promote stable polymorphisms if the functional alleles cause fitness disadvantages in environments with little disease [26]. Such fitness costs have been demonstrated experimentally for the Rpm1 locus in Arabidopsis [62] and both functional and nonfunctional alleles are maintained across broad geographic regions [63], [64]. In summary, increased disease pressure in the east with associated diversifying or balancing selection is one explanation for the trend in Rxo1 homolog diversity.
In the western prairies (Smoky Valley Ranch and Wilson Lake), drier conditions and more isolated plant populations would tend to result in lower disease pressure [41], [45] and dissimilarity in the Rxo1 homologs was disproportionately low relative to the neutral markers. Pathogens may be shared between tallgrass prairie and agricultural systems (e.g., [65]), increasing the connectivity of host systems for pathogens [66], but the extent to which this is an important factor for B. andropogonis is not known. Though the higher degree of isolation experienced by A. gerardii populations in the west could contribute to a decrease in overall diversity, the AFLP markers did not indicate any significant east-west trend. The increased similarity in Rxo1 homologs in the west could be due to selection for a single valuable allele or there could possibly be selection for a gene linked to Rxo1 that is important under western environmental conditions. Selection has been found at low-variability quantitative trait loci caused by selective sweeps in drought and saline adapted Helianthus annuus populations in Utah [67]. However, many recombination events are likely to have occurred in such a natural population over long periods of time, buffering any such linkage disequilibrium between Rxo1 and potential linked loci under selection. In summary, selection pressures in the western prairies including purifying selection is a second possible explanation for the observed trend of Rxo1 homolog diversity across environments.
Comparative sequence analysis of the Rxo1 homologous genes within and among natural populations of A. gerardii will allow assessment of what types of selection are controlling diversity at the Rxo1 homolog locus. It has been demonstrated in models that coevolutionary clines may result from multiple effects [68]. And the question remains whether the cline in Rxo1 diversity is adaptive. Sequence data for Rxo1 homologs across the different populations could help to elucidate whether or not the diversity cline is conferred by adaptive evolution. However, a substantial effort would be required to sequence the potentially multiple Rxo1 homologs in each individual. Functional analysis of the genes would also be valuable to more fully understand the molecular evolution of the Rxo1 homologs.
Andropogon gerardii is often found in populations of mixed ploidy [50], and our results indicate that, based on AFLP and Rxo1-RFLP dissimilarities, there was no differentiation between individuals of different ploidy. This is consistent with allozyme data suggesting that individuals of dissimilar ploidy level from the same plot were more similar than individuals of the same ploidy level from different plots [69]. We anticipated that the average number of AFLP fragments in enneaploids would be greater than the average number of fragments in hexaploids, since enneaploids have 50% more genomic DNA and AFLPs are dominant markers. However, we found no difference. Rxo1-RFLP band number was also the same across ploidy levels. While it is possible that the relationship between ploidy and band number could differ among populations of A. gerardii, this finding in the Boulder population suggests that relative proportions of hexaploid and enneaploid ramets in the other populations was not important for determining dissimilarity and diversity in our analyses.
Our finding of no difference in band number between the two ploidy levels has implications for the origin and establishment of enneaploids in mixed ploidy populations. Since enneaploids were not found to have an increase in heterozygosity, our data suggest that enneaploids may be produced through second division restitution (SDR) of hexaploid gametes. This process may be mediated by temperature, as chromosome doubling has been observed at high temperatures for maize and other species of the tribe Triticeae [70], [71]. Such temperature-dependent polyploidy may explain the origin and maintenance of enneaploids and explain the adaptive role of populations in the west with mixed ploidy levels, since enneaploids seem to be more vegetatively fit in marginal environments [9] where heat stress is more common, providing the opportunity for increased chromosome doubling. Experiments testing the hypothesis of temperature-dependent ploidy levels during meiosis could identify a unique mechanism for population-level genomic diversity.
We have demonstrated that resistance gene diversity in A. gerardii varies with precipitation, one of the most important environmental drivers of plant disease. Diversity of resistance phenotypes has been shown to vary with ecoregion previously in the Linum marginale – Melampsora lini pathosystem, where higher host phenotypic diversity (significantly associated with population mean resistance) was present in hill populations compared to bog populations [72]. However, hill populations of Linum marginale also showed higher diversity at neutral allozyme loci and were significantly distinct from bog populations as measured by these neutral markers. Also, the proportion of Aegilops tauschii resistance to stem and leaf rust was found to vary greatly by region, with the highest resistance exhibited by populations adjacent to the Caspian Sea [73]. Our study is unique in demonstrating a cline in resistance gene homolog diversity in natural plant communities that is independent of genomic diversity.
Diversity of important genes in native populations may prove to be particularly valuable if selection pressures shift rapidly due to changing climate [74]. Unfortunately, the diversity within native populations may not be adequate to allow adaptive change to keep up with the rate of climate change in the twenty-first century, as is apparently the case for Chamaecrista fasciculata populations in the Great Plains [75]. Though we did not find that genomic diversity changes across a precipitation gradient (and thus the genomic results provide no support for a gradient model of diversity), we found that the gradient model applies to resistance gene homolog diversity, indicating the importance of individual loci for adaptation to an environmental gradient. Our novel method for quantifying disease resistance gene diversity for an organism with a complex genome could enable studies of genes known or hypothesized to be important in other natural systems for which few genetic tools are available. Further studies integrating population-level dynamics with evolutionary history for disease resistance genes in dominant natural plant species will also help to inform more effective deployment of resistance genes in biofuel and other agricultural systems.
Supporting Information
This appendix provides additional details about the methods.
(DOCX)
Acknowledgments
We thank E. Akhunov, L. Johnson, J. Jurgenson, M. Nita, P. St. Amand, M. Ungerer, F. White, and anonymous reviewers for many helpful suggestions, and C. Anderson, S. Dendy, J. Farr, and C. Yaege for technical assistance. Access to prairies was provided by The Nature Conservancy, Kansas Department of Wildlife and Parks, Kansas State University, University of Missouri, The Land Institute, and City of Boulder Open Space and Mountain Parks.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors appreciate the support provided by The Land Institute (http://www.landinstitute.org/), The Ecological Genomics Institute at Kansas State University, the US National Science Foundation under Grants EF-0525712 (as part of the joint NSF-NIH Ecology of Infectious Disease program) and NSF DEB-0516046, and the Kansas Agricultural Experiment Station. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gaston KJ, Chown SL, Evans KL. Ecogeographical rules: elements of a synthesis. Journal of Biogeography. 2008;35:483–500. [Google Scholar]
- 2.Green DM, Sharbel TF, Kearsley J, Kaiser H. Postglacial range fluctuation, genetic subdivision and speciation in the western North American spotted frog complex, Rana pretiosa. Evolution. 1996;50:374–390. doi: 10.1111/j.1558-5646.1996.tb04500.x. [DOI] [PubMed] [Google Scholar]
- 3.Collinge JE, Hoffmann AA, McKechnie SW. Altitudinal patterns for latitudinally varying traits and polymorphic markers in Drosophila melanogaster from eastern Australia. Journal of Evolutionary Biology. 2006;19:473–482. doi: 10.1111/j.1420-9101.2005.01016.x. [DOI] [PubMed] [Google Scholar]
- 4.Weeks AR, McKechnie SW, Hoffman AA. Dissecting adaptive clinal variation: markers, inversions and size/stress associations in Drosophila melanogaster from a central field population. Ecology Letters. 2002;5:756–763. [Google Scholar]
- 5.Richards CL, Hamrick JL, Donovan LA, Mauricio R. Unexpectedly high clonal diversity of two salt marsh perennials across a severe environmental gradient. Ecology Letters. 2004;7:1155–1162. [Google Scholar]
- 6.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gustafson DJ, Gibson DJ, Nickrent DL. Random amplified polymorphic DNA variation among remnant big bluestem (Andropogon gerardii Vitman) populations from Arkansas' Grand Prairie. Molecular Ecology. 1999;8:1693–1701. doi: 10.1046/j.1365-294x.1999.00756.x. [DOI] [PubMed] [Google Scholar]
- 8.Gustafson DJ, Gibson DJ, Nickrent DL. Conservation genetics of two co-dominant grass species in an endangered grassland ecosystem. Journal of Applied Ecology. 2004;41:389–397. [Google Scholar]
- 9.Keeler KH. Impact of intraspecific polyploidy in Andropogon gerardii (Poaceae) populations. American Midland Naturalist. 2004;152:63–74. [Google Scholar]
- 10.Hobbs RJ. Setting effective and realistic restoration goals: Key directions for research. Restoration Ecology. 2007;15:342–357. [Google Scholar]
- 11.Travers SE, Smith MD, Bai JF, Hulbert SH, Leach JE, et al. Ecological genomics: making the leap from model systems in the lab to active populations in the field. Frontiers in Ecology and the Environment. 2007;5:19–24. [Google Scholar]
- 12.Travers SE, Tang Z, Caragea D, Garrett KA, Hulbert SH, et al. Variation in gene expression of Andropogon gerardii in response to altered environmental conditions associated with climate change. Journal of Ecology. 2010;98:374–383. [Google Scholar]
- 13.Ouborg NJ, Angeloni F, Vergeer P. An essay on the necessity and feasibility of conservation genomics. Conservation Genetics. 2010;11:Sp. Iss. 643–653. [Google Scholar]
- 14.Dinoor A. Sources of oat crown rust resistance in hexaploid and tetraploid wild oats in Israel. Canadian Journal of Botany. 1970;48:153–161. [Google Scholar]
- 15.Nevo E, Moseman JG, Beiles A, Zohary D. Correlation of ecological factors and allozymic variations with resistance to Erysiphe graminis hordei in Hordeum spontaneum in Israel: Patterns and application. Plant Systematics and Evolution. 1984;145:79–96. [Google Scholar]
- 16.Burdon JJ, Thrall PH. Spatial and temporal patterns in coevolving plant and pathogen associations. The American Naturalist. 1999;153:S15–S33. doi: 10.1086/303209. [DOI] [PubMed] [Google Scholar]
- 17.Caicedo AL. Geographic diversity cline of R gene homologs in wild populations of Solanum pimpinellifolium (Solanaceae). American Journal of Botany. 2008;95:393–398. doi: 10.3732/ajb.95.3.393. [DOI] [PubMed] [Google Scholar]
- 18.Laine A-L, Burdon JJ, Dodds PN, Thrall PH. Spatial variation in disease resistance: from molecules to metapopulations. Journal of Ecology. 2011;99:96–112. doi: 10.1111/j.1365-2745.2010.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, et al. The genome of Black Cottonwood Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 20.Bakker EG, Toomajian C, Kreitman M, Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. The Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Wang C, Ping L, Tian D, Yang S. Pattern of LRR nucleotide variation in plant resistance genes. Plant Science. 2007;173:253–261. [Google Scholar]
- 22.Leonard KJ. Genetic equilibria in host-pathogen systems. Phytopathology. 1969;59:1858–1863. [PubMed] [Google Scholar]
- 23.Leonard KJ. Selection pressures and plant pathogens. Annals of the New York Academy of Science. 1977;287:207–222. [Google Scholar]
- 24.Leonard KJ, Czochor RJ. Theory of genetic interactions among populations of plants and their pathogens. Annual Review of Phytopathology. 1980;18:237–258. [Google Scholar]
- 25.Leonard KJ. Stability of equilibria in a gene-for-gene coevolution model of host-parasite interactions. Phytopathology. 1994;84:70–77. [Google Scholar]
- 26.Bergelson J, Dwyer G, Emerson JJ. Models and data on plant-enemy coevolution. Annual Review of Genetics. 2001;35:49–499. doi: 10.1146/annurev.genet.35.102401.090954. [DOI] [PubMed] [Google Scholar]
- 27.Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- 28.Rose LE, Michelmore RW, Langley CH. Natural variation in the Pto disease resistance gene within species of wild tomato (Lycopersicon). II. Population genetics of Pto. Genetics. 2007;175:1307–1319. doi: 10.1534/genetics.106.063602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caicedo AL, Schaal BA. Heterogeneous evolutionary processes affect R gene diversity in natural populations of Solanum pimpinellifolium. Proceedings of the National Academy of Sciences of the USA. 2004;101:17444–17449. doi: 10.1073/pnas.0407899101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garrett KA, Mundt CC. Epidemiology in mixed host populations. Phytopathology. 1999;89:984–990. doi: 10.1094/PHYTO.1999.89.11.984. [DOI] [PubMed] [Google Scholar]
- 31.Garrett KA, Zúñiga LN, Roncal E, Forbes GA, Mundt CC, et al. Intraspecific functional diversity in hosts and its effect on disease risk across a climatic gradient. Ecological Applications. 2009;19:1868–1883. doi: 10.1890/08-0942.1. [DOI] [PubMed] [Google Scholar]
- 32.Mitchell CE, Tilman D, Groth JV. Effects of grassland plant species diversity, abundance, and composition on foliar fungal disease. Ecology. 2002;83:1713–1726. [Google Scholar]
- 33.Cheatham MR, Rouse MN, Esker PD, Ignacio S, Pradel W, et al. Beyond yield: Plant disease in the context of ecosystem services. Phytopathology. 2009;99:1228–1236. doi: 10.1094/PHYTO-99-11-1228. [DOI] [PubMed] [Google Scholar]
- 34.Tilman D, Hill J, Lehman C. Carbon-negative biofuels from low-input high-diversity grassland biomass. Science. 2006;314:1598–1600. doi: 10.1126/science.1133306. [DOI] [PubMed] [Google Scholar]
- 35.Zhao BY, Lin XH, Poland J, Trick H, Leach J, Hulbert S. A maize resistance gene functions against bacterial streak disease in rice. Proceedings of the National Academy of Sciences of the USA. 2005;102:15383–15388. doi: 10.1073/pnas.0503023102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y, Lin NC, Martin G. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell. 2002;109:589–598. doi: 10.1016/s0092-8674(02)00743-2. [DOI] [PubMed] [Google Scholar]
- 37.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 38.Claflin LE. Frederiksen RA, Odvody GN, editors. Diseases caused by prokaryotes. Compendium of Sorghum Diseases. 2000. pp. 5–7. American Phytopathological Society Press, Saint Paul, MN.
- 39.Morgan GW. Effects of spring burning and fungicide application on growth of warm season grasses and their pathogens in a native tallgrass prairie, greenhouse, and monoculture. 2003. M.S. thesis, Kansas State University, Manhattan, KS, USA.
- 40.Muriithi LM, Claflin LE. Genetic variation of grain sorghum germplasm for resistance to Pseudomonas andropogonis. Euphytica. 1997;98:129–132. [Google Scholar]
- 41.Huber L, Gillespie TJ. Modeling leaf wetness in relation to plant disease epidemiology. Annual Review of Phytopathology. 1992;30:553–577. [Google Scholar]
- 42.Claflin LE. White DG, editor. Bacterial stripe and leaf spot. Compendium of Corn Diseases. 1999. 7 American Phytopathological Society Press, Saint Paul, MN.
- 43.Duan YP, Sun X, Zhou LJ, Gabriel DW, Benyon LS, Gottwald T. Bacterial brown leaf spot of citrus, a new disease caused by Burkholderia andropogonis. Plant Disease. 2009;93:607–614. doi: 10.1094/PDIS-93-6-0607. [DOI] [PubMed] [Google Scholar]
- 44.Lane DR, Coffin DP, Lauenroth WK. Changes in grassland canopy structure across a precipitation gradient. Journal of Vegetation Science. 2000;11:359–368. [Google Scholar]
- 45.Alexander HM, Price S, Houser R, Finch D, Tourtellot M. Is there reduction in disease and pre-dispersal seed predation at the border of a host plant's range? Field and herbarium studies of Carex blanda. Journal of Ecology. 2007;95:446–457. [Google Scholar]
- 46.Sork VL, Stowe KA, Hochwender C. Evidence for local adaptation in closely adjacent subpopulations of Northern Red Oak (Quercus rubra L.) expressed as resistance to leaf herbivores. The American Naturalist. 1993;142:928–936. doi: 10.1086/285581. [DOI] [PubMed] [Google Scholar]
- 47.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- 48.Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Research. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R Development Core Team. R: A language and environment for statistical computing. 2010. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org.
- 50.Keeler KH. Distribution of polyploid variation in big bluestem (Andropogon gerardii Poaceae) across the tallgrass prairie region. Genome. 1990;33:95–100. [Google Scholar]
- 51.Keeler KH. Local polyploid variation in the native prairie grass Andropogon gerardii. American Journal of Botany. 1992;79:1229–1232. [Google Scholar]
- 52.Norrmann GA, Quarin CL, Keeler KH. Evolutionary implications of meiotic chromosome behavior, reproductive biology, and hybridization in 6× and 9× cytotypes of Andropogon gerardii (Poaceae). American Journal of Botany. 1997;84:201–207. [PubMed] [Google Scholar]
- 53.Norrmann GA, Keeler KH. Cytotypes of Andropogon gerardii Vitman (Poaceae): fertility and reproduction of aneuploids. Botanical Journal of the Linnean Society. 2003;141:95–103. [Google Scholar]
- 54.Hammerli A, Reusch TBH. Genetic neighbourhood of clone structures in eelgrass meadows quantified by spatial autocorrelation of microsatellite markers. Heredity. 2003;91:448–455. doi: 10.1038/sj.hdy.6800310. [DOI] [PubMed] [Google Scholar]
- 55.Kosman E, Leonard KJ. Similarity coefficients for molecular markers in studies of genetic relationships between individuals for haploid, diploid, and polyploid species. Molecular Ecology. 2005;14:415–424. doi: 10.1111/j.1365-294X.2005.02416.x. [DOI] [PubMed] [Google Scholar]
- 56.Vekemans X, Hardy OJ. New insights from fine-scale spatial genetic structure analyses in plant populations. Molecular Ecology. 2004;13:921–935. doi: 10.1046/j.1365-294x.2004.02076.x. [DOI] [PubMed] [Google Scholar]
- 57.National Oceanic and Atmospheric Administration (NOAA) Monthly normals of temperature, precipitation, and heating and cooling degree days, 1971–2000: Kansas and Missouri. 2003. Climatography of the United States, No. 81.
- 58.Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- 59.Clay K, Kover PX. The Red Queen Hypothesis and plant/pathogen interactions. Annual Review of Phytopathology. 1996;34:29–50. doi: 10.1146/annurev.phyto.34.1.29. [DOI] [PubMed] [Google Scholar]
- 60.Lythgoe KA, Read AF. Catching the Red Queen? The advice of Rose. Trends in Ecology and Evolution. 1998;13:473–474. doi: 10.1016/s0169-5347(98)01486-4. [DOI] [PubMed] [Google Scholar]
- 61.Dodds PN, Lawrence GJ, Catanzariti AM, Teh T, Wang CA, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proceedings of the National Academy of Sciences of the USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tian D, Traw MB, Chen JQ, Krietman M, Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- 63.Stahl EA, Dwyer G, Mauricio R, Krietman M, Bergelson J. Dynamics of disease resistance polymorphism at the Rpm1 locus of Arabidopsis. Nature. 1999;400:667–671. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 64.Tian DC, Araki H, Stahl E, Bergelson J, Kreitman M. Signature of balancing selection in Arabidopsis. Proceedings of the National Academy of Sciences of the USA. 2002;99:11525–11530. doi: 10.1073/pnas.172203599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saleh AA, Ahmed HU, Todd TC, Travers SE, Zeller KA, et al. Relatedness of Macrophomina phaseolina isolates from tallgrass prairie, maize, soybean, and sorghum. Molecular Ecology. 2010;19:79–91. doi: 10.1111/j.1365-294X.2009.04433.x. [DOI] [PubMed] [Google Scholar]
- 66.Margosian ML, Garrett KA, Hutchinson JMS, With KA. Connectivity of the American agricultural landscape: Assessing the national risk of crop pest and disease spread. BioScience. 2009;59:141–151. [Google Scholar]
- 67.Kane NC, Rieseberg LH. Selective sweeps reveal candidate genes for adaptation to drought and salt tolerance in common sunflower, Helianthus annuus. Genetics. 2007;175:1823–1834. doi: 10.1534/genetics.106.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nuismer SL, Thompson JN, Gomulkiewicz R. Coevolutionary clines across selection mosaics. Evolution. 2000;54:1102–1115. doi: 10.1111/j.0014-3820.2000.tb00546.x. [DOI] [PubMed] [Google Scholar]
- 69.Keeler KH, Williams CF, Vescio LS. Clone size of Andropogon gerardii Vitman (Big Bluestem) at Konza Prairie, Kansas. American Midland Naturalist. 2002;147:295–304. [Google Scholar]
- 70.Randolph LF. Some effects of high temperature on polyploidy and other variations in maize. Proceedings of the National Academy of Sciences of the USA. 1932;18:222–229. doi: 10.1073/pnas.18.3.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dorsey E. Induced polyploidy in wheat and rye. Journal of Heredity. 1937;27:154–160. [Google Scholar]
- 72.Thrall PH, Burdon JJ, Young A. Variation in resistance and virulence among demes of a host-pathogen metapopulation. Journal of Ecology. 2001;89:736–748. [Google Scholar]
- 73.Cox TS, Raupp WJ, Wilson DL, Gill BS, Leath S, et al. Resistance to foliar diseases in a collection of Triticum tauschii germ plasm. Plant Disease. 1992;76:1061–1064. [Google Scholar]
- 74.Garrett KA, Dendy SP, Frank EE, Rouse MN, Travers SE. Climate change effects on plant disease: genomes to ecosystems. Annual Review of Phytopathology. 2006;44:498–509. doi: 10.1146/annurev.phyto.44.070505.143420. [DOI] [PubMed] [Google Scholar]
- 75.Etterson JR, Shaw RG. Constraint to adaptive evolution in response to global warming. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This appendix provides additional details about the methods.
(DOCX)