Abstract
The centrosome in animal cells provides a major microtubule-nucleating site that regulates the microtubule cytoskeleton temporally and spatially throughout the cell cycle. We report the identification in Drosophila melanogaster of a large coiled-coil centrosome protein that can bind to calmodulin. Biochemical studies reveal that this novel Drosophila centrosome protein, centrosome protein of 309 kDa (CP309), cofractionates with the γ-tubulin ring complex and the centrosome-complementing activity. We show that CP309 is required for microtubule nucleation mediated by centrosomes and that it interacts with the γ-tubulin small complex. These findings suggest that the microtubule-nucleating activity of the centrosome requires the function of CP309.
INTRODUCTION
The microtubule cytoskeleton is essential for a variety of cellular processes, including cell movement, organelle transport, and cell division. The centrosome, consisting of a pair of centrioles and a pericentriolar material, provides the major microtubule-nucleating function in animal cells (Kellogg et al., 1994). Structural and biochemical studies have revealed that the pericentriolar material, which harbors the microtubule-nucleating activity, consists of a matrix-like structure (also referred to as the centrosome scaffold) and hundreds of γ-tubulin ring complexes (γTuRCs) (Zheng et al., 1995) that are tethered to the scaffold (Moritz et al., 1995a,b; Schnackenberg et al., 1998, 2000; Schnackenberg and Palazzo, 1999). γTuRCs are essential for microtubule nucleation from the centrosome (Felix et al., 1994; Stearns and Kirschner, 1994; Martin et al., 1998; Moritz et al., 1998; Zhang et al., 2000). Recent studies of γTuRC have led to a better understanding of its molecular composition and structure (Martin et al., 1998; Murphy et al., 1998, 2001; Tassin et al., 1998; Fava et al., 1999; Oegema et al., 1999; Gunawardane et al., 2000, 2003; Zhang et al., 2000). However, very little is known about how the γTuRC is assembled and tethered in the centrosome.
In addition to γTuRC, many other proteins are also localized to the centrosome with distinct localization characteristics (Andersen, 1999; Bornens, 2002). For example, although some proteins such as the γTuRC are localized to the centrosomes throughout the cell cycle, others are found at the centrosome only during certain stages of the cell cycle. The centrosome proteins can also be classified based on whether microtubules are required for their localization to the centrosomes. γTuRC belongs to the group of centrosome proteins whose centrosomal localization is independent of microtubules. Proteins in this group can be further divided into those that can be extracted from the centrosome by high salt and those that cannot. Many centrosome proteins examined to date, including γTuRC, can be extracted from the centrosome with high salt (Moritz et al., 1998). Thus far, other than the mammalian centriolar tubulin, only one mammalian centrosome protein, the outer dense fiber 2, has been shown to remain in the centrosome scaffold after high salt extraction (Nakagawa et al., 2001). Although the list of centrosome proteins continues to grow, little is known about how these proteins are assembled and organized in the centrosome.
By using isolated mammalian (Buendia et al., 1992) or Drosophila centrosomes (Moritz et al., 1998), two similar assays have been developed to study the microtubule-nucleating activity of the centrosome. Both assays involve extracting the centrosome proteins by using reagents that disrupt protein–protein interactions at the centrosome. In the case of Drosophila centrosomes, potassium iodide (KI) is used to extract and inactivate the centrosome, which leads to the loss of γTuRC, CP190, CP60, Centrosomin, and some unknown factors from the Drosophila centrosome scaffold (Moritz et al., 1998). On incubation with Drosophila embryo extracts, the inactivated centrosome scaffold is able to recruit the lost proteins and recover microtubule-nucleating activity. This centrosome-complementing assay provides us with an opportunity to identify the proteins required for microtubule nucleation from the centrosome.
Among the known Drosophila centrosome proteins that have been examined using the above-mentioned centrosome-complementing assay, only γTuRC is essential for centrosome-mediated microtubule nucleation (Moritz et al., 1998). Importantly, although essential, γTuRC alone is insufficient for complementing the salt-extracted centrosome scaffold (Moritz et al., 1998). Therefore, additional protein(s) in Drosophila embryo extracts must be required. The additional protein(s) may be required to recruit and tether γTuRC to the centrosome scaffold.
Studies in Saccharomyces cerevisiae revealed that the γ-tubulin complex, called the Tub4p complex, is tethered at the spindle pole bodies via Spc110 (Knop and Schiebel, 1997). The Tub4p complex is equivalent to the γ-tubulin small complex (γTuSC), a major building block of the γTuRC (Oegema et al., 1999; Gunawardane et al., 2000). Interestingly, Spc110 contains a calmodulin (CaM)-binding motif at its C terminus. Furthermore, it has been shown that CaM is required for proper localization of Spc110 at the yeast spindle pole bodies (Kilmartin and Goh, 1996; Stirling et al., 1996; Sundberg et al., 1996; Nguyen et al., 1998). By analogy, these studies suggest that CaM and CaM-binding proteins might be required for centrosome-mediated microtubule nucleation by tethering γTuRC to centrosomes in animal cells. Consistent with this idea, two mammalian centrosome proteins, Kendrin (also called pericentrin B) and CG-NAP (also called AKAP350 or AKAP450), were found to contain CaM binding motifs homologous to Spc110 (Takahashi et al., 1999; Diviani et al., 2000; Flory et al., 2000; Gillingham and Munro, 2000; Li et al., 2001; Takahashi et al., 2002). Both Kendrin and CG-NAP also bind to GCP2, a component of human γTuSC (Takahashi et al., 2002).
Although Kendrin and CG-NAP are good candidates for tethering γTuRC to the centrosome, it remains unclear whether they are essential for centrosome-mediated microtubule nucleation. For example, one study showed that depleting Kendrin did not affect the ability of Chinese hamster ovary cell lysates to complement KI-extracted centrosomes (Li et al., 2001). However, in another study, antibodies against Kendrin and CG-NAP modestly inhibited microtubule-nucleating activity of the intact centrosome (Takahashi et al., 2002). Clearly, additional studies are necessary to determine the role of Kendrin and CG-NAP in centrosome function.
Here, we report a novel Drosophila CaM-binding protein that is localized to centrosomes throughout the cell cycle. Our studies reveal that centrosome protein of 309 kDa (CP309) shares a number of similarities with Kendrin and CG-NAP. Importantly, CP309 binds to γTuSC and is required for microtubule nucleation mediated by Drosophila centrosomes in vitro. Our studies provide additional evidence to support the notion that proteins such as Kendrin and CG-NAP play roles in centrosome-mediated microtubule nucleation.
MATERIALS AND METHODS
Buffers and Protease Inhibitors
Buffers were as follows: BRB80, 80 mM K-PIPES, pH 6.8, 1 mM MgCl2, 1 mM EGTA; HB, 50 mM K-HEPES, pH 7.6, 1 mM MgCl2, 1 mM EGTA, and 1 mM β-mercaptoethanol; HB100Na, HB buffer containing 100 mM NaCl; HB100K, HB buffer containing 100 mM KCl; and HB3, HB buffer containing 100 mM KCl, 10% glycerol, protease inhibitor (1:100 dilution from protease inhibitor stock), 1 mM phenylmethylsulfonyl fluoride. Protease inhibitor stock contained 10 mM benzamidine-HCl, 0.1 mg/ml phenanthroline, 1 mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A in ethanol.
Preparation of Drosophila Embryo Extract and Centrosomes
Crude embryo extracts were made from Drosophila embryos (0–2 h) as described previously (Gunawardane et al., 2000) and clarified by centrifuging for 20 min at 50,000 rpm in a TLS55 or SW55 rotor at 4°C in the presence of nocodazole (final 100 μM). Drosophila centrosomes were isolated from 0 to 3.5 h Drosophila embryos on sucrose gradients as described previously (Moritz et al., 1995b).
Partial Purification of Centrosome Complementing Factor(s)
Polyethylene glycol 8000 (PEG8000) was added to the clarified embryo extract to a final concentration of 3% from a 30% stock in HB100Na. The mixture was incubated on ice for 20 min and centrifuged at 14,000 rpm for 15 min at 4°C. The precipitant was resuspended with HB100K containing 0.1 mM GTP and 0.05% NP-40 in 1/4 volume of the clarified extract, homogenized, and centrifuged at 14,000 rpm for 15 min at 4°C. Sucrose gradients (5–40%) were prepared as described previously in HB100K containing 0.1 mM GTP and 1:200 protease inhibitor stock (Oegema et al., 1999; Gunawardane et al., 2001). The resuspended and clarified PEG-precipitated proteins were loaded onto the gradient and centrifuged in a SW55 rotor at 50,000 rpm for 4 h at 4°C. Gradients were fractionated from the top. Fractions 11–13 were combined, filtered, and subjected to further purification on a Mono S column (HR 5/5; Pharmacia Amersham Biosciences, Piscataway, NJ) equilibrated with HB buffer. The column was washed with HB100Na, and bound proteins were eluted by a linear NaCl gradient (0.1–0.5 M).
Centrosome-complementing Assay
Embryo extracts or protein fractions were used to complement the salt-extracted centrosome scaffolds to assemble into functional centrosomes according to published methods with minor modifications (Moritz et al., 1998). Briefly, the isolated centrosomes were treated with 2 M KI on ice for 7 min. The KI-stripped centrosomes were first attached to the polylysine-coated coverslips and then incubated with 60 μl of embryo extract or 60 μl of different fractions from PEG precipitation, sucrose gradient sedimentation, or Mono S column chromatography for 15 min at 30°C. After incubation, soluble proteins were washed out and then a mixture of unlabeled and rhodamine-labeled tubulin was added and incubated for 10 min at 30°C. The number of microtubule asters nucleated from the reconstituted centrosomes was counted under a fluorescence microscope using a 60× objective.
Cloning of CP309
Drosophila mRNA was prepared from 0- to 2-h embryos by using a QuickPrep mRNA purification kit (Pharmacia Amerisham Biosciences, Piscataway, NJ). Three primers (gsp1, CTG CTC CTC CAG CCG ATT TTG; gsp2, TTG GAG GGT AGA AAT ACG CTG; and gsp3, TTC CTC CAT ACG ACC CTG CAG) corresponding to the 5′ region of the second exon of CG6735 gene were used for the 5′ rapid amplification of cDNA ends (RACE) by using a RACE kit (Roche Diagnostics, Indianapolis, IN. The amplified DNA fragments were subcloned into the pCRscript vector (Stratagene, La Jolla, CA) and sequenced. To obtain the additional 5′ sequences of CP309, another round of RACE was performed using three primers (gsp4, GCA CTT GAT TGC TCA TCC TTT GG; gsp5, CGC CTC CGA TAG AGA TTC CAG; and gsp6, CAC CTC ACT GAT CTC GAT GGC) corresponding to the middle region of the annotated first exon of the CG13459 gene (Figure 1A). The resulting RACE product contains the first ATG of CP309 and the 5′-untranslated region of the CG6945 gene.
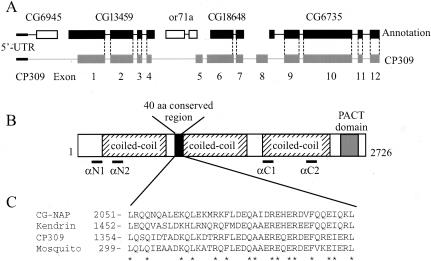
Figure 1.
CP309 is a large coiled-coil protein similar to Kendrin and CG-NAP. (A) Organization of the CP309 gene. The CP309 gene is composed of 12 exons (gray boxes). The corresponding annotated exons in the Drosophila genome database are shown as either black or white boxes. The transcript for CP309 contains three genes that are annotated separately in the Drosophila genome database. Two annotated genes, CG6945 and or71a, are spliced out from the mature transcript of CP309. The dotted lines represent the exonintron boundaries that are common between the annotated genes in the database and in CP309. (B) Schematic representation of the primary structure of CP309 (not drawn exactly to scale). CP309 contains three predicted coiled-coil regions, a 40-amino acid conserved region, and a conserved PACT domain. Regions of CP309 used for antibody production are indicated as αN1, αN2, αC1, and αC2. (C) Sequence alignment of the conserved 40 amino acids found in CG-NAP, Kendrin, CP309, and a partial mosquito protein (EAA07046) that are homologous to CP309. The sequence alignment of the CP309 PACT domain with other PACT domain proteins was described previously (Gillingham and Munro, 2000).
Antibodies
To generate rabbit polyclonal antibodies against the C terminus of CP309, fusion proteins were made between glutathione S-transferase (GST) and fragments of CP309 corresponding to amino acids 1636–1817 (GST-CP309C1), 1950–2109 (GST-CP309C2), or 2287–2442 (GST-CP309C3). The fusion proteins were purified, mixed, and injected into rabbits. Antibody αC1 was affinity purified against CP309C1, whereas antibody αC2 was affinity purified against a mixture of CP309C2 and CP309C3. Rabbit polyclonal antibody against the N terminus of CP309 was generated using the GST-fusion fragment of CP309 corresponding to amino acids 250–501 (αN1). The rabbit polyclonal antibody against amino acids 512–803 (αN2) was a gift from Dr. Jordan Raff (Wellcome/CR-UK Institute, Cambridge, United Kingdom). The antibodies against Drosophila γ-tubulin (DrosC) and Dgrips84 and 91 were described previously (Oegema et al., 1999). Monoclonal antibodies against γ-tubulin (GTU-88; Sigma-Aldrich, St. Louis, MO) and α-tubulin (DM1α; Sigma-Aldrich) were purchased.
CaM-binding Assay
Clarified Drosophila embryo extracts (200 μl) were mixed with 800 μl of HB100K containing 0.02% Triton X-100 and CaM agarose beads in the presence of 2.5 mM CaCl2 or 5 mM EGTA. After incubation for 1.5 h at room temperature, the beads were washed with the same buffer in the presence of 2.5 mM CaCl2 or 5 mM EGTA. The bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and Western blotting. To prepare CaM-free CP309, extracts were incubated with CaM-agarose beads in the presence of 2.5 mM CaCl2, and the beads were washed as described above. Bound CP309 was eluted from the CaM beads with 10 mM EGTA and passed through a desalting column to remove EGTA.
Immunodepletion and Immunoprecipitation
To deplete CP309 from the embryo extract, affinity-purified antibody αC1, αC2, or nonimmunized rabbit IgG (NR) was bound to protein A agarose beads for 1 h at room temperature. The beads were washed with phosphate-buffered saline and incubated with the clarified embryo extracts for 1 h at 4°C. The level of CP309 depletion was analyzed by Western blotting. To remove CP309-specific antibody from αC1 and αC2, the baculovirus expressed FLAG-tagged C-terminal half of CP309, FLAG-CP126, was bound to anti-FLAG M2 agarose beads (Sigma-Aldrich) and incubated with αC1 or αC2. Depletion of CP309-specific antibody was confirmed by Western blotting.
For immunoprecipitation, affinity-purified rabbit polyclonal antibodies against CP309, γ-tubulin, or Dgrip84 were bound to protein A agarose beads. The antibody-bound beads were blocked with bovine serum albumin (10 mg/ml) overnight at 4°C. The beads were washed with HB100K containing 0.1% Triton X-100 and 0.1 mM GTP and incubated with clarified Sf9 cell lysates made from cells expressing FLAG-tagged CP309, FLAG-tagged γ-tubulin, and untagged Dgrip84 and Dgrip91. After incubation for 2 h at 4°C, the beads were washed with the same buffer. Proteins bound to beads were analyzed by Western blotting.
Immunofluorescence Analysis
Drosophila embryos (0–2 h) were used for immunofluorescence according to previously published methods (Theurkauf, 1994). Briefly, embryos were blocked with 3% bovine serum albumin containing 0.1% Triton X-100 and then labeled with rabbit antibody against CP309 (1:500) and mouse antibody against α-tubulin (1:500) or γ-tubulin (1:500). RNase A (0.1 mg/ml) was added and incubated for 2 h at room temperature. Chromosomes were stained with 1 μM TOTO-3 (Molecular Probes, Eugene, OR) for 20 min at room temperature, followed by Alexa red-labeled anti-mouse (1:400) and Alexa green-labeled anti-rabbit (1:400) secondary antibodies (Molecular Probes). Images were taken on a confocal microscope. Embryos were treated with 5 mM colchicine for 20 min at room temperature to depolymerize microtubules.
Protein Expression in Insect Cells
FLAG-tagged γ-tubulin, untagged Dgrip84, and Dgrip91 were coexpressed in Sf9 cells as described previously (Gunawardane et al., 2000). FLAG-tagged CP309 and the FLAG-tagged transcript of CG6735 were cloned individually into the pFastBac1 vector (Invitrogen, Carlsbad, CA). The recombinant bacmids of CP309 and CG6735 were transfected into Sf9 cells. Baculoviruses were generated and recombinant proteins were expressed according to the manufacturer's instructions.
RESULTS
Identification of a Novel Drosophila Protein, CP309, That Contains the CaM-binding Motif
We reasoned that whether the CaM-binding proteins such as Kendrin and CG-NAP are important for centrosome function, similar protein(s) should exist in Drosophila. Database searches by using the full-length Kendrin or CG-NAP did not reveal any Drosophila proteins sharing overall sequence homology with Kendrin or CG-NAP. However, we found that several predicted Drosophila proteins contain the highly conserved CaM-binding motif that is found in Spc110, Kendrin, and CG-NAP. Interestingly, only one of these proteins, encoded by CG6735, is predicted to contain largely coiled-coil sequences 5′ to the C-terminal CaM-binding domain such as Spc110, Kendrin, and CG-NAP. Because CG6735 has previously been reported to encode a potential homolog of Kendrin and CG-NAP (Gillingham and Munro, 2000), we decided to analyze the gene product of CG6735 further.
Sequence analyses reveal that CG6735 encodes a complete open reading frame of 1109 amino acids. This predicted protein is significantly shorter than Kendrin (3246 amino acids) and CG-NAP (3899 amino acids). Therefore, we asked whether there is a longer splicing variant(s) of CG6735 in Drosophila by using RACE analyses starting from the 5′ end of CG6735. This led to the identification of a complete open reading frame that encodes a predicted protein of 2726 amino acids and 309 kDa (Figure 1, A and B). We will refer to this protein as CP309 (see below). This sequence is deposited in the GenBank under the accession number AY373570.
CP309 is similar to Kendrin and CG-NAP in the following three aspects. First, all three proteins contain the conserved pericentrin AKAP450 centrosome targeting (PACT) domain of ∼200 amino acids at their C termini (Figure 1B) (Gillingham and Munro, 2000). Second, they have similar coiled-coil organizations N-terminal to the PACT domain (Figure 1B) (Takahashi et al., 2002). Third, they share a conserved region of ∼40 amino acids located in the N-terminal half of these proteins (Figure 1, B and C). Based on these analyses, we suggest that CP309 is the Drosophila equivalent of Kendrin or CG-NAP.
CP309 Is Localized to Centrosomes in Drosophila Embryos
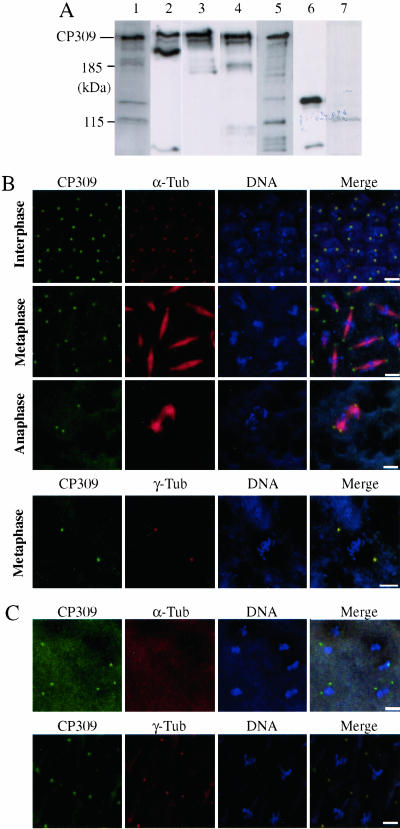
To determine whether both CP309 and CG6735 are expressed in Drosophila embryos, we obtained antibodies against sequences in either N-terminal (αN1 and αN2) or C-terminal (αC1 and αC2) halves of CP309 (Figure 1B). Western blotting analyses showed that CP309 expressed in Drosophila embryos migrated to a similar position as the CP309 expressed in Sf9 cells on SDS-PAGE (Figure 2A). In addition, although we could detect the baculovirus-expressed CG6735 protein in Sf9 cells, we failed to detect CG6735 protein in Drosophila embryo extracts by using either αC1 or αC2 antibodies (Figure 2A). These analyses demonstrate that the Drosophila embryo expresses CP309, but not CG6735.
Figure 2.
CP309 is a centrosome protein. (A) CP309, but not CG6735, is expressed in the embryos. Embryo extracts (lanes 1–4 and 7) were probed with αN1 (lane 1), αN2 (lane 2), αC1 (lane 3), αC2 (lane 4), or serum from a nonimmunized rabbit (lane 7). Sf9 cell lysates expressing FLAG-tagged CP309 protein (lane 5) and FLAG-tagged recombinant protein encoded by CG6735 (lane 6) were probed with αC2. (B) CP309 is localized to the centrosomes throughout the cell cycle. CP309 (green), microtubules (red), or γ-tubulin (red), and chromosome (blue) are shown. (C) CP309 (green), like γ-tubulin (red), localizes to the centrosomes in the absence of microtubules. Embryos were treated with colchicine before immunofluorescence staining. Bars, 5 μm.
We next determined the subcellular localization of CP309 in Drosophila embryos by immunofluorescence microscopy. CP309 is localized to centrosomes throughout the cell cycle (Figure 2B). Furthermore, the localization of CP309 to centrosomes is independent of microtubules (Figure 2C). These studies show that CP309, like γTuRC, is a core centrosome protein, whose localization to the centrosome is independent of microtubules.
CP309, Like γ-Tubulin, Can Be Extracted from the Centrosome by 2 M KI
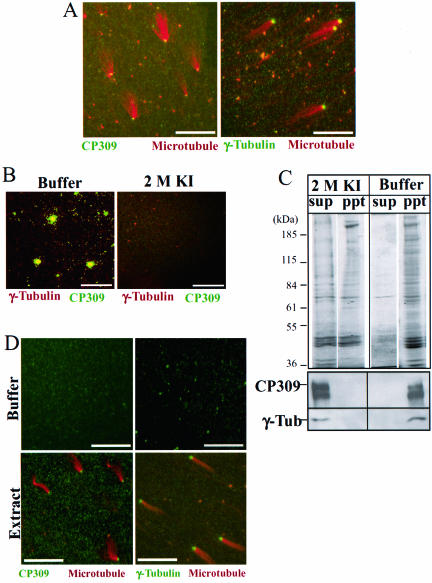
The core centrosome proteins can be further divided into two groups, those that can be extracted from the centrosome with 2 M KI and those that remain bound to the centrosome scaffold after extraction. Thus far, all known Drosophila centrosome proteins can be completely extracted from centrosomes by 2 M KI. We found that CP309, like γ-tubulin, could be detected on the isolated centrosomes by immunofluorescence staining (Figure 3, A and B). However, after the centrosomes were treated with 2 M KI, CP309 and γ-tubulin were no longer detected as colocalized spots, suggesting that both proteins were removed from centrosomes (Figure 3B). Consistent with this, Western blotting analysis showed that 2 M KI extracted both CP309 and γ-tubulin into supernatant fractions (Figure 3C). To determine whether CP309 can rebind to the KI-treated centrosomes, we incubated centrosome scaffolds with Drosophila embryo extracts followed by a microtubule nucleation assay (see MATERIALS AND METHODS). Immunofluorescence staining showed that the assembly of functional centrosomes from the inactive scaffolds (as judged by microtubule nucleation) is accompanied by the recruitment of both γ-tubulin and CP309 to the centrosome (Figure 3D). These studies show that CP309 behaves similarly to γ-tubulin.
Figure 3.
CP309, like γ-tubulin, is extracted from the centrosomes by KI. (A) CP309 (green), like γ-tubulin (green), localizes to isolated centrosomes that are used to nucleate microtubules (red) on glass coverslips. (B) CP309 (green) and γ-tubulin (red) colocalize to isolated centrosomes adhered on glass coverslips (buffer). The colocalization disappears after treating the centrosomes with 2 M KI (2 M KI). (C) Both CP309 and γ-tubulin can be extracted from the centrosome by 2 M KI. Isolated centrosomes were treated with 2 M KI or control buffer. The insoluble centrosome scaffolds were pelleted by centrifugation. The supernatant (sup) and pellet (ppt) were subjected to SDS-PAGE followed by either Coomassie Blue staining or Western blotting probing with antibodies against CP309 or γ-tubulin. (D) Both CP309 and γ-tubulin are recruited to the reconstituted centrosomes. KI extracted centrosome scaffolds do not have CP309 or γ-tubulin staining after incubating with buffer control (top two panels). However, after incubating with embryo extracts, the reconstituted centrosomes can nucleate microtubules (red), and these centrosomes contain both CP309 (green) and γ-tubulin (green). Bars, 10 μm.
CP309 Cofractionates with the Centrosome-complementing Activity
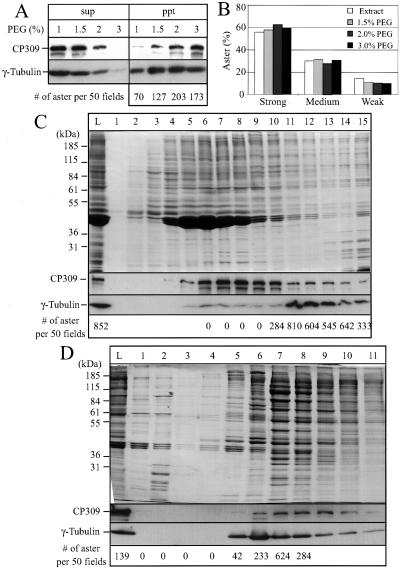
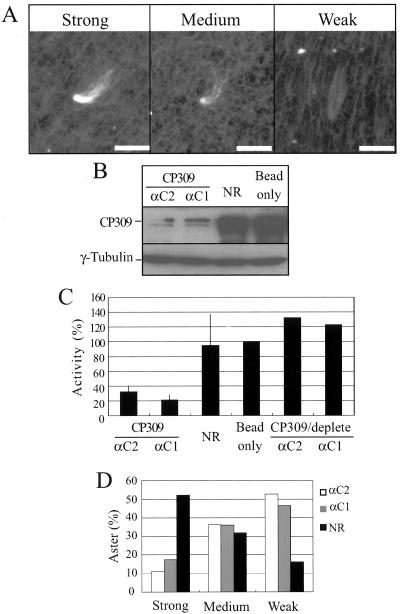
To study the function of CP309 biochemically, we determined whether CP309 cofractionated with the centrosome-complementing activity present in the embryo extracts. We subjected the Drosophila embryo extract to precipitation by using different concentrations of PEG8000 and separated the proteins into supernatant and pellet fractions. The pellet fractions were solubilized and passed through desalting columns to remove PEG. Western blotting showed that both CP309 and γ-tubulin were efficiently precipitated by 3% PEG (Figure 4A). To determine the complementing activity present in the PEG pellets, we incubated the protein mixture with the KI-inactivated centrosome scaffolds and then carried out the microtubule nucleation assay. The number of microtubule asters was counted per 50 random microscopy fields (Figure 4A). The examples of typical strong, medium, and weak microtubule asters are shown in Figure 5A. We found that the centrosome-complementing activity is present in the PEG pellets that contain both CP309 and γ-tubulin (Figure 4A). Furthermore, >80% of the asters formed in these complementing assays are either strong or medium type asters (Figures 4B and 5A).
Figure 4.
CP309 copurifies with γ-tubulin and the centrosome-complementing activity. (A) PEG precipitates CP309 and γ-tubulin together with the centrosome-complementing activity. Clarified embryo extracts were fractionated into supernatants and pellets at the indicated concentrations of PEG8000. The supernatant (sup) and pellet (ppt) were analyzed by Western blotting probing with antibodies against CP309 and γ-tubulin. For complementing assays, the pellet fractions were resuspended in buffer, clarified, and passed through desalting columns to remove PEG. The number of asters reconstituted using the pellet fractions was counted in 50 random microscopy fields. (B) Quality of the reconstituted microtubules asters. Microtubule asters are divided into strong, medium, and weak categories depending on the amount of microtubules nucleated (see Figure 5A). More than 80% of the microtubule asters reconstituted using PEG pellets belong to the strong or medium categories. (C) Sucrose gradient fractionation of the centrosome-complementing activity. The 3% PEG pellet fraction from above was further purified by 5–40% sucrose gradient sedimentation; see lane L for proteins loaded. Fractions were analyzed by SDS-PAGE and Coomassie Blue staining (top) or by Western blotting probing with antibodies against CP309 or γ-tubulin (middle two panels). The number of asters formed (bottom) was counted in 50 random microscopy fields. (D) Mono S column chromatography of centrosome-complementing activity. The active fractions from the sucrose gradient (fractions 11–13) were further purified on a Mono S column (HR 5/5), see lane L for proteins loaded. The eluted proteins were analyzed as in C. The peak of complementing activity (fraction 7) lies between the γ-tubulin peak (fraction 6) and CP309 peak (fraction 8/9).
Figure 5.
CP309 is required for microtubule nucleation from centrosomes in vitro. (A) Three typical asters with strong, medium, and weak microtubule nucleation are shown. Bars, 5 μm. (B) Depletion of CP309 from embryo extracts. Embryo extracts were depleted using two affinity-purified antibodies (αC1 or αC2) against different regions of CP309, mock-depleted using nonimmunized rabbit IgG (NR), or with beads alone. Western blotting analysis showed that 70–90% of the CP309 was depleted from embryo extracts, whereas the amount of γ-tubulin was unchanged. (C) Centrosome-complementing assays. Extracts were immunodepleted with NR, beads alone, or αC1, αC2, or αC1 and αC2 that were depleted of CP309-specific antibody. Each of these extracts was then used to reconstitute centrosomes from KI extracted centrosome scaffolds. Asterforming activity was quantified as described above and expressed as percentage of the mock-depleted embryo extracts (beads only). Error bars represent SD from four independent experiments. (D) Percentages of asters with strong, medium, or weak microtubule nucleation in the extracts depleted with NR, αC1, or αC2.
Because the above-mentioned studies showed that 3% PEG efficiently precipitated γ-tubulin and CP309, we further purified the PEG precipitated proteins on a 5–40% sucrose gradient (Figure 4C). Western blotting by using antibodies against CP309 and γ-tubulin showed that less CP309 comigrated with γTuRC (Figure 4C, fractions 11–13) than with γTuSC (Figure 4C, fractions 6–8). Using the centrosome-complementing assay, we found that the fractions containing both CP309 and γTuRC had robust aster-forming activity that gave rise to >80% strong or medium type asters, whereas fractions containing the γTuSC and the majority of CP309 had no activity (Figure 4C).
To further determine whether CP309 can copurify with the complementing activity, we pooled fractions 11, 12, and 13 from the sucrose gradient (Figure 4C) and subjected them to Mono S column chromatography. We found that the peak activity that produced >80% either strong or medium type asters is present in the fractions that contain both γTuRC and CP309 (Figure 4D). Interestingly, we consistently observed that the peak aster forming activity lies between the γ-tubulin peak fraction and the CP309 peak fraction (Figure 4D). These studies show that a fraction of CP309 copurifies with γTuRC and the centrosome-complementing activity. Therefore, CP309 may be required for microtubule nucleation mediated by centrosomes.
Unfortunately, any further purification by using various conventional fractionation methods led to a complete separation of CP309 from γTuRC and a complete loss of the complementing activity. Combining fractions that contain CP309 and γTuRC also failed to restore the activity (our unpublished data). It is possible that additional centrosome assembly factors might have been separated from γTuRC and CP309 in these further purifications. Alternatively, CP309, γTuRC, or an unknown factor might have lost its activity during the additional purifications.
CP309 Is Required for Centrosome-mediated Microtubule Nucleation
To further determine whether CP309 is required for microtubule nucleation from centrosomes, we immunodepleted CP309 from Drosophila embryo extracts by using either αC1 or αC2 antibodies. The CP309-depleted extracts, mock-depleted extracts with either random IgG (NR), or protein A beads alone were then used in the complementing assay. We found that ∼70–90% of the CP309 was depleted from the embryo extracts (Figure 5B), which led to a 60–90% reduction in the number of asters compared with the mock-depleted extracts (Figure 5C). Importantly, depletion of CP309 did not lead to γTuRC depletion (Figure 5B). Further analyses showed that the asters that did form in CP309-depleted extract nucleated fewer microtubules compared with those of mock-depleted extracts (Figure 5D).
We have attempted to rescue the CP309-depleted embryo extracts by using either baculovirus-expressed CP309 or the extract CP309 that cofractionated with γTuSC on sucrose gradients (Figure 4C, fractions 6–8). However, neither source of CP309 rescued the centrosome-complementing activity of the CP309-depleted extract. To further demonstrate that the CP309 antibodies specifically removed CP309, but not some unrelated proteins that are required for centrosome complementation, we used the baculovirus expressed C-terminal half of CP309 to deplete the CP309-specific antibodies in αC1 and αC2. We found that the depleted αC1 and αC2 failed to remove either CP309 (our unpublished data) or the centrosome-complementing activity from the egg extracts (Figure 5C). Together, these studies suggest that CP309 is required for centrosome-mediated microtubule nucleation in vitro.
CP309 Binds to CaM and γTuSC
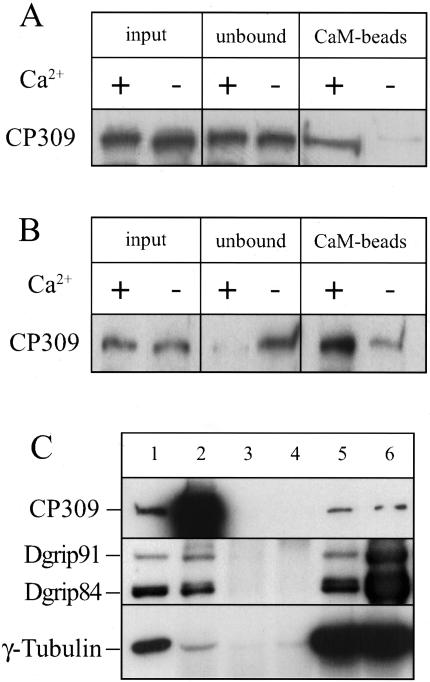
Because CP309 contains a CaM-binding motif, we examined whether CP309 can bind to CaM and whether this binding is Ca2+ dependent. We first incubated CaM-agarose beads with Drosophila embryo extracts in the presence of 2.5 mM Ca2+ or 5 mM EGTA (EGTA was used to chelate the Ca2+ in the extracts). Proteins bound to the CaM agarose beads were analyzed by Western blotting probing with antibodies against CP309. We found that significantly more CP309 in the extracts bound to CaM-agarose beads in the presence of Ca2+ than in the presence EGTA (Figure 6A). Next, we asked whether Ca2+ facilitates CaM-free CP309 binding to CaM. CaM-free CP309 was prepared by eluting CP309 from CaM-agarose beads with EGTA (see MATERIALS AND METHODS) and then used in the same binding assay as described above. We found that the CaM-free CP309 also binds to CaM-agarose beads more efficiently in the presence of Ca2+ than in the presence of EGTA (Figure 6B). Therefore, although CP309 can bind to CaM in the absence of Ca2+, Ca2+ significantly enhances the binding.
Figure 6.
CP309 binds CaM and γTuSC. (A) CP309 in embryo extracts binds to CaM efficiently in the presence of Ca2+. CaM-agarose beads were added into clarified embryo extracts containing either Ca2+ or EGTA. Significantly more CP309 binds to CaM beads in the presence of Ca2+ than in the presence of EGTA. (B) Ca2+ promotes the binding of CaM-free CP309 to CaM. CaM-free CP309 was incubated with CaM-agarose beads in the presence of Ca2+ or EGTA. Significantly more CaM-free CP309 is found on the CaM beads in the presence of Ca2+ than in the presence of EGTA. (C) CP309 binds to γTuSC. FLAG-CP309, FLAG-γ-tubulin, Dgrip84, and Dgrip91 were coexpressed in Sf9 cells by using baculovirus. Reciprocal immunoprecipitations were carried out in cell lysates (lane 1) by using αC1 antibody against CP309 (lane 2), two different nonimmunized rabbit sera (lanes 3 and 4), antibody against γ-tubulin (DrosC, lane 5), and antibody against Dgrip84 (lane 6). The immunoprecipitated proteins were analyzed by Western blotting probing with monoclonal antibody against FLAG to detect CP309 and γ-tubulin, and polyclonal antibodies to detect Dgrip91 and Dgrip84.
Previous studies have suggested that Kendrin and CG-NAP interact with a mammalian-γTuSC component (Takahashi et al., 2002). Consistent with this, our biochemical studies have shown that CP309 cofractionates with γTuSC and γTuRC in Drosophila embryo extracts (Figure 4). However, we found that γTuRC/γTuSC and CP309 do not coimmunoprecipitate with each other in the embryo extracts (our unpublished data). This suggests that the interaction between CP309 and γTuRC/γTuSC is weak. Because we could coexpress both CP309 and γTuSC in Sf9 cells to levels higher than those in the embryo extracts, we examined whether CP309 and γTuSC interact with each other in Sf9 cells. FLAG-tagged CP309, γ-tubulin, and untagged Dgrips84 and 91 were expressed in Sf9 cells, and reciprocal immunoprecipitations were carried out using antibodies against CP309, Dgrip84, and γ-tubulin. Either preimmune serum for CP309 antibody or random nonimmunized rabbit IgGs were used as control. We found that the antibody against CP309 immunoprecipitated all three γTuSC subunits. Because the antibodies against γ-tubulin and Dgrip84 also immunoprecipitated CP309 (Figure 6C), we conclude that CP309 interacts with γTuSC. We also carried out the above-mentioned experiments in the presence or absence of Ca2+ and CaM and found that they did not affect the interaction between CP309 and γTuSC (our unpublished data). Therefore, Ca2+ or CaM does not seem to regulate the interaction between CP309 and γTuSC in vitro.
DISCUSSION
D. melanogaster offers both genetic and biochemical means to study centrosome function. Due to the relative ease in obtaining large quantities of Drosophila embryo extracts, a convenient assay has been developed to study the proteins required for centrosome-mediated microtubule nucleation (Moritz et al., 1998). Using this assay, we studied the function of CP309, a novel Drosophila centrosome protein that shares a number of similarities with Kendrin and CG-NAP. Our findings suggest that CP309 is required for microtubule nucleation from centrosomes.
Although a number of centrosome proteins have been identified in Drosophila, only γTuRC has been found to play an essential role in centrosome-mediated microtubule nucleation. Three lines of evidences presented in this study suggest that CP309 is another centrosome protein required for centrosome-mediated microtubule nucleation. First, CP309 is a centrosome protein that can interact with γTuSC. Second, CP309 cofractionates with centrosome-complementing activity. Finally, depleting CP309 by using two antibodies raised against different regions of CP309 abolishes the ability of extracts to complement the KI-stripped centrosomes.
The inability to rescue centrosome-complementing activity by the add-back experiments could be because the CP309 used is not fully functional. For example, CP309 expressed in baculovirus might not be folded properly. Alternatively, the depletion of CP309 might have led to depletion of CP309 interacting protein(s) that are also required for centrosome assembly. If the CP309 we used for add-back does not have the interacting protein(s), we would not achieve complementation.
Unlike γTuRC components, CP309 consists of largely coiled-coil regions. Because the centrosome scaffold has been postulated to consist of mostly coiled-coil proteins, CP309 may be a structural component of the centrosome scaffold to which proteins such as γTuRC are tethered. Interestingly, we found that, like γTuRC, CP309 could be extracted from the centrosome by KI. Therefore, the centrosome scaffold may be further divided into regions that can be extracted with KI and regions that cannot. We suggest that CP309 represents the first member of the essential scaffold proteins that can be extracted by KI.
Using a baculovirus protein expression system, we found that CP309 interacts with γTuSC. However, immunoprecipitation by using various antibodies against CP309 or γTuRC components has failed to detect any interaction between CP309 and γTuRC or CP309 and γTuSC in the Drosophila embryo extracts. One explanation for this is that the interaction between CP309 and γTuSC/γTuRC is weak in the cytosol. Therefore, only when the amount of CP309 and γTuSC is increased by expression in baculovirus can we detect such kind of interaction. Consistent with this idea, a weak interaction between γTuSC and Kendrin/CG-NAP was also observed a previous study (Takahashi et al., 2002). The interaction between γTuSC/γTuRC and the PACT domain containing proteins such as CP309, Kendrin, and CG-NAP suggests that these proteins are involved in recruiting γTuRC to the centrosome. Consistent with this, we found that CP309 is essential for centrosome-mediated microtubule nucleation in vitro.
Because the PACT domain-containing proteins interact with γTuRC/γTuSC weakly in the cytosol, an important question is how these proteins may function as tethers for γTuRC at the centrosome. One possibility is that the PACT domain-containing proteins have to be first assembled with other proteins into a proper γTuRC-docking site at the centrosome to interact with γTuRC strongly. If only the properly assembled docking site has a high affinity for γTuRC, the individual components of the docking site would have a weak interaction with γTuRC in the cytosol. Alternatively, the PACT domain-containing proteins may be solely responsible for γTuRC docking at the centrosome. However, in the cytosol, these proteins may exist in conformations in which their γTuRC interaction sites are not completely exposed. The assembly of the PACT domain-containing proteins to the centrosome could lead to conformational changes, thereby completely exposing the binding site. Interestingly, CP309, Kendrin, and CG-NAP can all bind to CaM. Although we found that CaM and Ca2+ did not affect the interaction between CP309 and γTuSC/γTuRC in the cytosol, it is possible that CaM and Ca2+ might be involved in organizing the PACT domain proteins into the proper γTuRC docking site at the centrosome.
Finally, it is worth noting that two PACT domain proteins, Kendrin and CG-NAP, exist in mammalian cells. However, we found that CP309 is the only PACT domain-containing protein found in the D. melanogaster genome. It is possible that Kendrin and CG-NAP share redundant functions in tethering γTuRC to the centrosome in mammalian cells, whereas in Drosophila, γTuRC is tethered to the centrosome by CP309 alone. If so, this could explain why depleting Kendrin from CHO cell lysates did not block the lysate to support microtubule aster reconstitution (Li et al., 2001). Undoubtedly, further characterization of these CaM binding centrosomal proteins in different organisms should shed light on the organization of the microtubule-nucleating material at centrosomes.
Acknowledgments
We thank Dr. Jordan Raff for the anti-CP309 antibody, Dr. Sofia Lizzaraga for help with the centrosome-complementing assay, and the members of the Zheng laboratory for helpful discussions during the course of this work. This study was supported by National Institutes of Health grant RO1-GM56312-01 (to Y.Z.) and Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (to S.K.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–03–0191. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-03-0191.
Abbreviations used: CaM, calmodulin; CP309, centrosome protein of 309 kDa; Dgrip, Drosophila gamma ring protein; NR, nonimmunized rabbit IgG; γTuSC, γ-tubulin small complex; γTuRC, γ-tubulin ring complex.
References
- Andersen, S. (1999). Molecular characterization of the centrosome. Int. Rev. Cytol. 187, 24–34. [DOI] [PubMed] [Google Scholar]
- Bornens, M. (2002). Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Struct. Biol. 14, 24–34. [DOI] [PubMed] [Google Scholar]
- Buendia, B., Draetta, G., and Karsenti, E. (1992). Regulation of the microtubule nucleating activity of centrosomes in Xenopus egg extracts: role of cyclin A-associated protein kinase. J. Cell Biol. 116, 1431–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviani, D., Langeberg, L., Doxsey, S., and Scott, J. (2000). Pericentrin anchors protein kinase A at the centrosome through a newly identified RII-binding domain. Curr. Biol. 10, 417–420. [DOI] [PubMed] [Google Scholar]
- Fava, F., Raynaud, M.B., Leung, T.J., Mazzolini, L., Li, M., Guillemot, J., Cachot, D., Tollon, Y., Ferrara, P., and Wright, M. (1999). Human 76p: A new member of the γ-tubulin-associated protein family. J. Cell Biol. 147, 857–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix, M.A., Antony, C., Wright, M., and Maro, B. (1994). Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopus sperm aster formation. J. Cell Biol. 124, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory, M.R., Moser, M.J., Monnat, R.J., Jr., and Davis, T.N. (2000). Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc. Natl. Acad. Sci. USA 97, 5919–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham, A.K., and Munro, S. (2000). The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1, 524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, N.R., Martin, O.C., Cao, K., Zhang, L., Dej, K., Akihiro, I., and Zheng, Y. (2000). Characterization and reconstitution of Drosophila gamma tubulin ring complex subunits. J. Cell Biol. 151, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R.N., Martin, O.C., and Zheng, Y. (2003). Characterization of a New γTuRC subunit with WD repeats. Mol. Biol. Cell 14, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R.N., Zheng, Y., Oegema, K., and Wiese, C. (2001). Purification and reconstitution of Drosophila gamma tubulin complexes. Methods Cell Biol. 67, 1–25. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., Moritz, M., and Alberts, B.M. (1994). The centrosome and cellular organization. Annu. Rev. Biochem. 63, 639–674. [DOI] [PubMed] [Google Scholar]
- Kilmartin, J.V., and Goh, P.Y. (1996). Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 15, 4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and Schiebel, E. (1997). Spc98p and Spc97p of the yeast γ-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16, 6985–6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q., Hansen, D., Killilea, A., Joshi, H.C., Palazzo, R.E., and Balczon, R. (2001). Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J. Cell Sci. 114, 797–809. [DOI] [PubMed] [Google Scholar]
- Martin, O.C., Gunawardane, R.N., Iwamatsu, A., and Zheng, Y. (1998). Xgrip 109, a γ-tubulin-associated protein with an essential role in γ-tubulin ring complex (γTuRC) assembly and centrosome function. J. Cell Biol. 141, 675–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M., Sedat, J., Alberts, B., and Agard, D. (1995a). γ-Tubulin-containing rings in the centrosome. Nature 378, 638–640. [DOI] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M.B., Fung, J.C., Sedat, J.W., Alberts, B.M., and Agard, D.A. (1995b). 3D structural characterization of centrosomes from early Drosophila embryos. J. Cell Biol. 130, 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Zheng, Y., Alberts, B., and Oegema, K. (1998). Recruitment of the γ-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J. Cell Biol. 142, 775–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S., Urbani, L., and Stearns, T. (1998). The mammalian gammatubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J. Cell Biol. 141, 663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, S.M., Preble, A.M., Patel, U.K., O'Connell, K.L., Dias, D.P., Moritz, M., Agard, D., Stults, J.T., and Stearns, T. (2001). GCP5 and GCP 6, two new members of the human gamma-tubulin complex. Mol. Biol. Cell 12, 3340–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa, Y., Yamane, Y., Okanoue, T., and Tsukita, S. (2001). Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell 12, 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T., Vinh, D.B., Crawford, D.K., and Davis, T.N. (1998). A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol. Biol. Cell 9, 2201–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Wiese, C., Martin, O.C., Milligan, R.A., Iwamatsu, A., Mitchison, T.J., and Zheng, Y. (1999). Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg, B., Khodjakov, A., Rieder, C., and Palazzo, R. (1998). The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl. Acad. Sci. USA 95, 9295–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg, B.J., Hull, D.R., Balczon, R.D., and Palazzo, R.E. (2000). Reconstitution of microtubule nucleation potential in centrosomes isolated from Spisula solidissima oocytes. J. Cell Sci. 113, 943–953. [DOI] [PubMed] [Google Scholar]
- Schnackenberg, B.J., and Palazzo, R.E. (1999). Identification and function of the centrosome centromatrix. Biol. Cell 91, 429–438. [PubMed] [Google Scholar]
- Stearns, T., and Kirschner, M. (1994). In vitro reconstitution of centrosome assembly and function: the role of γ-tubulin. Cell 76, 623–637. [DOI] [PubMed] [Google Scholar]
- Stirling, D.A., Rayner, T.F., Prescott, A.R., and Stark, M.J. (1996). Mutations which block the binding of calmodulin to Spc110p cause multiple mitotic defects. J. Cell Sci. 109, 1297–1310. [DOI] [PubMed] [Google Scholar]
- Sundberg, H.A., Goetsch, L., Byers, B., and Davis, T.N. (1996). Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J. Cell Biol. 133, 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, M., Shibata, H., Shimakawa, M., Miyamoto, M., Mukai, H., and Ono, Y. (1999). Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosomes and the Golgi apparatus. J. Biol. Chem. 274, 17267–17274. [DOI] [PubMed] [Google Scholar]
- Takahashi, M., Yamagiwa, A., Nishimura, T., Mukai, H., and Ono, Y. (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol. Biol. Cell 13, 3235–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin, A., Celati, C., Moudjou, M., and Bornens, M. (1998). Characterization of the human homologue of the yeast spc98p and its association with gammatubulin. J. Cell Biol. 141, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf, W. (1994). Immunofluorescence analysis of the cytoskeleton during oogenesis and early embryogenesis. Methods Cell Biol. 44, 489–505. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Keating, T.J., Wilde, A., Borisy, G.G., and Zheng, Y. (2000). The role of Xgrip210 in gamma tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151, 1525–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Wong, M., Alberts, B., and Mitchison, T. (1995). Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583. [DOI] [PubMed] [Google Scholar]