Abstract
In most eukaryotes, genes encoding ribosomal RNAs (rDNA) are clustered in long tandem head-to-tail repeats. Studies of Saccharomyces cerevisiae have indicated that rDNA copy number is maintained through recombination events associated with site-specific blockage of replication forks (RFs). Here, we describe two Schizosaccharomyces pombe proteins, homologs of S. cerevisiae Slx1 and Slx4, as subunits of a novel type of endonuclease that maintains rDNA copy number. The Slx1-Slx4–dependent endonuclease introduces single-strand cuts in duplex DNA on the 3′ side of junctions with single-strand DNA. Deletion of Slx1 or Rqh1 RecQ-like DNA helicase provokes rDNA contraction, whereas simultaneous elimination of Slx1-Slx4 endonuclease and Rqh1 is lethal. Slx1 associates with chromatin at two foci characteristic of the two rDNA repeat loci in S. pombe. We propose a model in which the Slx1–Slx4 complex is involved in the control of the expansion and contraction of the rDNA loci by initiating recombination events at stalled RFs.
INTRODUCTION
Accurate duplication of a eukaryotic genome depends on highly proficient DNA replication machinery acting in conjunction with DNA repair and checkpoint signaling pathways (Boddy and Russell, 2001; Osborn et al., 2002). Repair and checkpoint systems are vital because replisomes stall when they encounter DNA damage, protein complexes, or torsional stress. Arrested replication forks (RFs) are prone to rearrangement or collapse. Studies of bacteria have shown that stalled forks can be rescued through either recombinogenic or nonrecombinogenic pathways (Seigneur et al., 1998; Cox et al., 2000; Seigneur et al., 2000; Cox, 2001; McGlynn and Lloyd, 2002).
Recent studies have indicated that eukaryotes rescue stalled forks by mechanisms similar to those proposed to act in prokaryotes (Doe et al., 2000; Boddy et al., 2001; Cox, 2001). A conserved family of eukaryotic DNA helicases related to Escherichia coli RecQ helicase seems to play a central role in the rescue of stalled forks through a nonrecombinogenic mechanism (Doe et al., 2000; Boddy et al., 2001; Cox, 2001; Hickson et al., 2001). The crucial role of these RecQ-related helicases in maintenance of genome integrity is underscored by the pleiotropic phenotypes of the human Bloom, Werner, and Rothmund-Thomson syndromes associated with defects in BLM, WRN, and RecQ4 proteins, respectively. These hereditary disorders are characterized by high genomic instability, cancer predisposition and, in the case of Werner syndrome, also premature aging (Shen and Loeb, 2000). In budding yeast, Saccharomyces cerevisiae and fission yeast Schizosaccharomyces pombe, mutations in the genes encoding the RecQ-related helicases Sgs1 and Rqh1, respectively, are associated with hyper-recombination phenotypes. The sgs1 phenotype is characterized by an increase in intra- and interchromosomal recombination, especially at the tandem repeated ribosomal DNA (rDNA) loci (Watt et al., 1995; Sinclair and Guarente, 1997), whereas rqh1 mutants are unable to segregate their chromosomes properly under conditions that stall replication (Stewart et al., 1997). Interestingly, Sgs1 and Rqh1 are essential for viability in the absence of the Mus81–Mms4 and Mus81–Eme1 complexes, respectively (Boddy et al., 2000; Mullen et al., 2001). Mus81 complexes are structure-specific endonucleases that process recombination intermediates formed during meiotic recombination or that arise from fork arrest or collapse (Boddy et al., 2001; Kaliraman et al., 2001). Although much uncertainty remains about the processing of stalled forks, it is generally thought that Sgs1 and Rqh1 act at the interface between DNA replication and recombination where they seem to prevent initiation of recombination by structure-specific nucleases.
A recent genetic screen carried out in S. cerevisiae identified six proteins (Mus81, Mms4, Slx1, Slx4, Slx5, and Slx8) that are essential in sgs1 cells (Mullen et al., 2001). Slx5 and Slx8 associate in vivo and are required for full resistance to the replication inhibitor hydroxyurea (HU) and efficient sporulation. Slx1 and Slx4 also interacted in vivo, but surprisingly, slx1 and slx4 mutants displayed no obvious phenotypes and were not abnormally sensitive to genotoxic agents. Slx1 shares sequence similarity with the UvrC-Intron-Type (URI) endonuclease family (Aravind and Koonin, 2001), suggesting a role in DNA cleavage. Importantly, Slx4 and Sgs1 were recently shown to have overlapping roles in maintenance of rDNA structure during replication (Kaliraman and Brill, 2002). Copy number of rDNA repeats seems to be maintained by regulated recombination controlling both contraction and expansion of the rDNA repeat locus (Kobayashi et al., 1998). An important protein controlling rDNA repeat contraction and expansion is Fob1, which is required for polar RF-blocking (RFB) activity at the RFB site in the rDNA repeat in S. cerevisiae. The RFB site is located near the rDNA transcription termination site, and one of its functions may be to prevent collisions between transcription and replication machineries. However, its most important function seems to be to control site-specific recombination. It has been proposed that RFs blocked at RFB are subject to endonuclease cleavage that triggers recombination (Kobayashi et al., 1998). The identity of the proposed endonuclease is unknown.
To gain additional insight into Slx1–Slx4 complex and its role in genome maintenance, and to assess evolutionary conservation of this complex, we undertook the identification and functional characterization of Slx1 and Slx4 homologs in S. pombe. In vitro nuclease assays have allowed us to precisely analyze the enzymatic activity associated with the endogenous Slx1–Slx4 complex isolated from fission yeast. In conjunction with recent biochemical studies of a recombinant form of budding yeast Slx1–Slx4 complex expressed and purified from E. coli (Fricke and Brill, 2003), these cell and molecular biological experiments define Slx1-Slx4 as an evolutionary conserved structure-specific endonuclease that plays an important role in the maintenance of rDNA.
MATERIALS AND METHODS
General Techniques
S. pombe methods and media have been described previously (Moreno et al., 1991).
Strains and Plasmids
Strains used in this study are ura4-D18 and leu1-32 unless otherwise stated: PR109, wild-type; SC3250, rqh1::ura4; SC3240, slx1::kanMx6; SC3241, slx1-TAP::kanMx6; SC3242 slx1-13myc::kanMx6; SC3243, slx4::kanMx6; SC3244, slx4-13myc::kanMx6; SC3245, slx1::kanMx6 slx4::kanMx6; SC3251, slx1-TAP::kanMx6 slx4::kanMx6; SC3246, slx1-TAP::kanMx6 slx4-13myc::kanMx6; SC3281, slx4-TAP::kanMx6; SC3282, slx4-TAP::kanMx6 slx1-13myc::kanMx6; SC3247, slx1::kanMx6 slx1wt-TAP:LEU1; SC3248 slx1::kanMx6 slx1R34A,E74A-TAP:LEU1; SC3249, slx1::kanMx6 slx1R34A-TAP:LEU1; SC3284, rqh1::ura4 slx1-13myc::kanMx6.
Identification of Slx4
Identification of Slx4 by MudPIT was performed as described previously (Link et al., 1999; Boddy et al., 2001; Washburn et al., 2001). Three peptides (VAEDNDVLLSR, IVESCLDAIDSR, and TVLEFDDIVTQTHR) covering 8.8% of SPAC688.06c were identified (Figure 4A). BLAST analysis identified potentially significant sequence similarity of SPAC68806C to S. cerevisiae Slx4.
Figure 4.
Identification of S. pombe Slx4 homolog. (A) Sequence alignments of Slx4 homologs from S. cerevisiae (Sc), S. pombe (Sp; SPAC688.06c), and C. albicans (C.al; Orf6.4697). Dark gray and light gray boxes indicate identical and similar residues, respectively. The peptides identified by MudPIT analysis of Slx1-TAP are underlined. (B) Fivefold serial dilutions of mutant slx1, slx4, or slx1 slx4 cells were incubated on YES agar medium supplemented with indicated amounts of MMS or HU at 30°C. To determine UV survival, cells were ascertained using a hemocytometer, plated on YES agar medium, and exposed to the indicated doses of UV. The agar plates were incubated at 30°C, and percentage of survival was calculated as the number of colonies in irradiated versus nonirradiated control. Data represent the average of two independent trials. (C). Slx4 is essential in rqh1 cells. Left, tetrad analysis of mating between slx4::kanMX6 and rqh1::ura4+ strains. An example of germination products from a slx4 rqh1 spore is presented in right panel. S, slx4; R, rqh1; RS, slx4 rqh1.
In Situ Chromatin Binding Assay and Indirect Immunofluorescence Microscopy
In situ chromatin binding assays were carried out as described previously (Kearsey et al., 2000). PR109/pREP1-slx1-13myc cells were incubated with anti-myc 9E10 antibody and then with Alexa Fluor 488 anti-mouse (Molecular Probes, Eugene, OR) as described previously (Lopez-Girona et al., 1999). DNA was stained with 4,6-diamidino-2-phenylindole (1 μg/ml). Samples were analyzed using a Nikon Eclipse E800 microscope equipped with a Photometrics Quantix charge-coupled device camera. Images were acquired with IPlab Spectrum software (Signal Analytics, Vienna, VA).
Sequence Alignments
Sequences were aligned using PIMA 1.4, pattern-induced (local) multiple alignment matrix and BOXSHADE programs.
Structure-specific DNA Substrates
Stem loop oligonucleotides were SL (5′-GCCAGCGCTCGG[T22]CCGAGCGCTGGC), SL2 (5′-GCCAGCGCTCGGA[T22]CCGAGCGCTGGC), SL3 (5′-GCCAGCGCTCGGAC[T22]GTCCGAGCGCTGGC), SL4 (5′-GCCAGCGCTCGGACA[T22]GTCCGAGCGCTGGC), SL16T (5′-GCCAGCGCTCGG[T16]CCGAGCGCTGGC), and SL10T (5′-GCCAGCGCTCGG[T10]CCGAGCGCTGGC). Splayed arm Y structure oligonucleotides were y1 (5′-GCCAGCGCTCGG[T11]) and y2 (5′-[T11]CCGAGCGCTGGC).
The various SL substrates were made with the partially self-complementary oligonucleotides. Each oligonucleotide was 5′-32P labeled. The Y structure was obtained by annealing oligonucleotides y1 and y2.
X12 and X0 substrates were prepared as described previously (Boddy et al., 2001). Annealing was achieved by incubating oligonucleotides for 3 min at 95°C, followed by subsequent 10-min incubations at 65°C, 37°C, room temperature, and 0°C. Labeled substrates were purified after separation by electrophoresis in a nondenaturing 10% polyacrylamide gel and stored in a TE (pH 7.5), 50 mM NaCl buffer.
Nuclease and Resolution Assays
Preparation of TEV-Slx1 and TEV-Slx4 was done accordingly to previously published methods (Rigaut et al., 1999; Boddy et al., 2001). All in vitro nuclease assays were set up as published previously (Boddy et al., 2001).
Pulsed-Field gel Electrophoresis
For pulsed-field gel electrophoresis, samples were prepared as described previously (Nakamura et al., 2002). To observe whole chromosomes, a 1% agarose gel was subjected to electrophoresis with constant circulation of Tris-borate EDTA 0.5× buffer at 14°C for 48 h (2 V/cm, included angle 106°, initial and final switch time 30 min). The gel was stained with ethidium bromide, transferred to a nylon membrane, and hybridized to a rDNA probe or to a probe specific of chromosome 3. The rDNA probe was created with Prime-IT II labeling kit (Stratagene, La Jolla, CA) with purified PCR fragments of 25s gene amplified with the forward primer 5′TTGAACGCACATTGCGCCTTTGGGTTCTAC and the reverse primer 3′TGGTTCCTTAATGGAACAAATCTTGGGAAC as template.
RESULTS
Identification of S. pombe Slx1
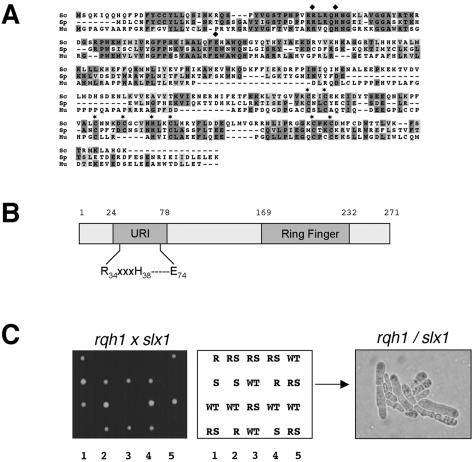
An S. pombe Slx1 homolog (SPAP27G11.15) was identified in a BLAST search of proteins encoded by the S. pombe genome (Figure 1A). S. pombe Slx1 shares 32% identity and 48% similarity to S. cerevisiae Slx1. Like its S. cerevisiae counterpart, the N-terminal half of S. pombe Slx1 has sequence similarity to the URI endonuclease domain (Figure 1B). The URI domain, characterized by the RXXX(YH)—E motif found in proteins from bacteria to humans, is required for the nuclease activity of the homing endonuclease I-TevI and the structure-specific DNA endonuclease UvrC that carries out nucleotide excision repair in E. coli (Derbyshire et al., 1997; Verhoeven et al., 2000; Aravind and Koonin, 2001). Slx1 also has a C-terminal Ring Finger domain (Cys4-H-Cys3 motif) predicted to bind two atoms of zinc. This type of Ring Finger is thought to mediate protein–protein or protein–DNA interactions (Putilina et al., 1999).
Figure 1.
Identification of S. pombe Slx1. (A) Sequence alignment of Slx1proteins from S. cerevisiae (Sc), S. pombe (Sp; SPAP27G11.15), and human (Hu; MGC5178). Dark gray and light gray boxes indicate identical and similar residues, respectively. Conserved residues of the URI and Ring Finger domains are annotated by black squares and black stars, respectively. (B) Schematic representation of S. pombe Slx1. Positions of the URI domain and Ring finger domain are shown. (C) Genetic interaction between slx1 (S) and rqh1 (R). A slx1::kanMX6 haploid strain was mated with a rqh1::ura4+ haploid strain, and the cells were induced to undergo meiosis and sporulation. Tetrads were dissected onto YES agar. Colonies resulting from five such tetrads were photographed after 3 days growth at 30°C (left). The genotypes of these segregants were determined by replica plating. Double mutant slx1 rqh1 (RS) spores were unable to form colonies. A photomicrograph of cells arising from a germinated slx1 rqh1 spore is shown in the right panel.
We found that a slx1 deletion mutant was viable, displayed no obvious growth defect, and was not abnormally sensitive to genotoxic agents such as HU, UV, MMS, camptothecin, or mitomycin (Figure 4B; our unpublished data). Moreover, we found that a slx1 deletion mutant did not display any modifications in telomere length and did not have any sporulation defects (our unpublished data). To evaluate genetic interactions between Slx1 and Rqh1, forty tetrads from a slx1::kanMX6 X rqh1::ura4 mating were dissected. Single mutant spores germinated and formed colonies (Figure 1C). In contrast, slx1 rqh1 spores germinated but stopped growth within a few divisions. A representative slx1 rqh1 microcolony is shown in Figure 1C, right. Typical slx1 rqh1 cells were elongated and swollen. These findings showed that Slx1 is essential in the absence of Rqh1, consistent with the relationship between Slx1 and Sgs1 in S. cerevisiae (Mullen et al., 2001).
Structure-specific DNA Endonuclease Activity Associated with Slx1
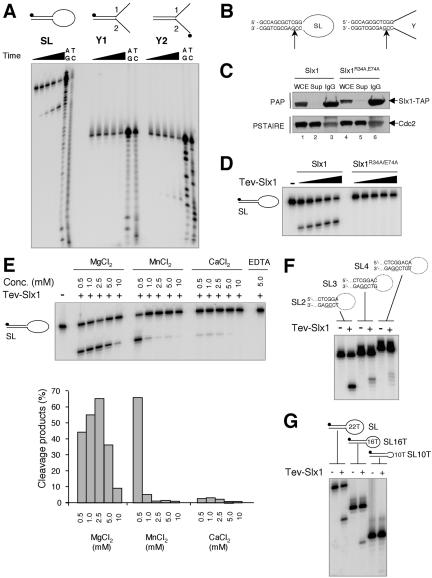
The presence of a URI nuclease motif suggested that Slx1 possesses DNA endonuclease activity. Accordingly, endonuclease activity of Slx1 was tested on various model DNA substrates (Figure 2A). Genomic slx1+ was modified to encode Slx1 with a C-terminal TAP tag. A slx1-TAP rqh1 strain was viable, indicating that Slx1-TAP retained function (our unpublished data). Immobilized Slx1, bound to IgG-Sepharose through the protein A portion of the TAP tag, was eluted by cleavage at the TEV protease site located in the tag (Rigaut et al., 1999). The partially purified eluate, resulting from TEV cleavage, hereafter referred to as TEV-Slx1, was incubated with various model DNA substrates under defined reaction conditions. A stem-loop structure (SL) consisting of a 22-nucleotide (nt) single-strand (ss) loop and a duplex stem of 12 base pairs was obtained with a partially self-complementary oligonucleotide (Sijbers et al., 1996). This type of structure allows detection of ss incisions made in duplex DNA on the 5′ or 3′ side of a junction with ss DNA. Single-strand specific nuclease activities would also be detected with such a structure. Some structure-specific nucleases such as FEN-1 require a free ss end to introduce an ss cut in duplex DNA at the junction with the ss flap (Harrington and Lieber, 1994). Therefore, we also designed a splayed arm Y structure consisting of the same 12 base pairs duplex stem in SL but with two 11 nt ss 3′ and 5′ flaps instead of a 22-nt loop. The Y1 substrate contained a 5′-labeled oligonucleotide 1, whereas Y2 contained a 5′-labeled oligonucleotide 2 for detection of incisions made on the 5′ or 3′ side of the junction, respectively. Linear double-strand (ds) and ss substrates were also used as nonstructured DNA controls.
Figure 2.
Structure-specific DNA endonuclease activity associated with Slx1. (A) Three different substrates were incubated for 30 min at 30°C with 3 μl of TEV-Slx1 for 0 s, 30 s, 2.5 min, 10 min, and 50 min. A+G and C+T sequencing ladders were derived from the SL, y1, and y2 oligonucleotides. Reaction products were analyzed by denaturing PAGE. (B) The sites of cleavage were determined from the Maxam-Gilbert sequencing ladders and are indicated by arrows on each DNA structure. (C) Whole cell extract (WCE) derived from strains slx1::kanMx6 slx1wt-TAP:LEU1 and slx1::kanMx6 slx1R34A,E74A-TAP:LEU1 were incubated with IgG-Sepharose beads. TAP-Slx1 and TAP-Slx1R34A,E74A were detected in the WCE, supernatant (Sup), and the pulled down IgG-Sepharose beads (IgG) by Western blotting by using peroxidase-anti-peroxidase antibody (PAP; Sigma-Aldrich, St. Louis, MO). Cdc2 detected with an antibody directed against the Cdc2 PSTAIRE motif was used as a loading control. (D) SL was incubated with 1, 2, 4, 6, and 9 μl of TEV-eluate obtained from slx1/slx1wt-TAP::LEU1 and slx1/slx1R34A,E74A-TAP::LEU1 strains for 30 min at 30°C. Reaction products were analyzed by denaturing PAGE. (E) SL was incubated with 3 μl of TEV-Slx1 in the presence of various amounts of Mg2+, Mn2+, Ca2+, or EDTA and reaction products analyzed by denaturing PAGE. The relative amount of cleavage products was quantified by phosphorimager analysis. (F) The various SL substrates indicated above the gel were incubated with 3 μl of TEV-Slx1 for 30 min at 30°C. (G) Three microliters of TEV-Slx1 was incubated with SL containing ss loops of various sizes as indicated.
Analysis of reaction products by denaturing PAGE showed that defined cleavage products were detected with the SL and Y2 substrates but not with the Y1 substrate (Figure 2A) or the ds and ss control substrates (our unpublished data). These results suggested that the Slx1-dependent nuclease was acting on the 3′ side of the ds/ss junction with respect to the ss moving 3′ to 5′ away from the junction. This activity was consistent with the analysis of UvrC in E. coli (Verhoeven et al., 2000), in which the N-terminal URI domain of UvrC is required for the 3′ incision of nucleotide excision repair, whereas the 5′ incision depends on a C-terminal domain. The position of the cleavage products was compared with Maxam-Gilbert sequencing reactions obtained with oligonucleotides SL and Y2 (Figure 2A). The main cleavage site was mapped in the duplex stem 2 nt to the 3′ side of the junction with respect to the ss DNA moving 3′ to 5′ away from the junction.
To confirm that the nuclease activity was Slx1 dependent and to assess the requirement of an intact URI domain, we engineered point mutations at the conserved arginine 34 (R34) and glutamic acid 74 (E74) residues within the URI domain. The equivalent arginine residue in the URI domain of UvrC is essential for its 3′ nuclease activity (Verhoeven et al., 2000). We mutated the conserved glutamic acid residue because negatively charged residues often play an important role in metal ion coordination and as nucleophiles in catalysis. The wild-type (WT) and the mutated versions of slx1 were integrated at the leu1-32 locus in a strain deleted for endogenous slx1. Expression levels of TAP-tagged Slx1 and Slx1R34A,E74A were comparable (Figure 2C). TAP-Slx1R34A,E74A was pulled down by IgG-Sepharose beads as well as TAP-Slx1 (Figure 2C). TEV-eluates were tested for activity on the SL substrate. Whereas TEV-Slx1 was active, no activity was detected with TEV-Slx1R34A,E74A (Figure 2D). TEV-Slx1 obtained from a strain that expressed slx1-R34A was also inactive (our unpublished data). These results confirmed that the nuclease activity was Slx1 dependent and that it required an intact URI domain.
To establish whether the synthetic lethal phenotype of the slx1 rqh1 double-mutant is due to the loss of Slx1-Slx4 endonuclease activity, we crossed slx1 slx1-R34A and slx1 slx1-R34A,E74A strains with the rqh1 mutant. In three independent experiments, we were unable to obtain viable rqh1 slx1 slx1-R34A or rqh1 slx1 slx1-R34A,E74A clones (our unpublished data). From these results, we concluded that the loss of endonuclease activity is responsible for the synthetic lethal phenotype of the slx1 rqh1 double mutant.
Cation titration experiments performed with Mg2+, Mn2+, or Ca2+ showed that Mg2+ was the favored divalent cation with efficient cutting over a broader concentration range and optimum cutting at 2.5 mM Mg2+ (Figure 2E). The importance of metal chelation for the nuclease activity was confirmed by the total inhibition of the reaction by EDTA (Figure 2E).
SL substrates in which one, two, or three extra base pairs were inserted between the cleaved sequence and the duplex/ss junction were designed to assess the structural and sequence requirements of the enzyme. Reduced and less site-specific nuclease activity was detected when the GC phosphodiester bond was moved further than three nucleotides away from the junction (Figure 2F). Interestingly, the main cleavage site was shifted away from the junction along with the GC sequence. These results indicate that the enzyme has some sequence preference combined with a structure dependence. When we compared the processing of SL substrates with 16 and 10 poly-T loops to that of the original 22 poly-T loop substrate (Figure 2G), reduced cutting was detected with the 16 poly-T loop and no cutting was detected with the 10 poly-T loop substrate, further confirming that specific structural features are required for the enzyme to act.
Slx1-dependent Nuclease Cuts but Does Not Resolve Holliday Junctions
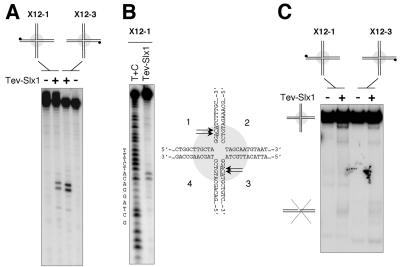
DNA helicases and Holliday Junctions (HJ) resolvases are thought to participate in alternative pathways of recovery from regressed RFs. The genetic interaction between Slx1 and Rqh1 suggested that Slx1 is required for a recombination pathway that is essential in the absence of Rqh1. We therefore investigated whether the Slx1-dependent nuclease was able to resolve HJs. We used the X12 structure obtained by annealing four complementary oligonucleotides. X12 contains a 12-base pairs homology core in its center that allows the point of crossover to branch migrate spontaneously (Parsons et al., 1990). We used two X12 substrates, X12-1 and X12-3, labeled on opposite strands 1 and 3, respectively. Slx1-complex cleaved X12-1 and X12-3, generating identical products (Figure 3A). Side-by-side comparisons indicated that these substrates were cut about eightfold less efficiently than the SL substrate (our unpublished data). The cleavage sites were mapped by comparing the position of the cleavage products to Maxam-Gilbert sequencing ladders (Figure 3B; our unpublished data).
Figure 3.
The Slx1-dependent nuclease cuts but does not resolve HJs. (A) X12-1 and X12-3 substrates were incubated with 3 μl of TEV-Slx1 for 30 min at 30°C. Reaction products were analyzed by denaturing PAGE. (B) The position of the cuts were determined from the Maxam-Gilbert sequencing ladder. (C) Reaction products from reactions carried out in A were analyzed by native PAGE to monitor the conversion of the four-way branched structures into linear duplex products.
To monitor the ability of the Slx1 complex to resolve the migratable X12 HJ into defined linear duplex products, part of the reactions carried out with X12-1 and X12-3 substrates in Figure 3A were analyzed by native PAGE. Despite the introduction of symmetric cuts on opposed strands across the junction, no significant duplex products were detected indicating that the Slx1-complex is not a HJ resolvase (Figure 3C).
Identification of S. pombe Slx4
Having defined a nuclease activity associated with Slx1, we turned our attention to the identification of an Slx4 homolog in S. pombe. Despite the substantial sequence similarity between Slx1 homologs in S. pombe and S. cerevisiae, BLAST searches failed to identify an obvious Slx4 homolog in S. pombe. We therefore used multidimensional protein identification technology (MudPIT) to identify proteins that coprecipitated with Slx1-TAP. MudPIT uses multidimensional liquid chromatography, tandem mass spectrometry, and sophisticated database searching to identify components of protein complexes. One of the proteins identified by Mud-PIT analysis of the Slx1-TAP sample was encoded by the SPAC688.06c open reading frame (peptides listed in MATERIALS AND METHODS). Integration of BLAST searches and MudPIT data suggested that SPAC688.06c might encode a Slx4 homolog. We subsequently named this gene slx4. Slx4 proteins from S. cerevisiae and S. pombe have weak sequence similarity but both share significant homology (20 and 25% identity, respectively) with Candida albicans Orf6.4697 protein (Figure 4A). Interestingly, S. cerevisiae Slx4 (748 residues) has a large N-terminal region absent in S. pombe Slx4 (419 residues) and C. albicans Orf6.4697 (351 residues).
Deletion of S. pombe slx4+ revealed that it is a nonessential gene and its loss caused no obvious phenotype. No genetic interaction was observed in a slx1 slx4 double mutant. Like the slx1 mutant, the slx4 and slx1 slx4 mutants were not abnormally sensitive to HU, MMS, and UV (Figure 4B). Surprisingly, slx1, slx4, and slx1 slx4 mutants seem slightly less sensitive to UV than a WT strain. Future studies will determine whether this is significant. Tetrad analysis showed that Slx4 was essential in the absence of Rqh1 (Figure 4C). The phenotypes of the slx4 rqh1 double mutant were indistinguishable from the slx1 rqh1 double mutant (Figure 4C). Microscopic observations indicated that like slx1 rqh1, slx4 rqh1 mutants always died between the first and the fifth cell generation.
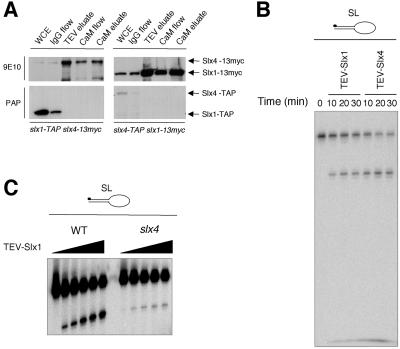
Coprecipitation studies were performed to confirm that Slx1 and Slx4 associate in vivo. Genomic slx1+ and slx4+ were modified to encode Slx1 and Slx4 with a C-terminal 13-myc epitope tag. The slx1-13myc rqh1 strain and slx4-13myc rqh1 strain were viable, indicating that 13myc-tagged Slx1 and Slx4 retained function (our unpublished data). Slx1-TAP and Slx4-TAP were partially purified from slx1-TAP slx4-13myc and slx4-TAP slx1-13myc strains, respectively. Copurification of Slx4-13myc and Slx1-13myc was confirmed by immunoblotting (Figure 5A). Together, these results strongly suggested that Slx1 and Slx4 physically interact in vivo and form a stable complex that is essential in absence of Rqh1.
Figure 5.
Slx4 associates with Slx1 to form an active endonuclease. (A) Slx1-TAP and Slx4-TAP fusion proteins were expressed in slx4-myc and slx1-myc strains, respectively. WCE, whole cell extract; IgG flow, flow through from WCE passed over IgG affinity column; TEV eluate, TEV cleavage products from IgG column; CaM flow, flow through from TEV eluate passed over calmodulin affinity column; CaM eluate, eluate from the calmodulin column. These fractions were subjected to immunoblotting revealed by 9E10 anti-myc monoclonal antibody and peroxidase-anti-peroxidase antibody (PAP; Sigma-Aldrich). Note: The loss of the TAP signal in the TEV-eluates and the following fractions is due to the loss of the protein A portion of the TAP-tag after TEV cleavage. (B) The SL substrate was incubated at 30°C for the indicated time with 3 μl of TEV-Slx1 or TEV-Slx4 obtained from slx1-TAP and slx4-TAP strains, respectively. Reaction products were analyzed by denaturing PAGE. (C) The SL substrate was incubated with 1, 2, 4, 6, and 9 μl of TEV-Slx1 obtained from slx1-TAP and slx1-TAP slx4 strains, for 30 min at 30°C, and reaction products were analyzed by denaturing PAGE.
Structure-specific DNA Endonuclease Activity Associated with Slx1–Slx4 complex
These studies suggested that Slx1-Slx4 might function as a heterodimeric DNA nuclease. This possibility was evaluated by assaying nuclease activity of a TEV-eluate obtained from the slx4-TAP strain on a SL substrate (Figure 5B). A nuclease activity identical to that observed with TEV-Slx1 was detected with the TEV-Slx4 eluate. To further understand the functional significance of the association between Slx1 and Slx4 and to determine whether Slx4 is required for catalysis, we undertook nuclease assays with TEV-Slx1 obtained from an slx1-TAP slx4 strain. As shown in Figure 5C, weak nuclease activity was detected with TEV-Slx1 obtained from the slx4 mutant. This activity was greater than that of TEV-Slx1R34A,E74A (Figure 2D) and generated identical cleavage products compared with those made by TEV-Slx1 obtained from a slx4+ strain. These findings indicated that Slx4 might not be required for the nuclease reaction per se but instead plays an important role in stimulating the action of the Slx1-dependent nuclease. The potential implications of these findings are discussed below.
Slx1 Binds to Chromatin In Vivo
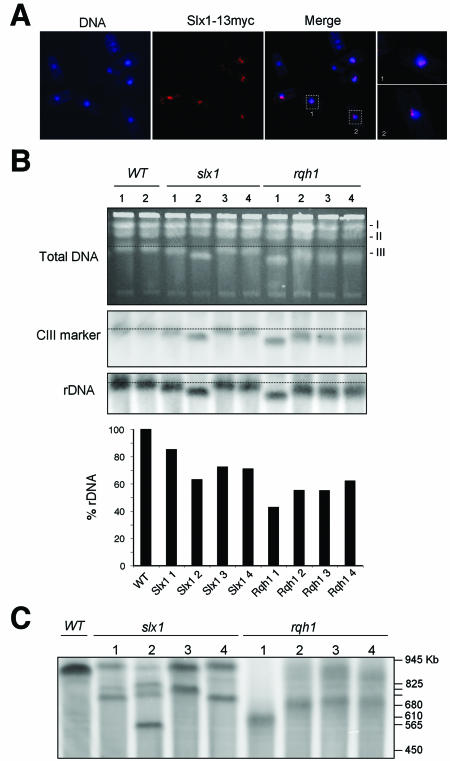
If Slx1-Slx4 is specifically involved in rDNA maintenance, the protein subunits might be expected to specifically associate with the rDNA repeats in vivo. We were unable to localize Slx1-13myc and Slx1-GFP expressed at endogenous levels, an observation consistent with immunoblotting studies that suggested that Slx1 is a very low abundance protein. We therefore placed a slx1-13myc construct under the control of the thiamine repressible nmt1 promoter. A pREP1 plasmid containing this construct was transformed into the PR109 strain. Full induction of the nmt1promoter occurs 16–20 h after thiamine depletion at 25°C. After 10 h, we detected an anti-myc signal that was strongly localized to the nucleus (our unpublished results). In situ chromatin binding assays (Kearsey et al., 2000) were performed to evaluate whether Slx1–13myc bound to chromatin could be detected after Triton extraction. The fluorescence signal of Slx1-13myc was reduced to two, or in some cases, one spot near the nucleus (Figure 6A). (Some cells did not stain at all because of plasmid loss). Merging pictures with 4,6-diamidino-2-phenylindole staining demonstrated that these spots were localized in most cases at the periphery of the nucleus, probably in the nucleolus (Figure 6A, bottom right). DNaseI treatment of these samples eliminated all Slx1-13myc signal (our unpublished data), confirming that the Slx1-13myc detected after Triton extraction was bound to chromatin. Thus, it seems that Slx1 is recruited to chromatin at one or two specific sites on chromosomes. The single or double-spot appearance and their specific localization at the periphery of the nucleus recalled the rDNA protrusions (Uzawa and Yanagida, 1992). The S. pombe rDNA consists of 100–150 10.9-kb tandem head-to-tail repeats in two clusters located at each end of chromosome III. Each repeat contains a nontranscribed spacer that includes regulatory sequences and promoter of genes encoding for 18S, 5.8S, and 25S rRNA molecules. These repeat regions protrude into the nucleolus (Uzawa and Yanagida, 1992).
Figure 6.
Slx1 complex binds to chromatin and is involved in rDNA maintenance. (A) In situ chromatin binding assays performed with a strain overexpressing Slx1-13myc. Slx1-13myc was detected as single or double spots at the periphery of the nucleus. (B) Genomic DNA samples of WT and strains that were freshly deleted for the slx1 or rqh1 genes were subjected to PFGE. The gel was stained with ethidium bromide before Southern blotting. The membrane was hybridized with an rDNA probe and with a probe specific for a region located outside of the rDNA loci on CIII. The signal quantification allowed for determination of the rDNA percentage in each sample taking wild-type templates as references. (C) Genomic DNA samples described in B were digested with SfiI before PFGE and hybridization to an rDNA probe. The SfiI fragments containing the rDNA loci were smaller in the slx1 strains and diffuse in rqh1 strains. The size of S. cerevisiae marker chromosomes is indicated.
Slx1 and Rqh1 Maintain rDNA
These observations suggest that Slx1-Slx4 endonuclease may function in maintenance of rDNA repeats. This possibility is consistent with studies in S. cerevisiae indicating that simultaneous loss of Sgs1 and Slx4 leads to the formation of aberrant rDNA structures during DNA replication (Kaliraman and Brill, 2002). We used pulsed-field gel electrophoresis (PFGE) to examine the chromosome structure in parental strains and eight independent strains that were freshly deleted for slx1 or rqh1. DNA plugs were prepared from single colonies (Figure 6B). Chromosomes I and II seemed normal in all strains, whereas chromosome III (CIII) was consistently shorter in the slx1 and rqh1 strains. There was considerable variation in the actual length of CIII in the mutant strains, particularly in the rqh1 strains. To evaluate whether these effects were associated with loss of rDNA repeats, the PFGE gel was analyzed with a probe specific of the rDNA (Figure 6B). Each rDNA signal was normalized to that of a unique sequence located outside of the rDNA loci on CIII (internal marker control) and standardized to the WT control. The quantification indicated a substantial decrease in the rDNA signal in all rqh1 and slx1 strains. This decrease was particularly accentuated in slx1#2 and rqh1#1 strains in which the length of CIII was most severely reduced.
These data indicated that the reduction of CIII length in rqh1 and slx1 strains was caused, at least in part, by loss of rDNA repeats. This possibility was investigated in more detail by digesting chromosomal DNA with SfiI restriction enzyme before PFGE. SfiI restriction enzyme has been chosen because of its ability to introduce three cuts in CIII outside of rDNA loci. After SfiI digestion, we monitored by Southern blotting the size of the chromosomal rDNA fragments (Fan et al., 1991; Mizukami et al., 1993) (Figure 6C). The rDNA fragments generated by SfiI digestion of slx1 genomic DNA were smaller than those obtained with wild-type genomic DNA, particularly for the slx1#2 strain. Curiously, multiple rDNA bands were observed in this strain. A likely explanation is that a subpopulation, which displayed structurally divergent rDNA loci occurred during cell growth. The presence of this subpopulation could lead to the appearance of extra rDNA bands. However, we cannot exclude that unexpected rDNA rearrangements in slx1#2 strain could have been at the origin of its specific phenotype. The rDNA bands in rqh1 samples were diffuse (heterogeneous size of rDNA fragments could generate this pattern) and smaller than those observed in WT, indicating that loss of Rqh1 leads to a high rate of rearrangements and loss in the rDNA repeats. Importantly, the size reduction of rDNA fragments monitored by SfiI digestion of genomic DNA from slx1 and rqh1 strains correlated with the reduction of the length of CIII and with the reduction in the intensity of the rDNA repeats signal (Figure 6B). Together, these observations support the evidence that deletion of rqh1 or slx1 leads to loss of rDNA repeats and affects rDNA organization.
DISCUSSION
We have identified and characterized Slx1 and Slx4 proteins in S. pombe. These proteins form a complex that is essential in the absence of Rqh1 DNA helicase. Loss of either Slx1-Slx4 or Rqh1 leads to contraction of the rDNA repeats. Chromatin bound Slx1 localizes to two foci that are reminiscent of rDNA protrusions into the nucleolus. Importantly, we have shown that partially purified Slx1–Slx4 complex is a structure-specific endonuclease that introduces an ss cut in duplex DNA on the 3′ side of a ds/ss junction with respect to ss moving 3′ to 5′ away from the junction. Together with evidence that Slx1–Slx4 complex is not required for survival of DNA damage, these findings strongly suggest that Slx1– Slx4 complex is dedicated to a specific DNA cleavage event that is involved in rDNA maintenance. The functional conservation of S. pombe and S. cerevisiae Slx1–Slx4 complexes, and the presence of Slx1 homologs in sequenced eukaryotic genomes, suggests that Slx1-Slx4 function is likely to be conserved in humans and other complex eukaryotes.
Slx1–Slx4 Complex: A New Structure-specific Endonuclease
Whereas S. pombe Slx1 was easily identified by its sequence similarity to S. cerevisiae Slx1, the identification of Slx4 required a more elaborate approach by using MudPIT analysis. S. pombe Slx4 curiously lacks a large N-terminal region present in S. cerevisiae Slx4. It will be interesting to determine whether this N-terminal region is needed for the function of S. cerevisiae Slx4 function and if so, whether another protein provides this function in S. pombe.
Despite the striking structural differences between the S. cerevisiae and S. pombe Slx4, we found that the overt phenotypes of slx1 and slx4 mutants in S. pombe, as well as their genetic and physical interactions, were very similar to their S. cerevisiae counterparts (Mullen et al., 2001). Most notably, Slx1 and Slx4 associated in vivo and were essential in the absence of Rqh1, matching the relationships between Slx1-Slx4 and Sgs1 in S. cerevisiae. These observations attest not only to conservation of Slx1-Slx4 but also to the functional equivalency of Rqh1 and Sgs1.
In vitro nuclease assays carried out with partially purified Slx1 and Slx4 complexes from S. pombe showed that Slx1 and Slx4 are components of a new type of structure-specific endonuclease that introduces ss cuts in duplex DNA close to junctions with ss DNA. Cuts are introduced with a defined polarity only on the 3′ side of the junction with respect to the ss DNA moving 3′ to 5′ away from the junction. Specific point mutations in the URI endonuclease domain abolish Slx1-dependent nuclease activity. The URI domain of bacterial UvrC is essential for its 3′ nuclease activity, but a distinct coiled-coil domain involved in binding to its UvrB partner is also required for the 3′ incision (Orren and Sancar, 1989; Verhoeven et al., 2000). UvrB is thought to stabilize the UvrC-DNA contacts. A similar situation may apply to the Slx1–Slx4 complex. We found that Slx4 was not essential for Slx1 nuclease activity per se, but it was essential for efficient processing of DNA substrates. It is tempting to speculate that Slx4 plays a similar role to UvrB by targeting and stabilizing the interaction of Slx1 with its substrate. These questions may be best answered with recombinant Slx1-Slx4.
Although the polarity of action of the Slx1–Slx4 complex was identical to that of eukaryotic FEN1 and XPG nucleases, both involved in general repair pathways, its DNA sequence-preference contrasted with the capacity of these nucleases as well as that of UvrC to act on specific DNA structures irrespective of sequence. In addition, unlike rqh1 mutants, slx1 and slx4 mutants were not abnormally sensitive to DNA damaging agents. This suggests a more specialized role of the Slx1–Slx4 complex, perhaps limited to a specific region of the genome.
The exact nature of the in vivo substrate of Slx1-Slx4 remains to be determined. We found that the Slx1–Slx4 complex processed various types of DNA structures ranging from SL to HJs. However, processing of HJs did not produce linear duplex products, thus Slx1-Slx4 is not a HJ resolvase. The favored substrates were SL and splayed arm Y structures that contain well defined ds/ss junctions. It is probable that the in vivo substrate contains a similar type of junction.
The Slx1–Slx4 Endonuclease Complex Is Important for Maintenance of rDNA
Slx1 is most tightly bound to chromatin in two limited regions of the genome localized at the periphery of the nucleus, reminiscent of the rDNA protrusions in the nucleolus (Uzawa and Yanagida, 1992). Rqh1 was recently shown to also display a predominantly nucleolar distribution (Caspari et al., 2002). These observations suggest that the function of Slx1–Slx4 complex may be related to a specific role of Rqh1 in the maintenance of the rDNA, independent to Rqh1's general role in the rescue of stalled RFs.
In S. cerevisiae, a central component of rDNA maintenance is the Fob1 protein that plays an important role in the control of the expansion and contraction of the rDNA copy number (Kobayashi et al., 1992, 1998; Kobayashi and Horiuchi, 1996; Defossez et al., 1999). Fob1 binds at the polar Replication Fork Barrier (RFB) where it blocks RFs from moving into an adjacent rDNA repeat in a direction opposite to that of rDNA transcription. Current models propose that RFs stalled by Fob1 at the RFB are collapsed under the action of a yet undefined enzymatic activity (Kobayashi et al., 1998). The resulting ds end would be processed into a 3′-ss end by 5′ to 3′ resection and an active RF reconstructed after the free 3′ end reinvades the intact chromatid. Downstream or upstream strand invasion of the sister chromatid leads to contraction or expansion, respectively, of the rDNA repeats (Kobayashi et al., 1998; Defossez et al., 1999; Johzuka and Horiuchi, 2002), whereas invasion within the same chromatid results in formation of extrachromosomal DNA circles. Interestingly, the absence of Sgs1 results in an accumulation of extrachromosomal DNA circles in a Fob1-independent manner, suggesting that Sgs1 is also required for the nonrecombinogenic rescue of RFs that have stalled within the rDNA loci but not at the RFB (Sinclair and Guarente, 1997; Park et al., 1999). In the absence of Sgs1, the stalled forks could be rescued through a recombinogenic mechanism that may proceed in a similar way to that described above. Recently, Versini et al. (2003) showed that DNA replication is specifically retarded at the rDNA locus in sgs1 cells and suggested that this could be due to their inability to prevent recombination at stalled forks, abundant in that region. It remains to be seen whether these proposed mechanisms of rDNA maintenance can be extrapolated to S. pombe and other eukaryotes. In particular, Fob1 seems to be a sequence orphan that is unique to S. cerevisiae. However, similarities between the two yeast models have been established. In particular, S. pombe has a pausing site at the 3′ end of the 35S gene acting like the RFB (Sanchez et al., 1998). This pausing site enforces unidirectional replication of rDNA in S. pombe.
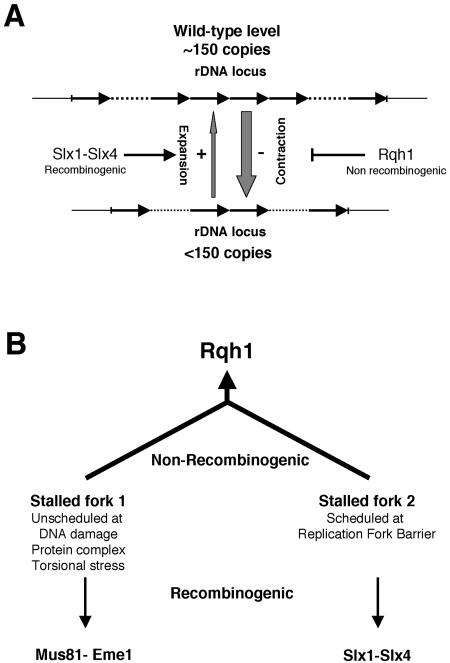
Our studies showed that loss of Slx1 or Rqh1 leads to contraction of the rDNA loci. These observations provide the first direct evidence that Rqh1 is involved in the maintenance of rDNA, like its S. cerevisiae counterpart Sgs1, and suggest that similar mechanisms may be involved in the maintenance of rDNA in the two yeasts. They also provide the first evidence on the identity of a structure-specific endonuclease that seems to play an active role in the control of the expansion of the rDNA repeats. Based on these results we propose that the Slx1–Slx4 complex may be involved in a recombination pathway that maintains or expands the rDNA repeats, whereas Rqh1 would be required in a parallel pathway to prevent the loss of rDNA copies (Figure 7A). Whether the Slx1–Slx4 complex could also be involved in the maintenance of other specific regions of the genome cannot be ruled out at this stage and is currently under investigation.
Figure 7.
Schemes rDNA copy number maintenance. (A) The Rqh1-dependent nonrecombinogenic pathway limits the contraction of the rDNA loci due to unscheduled recombination events. The Slx1–Slx4 complex initiates a scheduled recombination process that leads to the expansion of the rDNA loci. (B) Rqh1 would act in a main nonrecombinogenic pathway that rescues two types of stalled forks. Unscheduled stalling of a RF can occur at DNA damage, protein complexes, and torsional stress (fork 1). Another type of stalled fork is generated by a scheduled mechanism that occurs at the RFB (fork 2). In absence of Rqh1, rescue of the stalled forks 1 requires Mus81-Eme1, whereas rescue of fork 2 will necessitate the action of the Slx1–Slx4 complex.
Possible Overlapping and Distinct Roles of Slx1-Slx4 and Rqh1 in rDNA Maintenance
If Rqh1 and Slx1-Slx4 define separate pathways of maintaining rDNA repeats, why cannot one pathway fully compensate for loss of the other? One possible explanation is that loss of rqh1 leads to two types of stalled RFs (Figure 7B). One type may arise anywhere in the genome, including the rDNA loci, due to DNA damage, protein–DNA complexes, or torsional stress. These would be rescued by the Mus81–Eme1 complex through a general recombinogenic pathway (Boddy et al., 2001). Based on our observations, this general mechanism would lead mainly to a loss of rDNA copies resulting in the shortening of chromosome III. The second type of stalled RF would be forks that stalled in a programmed and controlled manner at RFBs. These may be rescued by a mechanism that involves Slx1–Slx4 complex acting on particular DNA structures specifically formed at RFBs. Analysis in S. cerevisiae of the architecture of a RF stalled at the RFB revealed an unusual structure where only limited stretches of ss DNA were exposed and the nascent lagging strand was extended three base pairs farther than the nascent leading strand (Gruber et al., 2000). This structure differs from that of RFs stalled after HU treatment where the extent of exposed ss DNA is increased (Sogo et al., 2002). The lack of genetic interactions involving Slx1–Slx4 and Mus81 complexes in S. cerevisiae (Mullen et al., 2001) and in S. pombe (our unpublished data) is consistent with the idea that the two structure-specific nucleases act in independent mechanisms where they process different types of DNA structures. The recombinogenic rescue of RFB stalled forks would lead to a gain of rDNA copies, partly compensating for the loss of copies due to the rescue of the first type of stalled fork.
The acute synthetic lethality of rqh1 and slx1 or slx4 mutations cannot be explained only by the loss of rDNA copies. One explanation could be that the DNA structures formed at RFB stalled forks need to be processed by either Rqh1 or Slx1-Slx4 for the replication of the rDNA locus to be completed and to allow proper chromosome segregation. Kaliraman and Brill (2002) recently reported that in S. cerevisiae, the SGS1-ts SLX4 double mutant is unable to undergo S phase at restrictive temperature and arrests in late S-G2 phase. These observations together with our data suggest that in the absence of Rqh1, the Slx1–Slx4 complex is absolutely required to process DNA structures formed at the RFB to allow complete replication of the rDNA loci and proper chromosome segregation.
Conserved Slx1–Slx4 Endonuclease Activity in Fission and Budding Yeasts
Recent studies carried out with a recombinant form of budding yeast Slx1–Slx4 complex expressed and purified from bacteria showed that Slx1–Slx4 nuclease cleaves a variety of branched DNA substrates but shows the greatest preference for a 5′-flap structure (Fricke and Brill, 2003). Recombinant Slx1-Slx4 cleaves the 5′-flap near the junction, consistent with the way that endogenous fission yeast Slx1–Slx4 complex cleaves stem-loops. These studies with recombinant enzyme suggest that the nuclease activity of endogenous Slx1–Slx4 complexes is intrinsic to the heterodimeric enzyme and does not require additional subunits or posttranslational modifications. These studies also lend further support to the notion that Slx1–Slx4 enzymatic functions are similar between the two highly divergent yeast and strengthen the likelihood that these functions are generally conserved among many eukaryotic organisms, including humans.
Acknowledgments
We thank all members of the Scripps Cell Cycle groups for encouragement and stimulating discussions, in particular T. Nakamura. We also thank C. McGowan for critical reading of the manuscript and are grateful to T. Nakamura for advice on PFGE. S.C. was a recipient of a fellowship from the Association pour la Recherche contre le Cancer. P.-H.L.G. is a Research Special Fellow of the Leukemia and Lymphoma Society. J.R.Y. is supported by RO1 EY1328801, MERK-MGRI-241, and CA81665RR11823. This work wasfunded by National Institutes of Health grants CA77325 and GM59477 awarded to P.R.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–08–0586. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-08-0586.
References
- Aravind, L., and Koonin, E.V. (2001). Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 11, 1365–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., Gaillard, P.H., McDonald, W.H., Shanahan, P., Yates, J.R., III, and Russell, P. (2001). Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107, 537–548. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Lopez-Girona, A., Shanahan, P., Interthal, H., Heyer, W.D., and Russell, P. (2000). Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell Biol. 20, 8758–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy, M.N., and Russell, P. (2001). DNA replication checkpoint. Curr. Biol. 11, R953–R956. [DOI] [PubMed] [Google Scholar]
- Caspari, T., Murray, J.M., and Carr, A.M. (2002). Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 16, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, M.M. (2001). Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet. 35, 53–82. [DOI] [PubMed] [Google Scholar]
- Cox, M.M., Goodman, M.F., Kreuzer, K.N., Sherratt, D.J., Sandler, S.J., and Marians, K.J. (2000). The importance of repairing stalled replication forks. Nature 404, 37–41. [DOI] [PubMed] [Google Scholar]
- Defossez, P.A., Prusty, R., Kaeberlein, M., Lin, S.J., Ferrigno, P., Silver, P.A., Keil, R.L., and Guarente, L. (1999). Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3, 447–455. [DOI] [PubMed] [Google Scholar]
- Derbyshire, V., Kowalski, J.C., Dansereau, J.T., Hauer, C.R., and Belfort, M. (1997). Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J. Mol. Biol. 265, 494–506. [DOI] [PubMed] [Google Scholar]
- Doe, C.L., Dixon, J., Osman, F., and Whitby, M.C. (2000). Partial suppression of the fission yeast rqh1(–) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19, 2751–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, J.B., Grothues, D., and Smith, C.L. (1991). Alignment of Sfi I sites with the Not I restriction map of Schizosaccharomyces pombe genome. Nucleic Acids Res. 19, 6289–6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke, W.M., and Brill, S.J. (2003). Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev. 17, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber, M., Wellinger, R.E., and Sogo, J.M. (2000). Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol. Cell Biol. 20, 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, J.J., and Lieber, M.R. (1994). The characterization of a mammalian DNA structure-specific endonuclease. EMBO J. 13, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson, I.D., Davies, S.L., Li, J.L., Levitt, N.C., Mohaghegh, P., North, P.S., and Wu, L. (2001). Role of the Bloom's syndrome helicase in maintenance of genome stability. Biochem. Soc. Trans. 29, 201–204. [DOI] [PubMed] [Google Scholar]
- Johzuka, K., and Horiuchi, T. (2002). Replication fork block protein, Fob1, acts as an rDNA region specific recombinator in S. cerevisiae. Genes Cells 7, 99–113. [DOI] [PubMed] [Google Scholar]
- Kaliraman, V., and Brill, S.J. (2002). Role of SGS1 and SLX4 in maintaining rDNA structure in Saccharomyces cerevisiae. Curr. Genet. 41, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman, V., Mullen, J.R., Fricke, W.M., Bastin-Shanower, S.A., and Brill, S.J. (2001). Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 15, 2730–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey, S.E., Montgomery, S., Labib, K., and Lindner, K. (2000). Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Heck, D.J., Nomura, M., and Horiuchi, T. (1998). Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 12, 3821–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Hidaka, M., Nishizawa, M., and Horiuchi, T. (1992). Identification of a site required for DNA replication fork blocking activity in the rRNA gene cluster in Saccharomyces cerevisiae. Mol. Gen. Genet. 233, 355–362. [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., and Horiuchi, T. (1996). A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1, 465–474. [DOI] [PubMed] [Google Scholar]
- Link, A.J., Eng, J., Schieltz, D.M., Carmack, E., Mize, G.J., Morris, D.R., Garvik, B.M., and Yates, J.R., III. (1999). Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17, 676–682. [DOI] [PubMed] [Google Scholar]
- Lopez-Girona, A., Furnari, B., Mondesert, O., and Russell, P. (1999). Nuclear localization of Cdc25 regulated by DNA damage and 14–3-3 protein. Nature 397, 172–175. [DOI] [PubMed] [Google Scholar]
- McGlynn, P., and Lloyd, R.G. (2002). Genome stability and the processing of damaged replication forks by RecG. Trends Genet. 18, 413–419. [DOI] [PubMed] [Google Scholar]
- Mizukami, T., et al. (1993). A. 13 kb resolution cosmid map of the 14 Mb fission yeast genome by nonrandom sequence-tagged site mapping. Cell 73, 121–132. [DOI] [PubMed] [Google Scholar]
- Moreno, S., Klar, A., and Nurse, P. (1991). Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Mullen, J.R., Kaliraman, V., Ibrahim, S.S., and Brill, S.J. (2001). Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, T.M., Moser, B.A., and Russell, P. (2002). Telomere binding of checkpoint sensor and DNA repair proteins contributes to maintenance of functional fission yeast telomeres. Genetics 161, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren, D.K., and Sancar, A. (1989). The (A)BC excinuclease of Escherichia coli has only the UvrB and UvrC subunits in the incision complex. Proc. Natl. Acad. Sci. USA 86, 5237–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, A.J., Elledge, S.J., and Zou, L. (2002). Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12, 509–516. [DOI] [PubMed] [Google Scholar]
- Park, P.U., Defossez, P.A., and Guarente, L. (1999). Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol. Cell Biol. 19, 3848–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, C.A., Kemper, B., and West, S.C. (1990). Interaction of a four-way junction in DNA with T4 endonuclease VII. J. Biol. Chem. 265, 9285–9289. [PubMed] [Google Scholar]
- Putilina, T., Wong, P., and Gentleman, S. (1999). The DHHC domain: a new highly conserved cysteine-rich motif. Mol. Cell Biochem. 195, 219–226. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032. [DOI] [PubMed] [Google Scholar]
- Sanchez, J.A., Kim, S.M., and Huberman, J.A. (1998). Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 238, 220–230. [DOI] [PubMed] [Google Scholar]
- Seigneur, M., Bidnenko, V., Ehrlich, S.D., and Michel, B. (1998). RuvAB acts at arrested replication forks. Cell 95, 419–430. [DOI] [PubMed] [Google Scholar]
- Seigneur, M., Ehrlich, S.D., and Michel, B. (2000). RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol. Microbiol. 38, 565–574. [DOI] [PubMed] [Google Scholar]
- Shen, J.C., and Loeb, L.A. (2000). The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 16, 213–220. [DOI] [PubMed] [Google Scholar]
- Sijbers, A.M., et al. (1996). Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86, 811–822. [DOI] [PubMed] [Google Scholar]
- Sinclair, D.A., and Guarente, L. (1997). Extrachromosomal rDNA circles–a cause of aging in yeast. Cell 91, 1033–1042. [DOI] [PubMed] [Google Scholar]
- Sogo, J.M., Lopes, M., and Foiani, M. (2002). Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602. [DOI] [PubMed] [Google Scholar]
- Stewart, E., Chapman, C., Al-Khodairy, F., Carr, A., and Enoch, T. (1997). rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO J. 16, 2682–2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa, S., and Yanagida, M. (1992). Visualization of centromeric and nucleolar DNA in fission yeast by fluorescence in situ hybridization. J. Cell Sci. 101, 267–275. [DOI] [PubMed] [Google Scholar]
- Verhoeven, E.E., van Kesteren, M., Moolenaar, G.F., Visse, R., and Goosen, N. (2000). Catalytic sites for 3′ and 5′ incision of Escherichia coli nucleotide excision repair are both located in UvrC. J. Biol. Chem. 275, 5120–5123. [DOI] [PubMed] [Google Scholar]
- Versini, G., Comet, I., Wu, M., Hoopes, L., Schwob, E., and Pasero, P. (2003). The yeast Sgs1 helicase is differentially required for genomic and ribosomal DNA replication. EMBO J. 22, 1939–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, M.P., Wolters, D., and Yates, J.R., III. (2001). Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247. [DOI] [PubMed] [Google Scholar]
- Watt, P.M., Louis, E.J., Borts, R.H., and Hickson, I.D. (1995). Sgs 1, a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell 81, 253–260. [DOI] [PubMed] [Google Scholar]