Abstract
Anatomy predicts that mammalian retinas should have in excess of 12 physiological channels, each encoding a specific aspect of the visual scene. Although several channels have been correlated with morphological cell types, the number of morphological types generally exceeds the known physiological types. Here, we attempted to sort the ganglion cells of the mouse retina purely on a physiological basis. The null hypothesis was that the outputs of the ganglion cells form a continuum or should be divided into only a few types. We recorded the spiking output of 471 retinal ganglion cells on a multielectrode array while presenting 4 classes of visual stimuli. Five parameters were chosen to describe each cell's response characteristics, including relative amplitude of the ON and OFF responses, response latency, response transience, direction selectivity, and the receptive field surround. We compared the results of four clustering routines and judged the results using the relevant validation indices. The optimal partition was the 12-cluster solution of the Fuzzy Gustafson-Kessel algorithm. This classification contained three visual channels that carried predominately OFF responses, six that carried ON responses, and three that carried both ON and OFF information. They differed in other parameters as well. Other evidence suggests that the true number of cell types in the mouse retina may be somewhat larger than 12, and a definitive typology will probably require broader stimulus sets and characterization of more response parameters. Nonetheless, the present results do allow us to reject the null hypothesis: it appears that in addition to well-known cell types (such as the ON-OFF direction selectivity cells) numerous other cell classes can be identified in the mouse retina based solely on their responses to a standard set of simple visual stimuli.
Keywords: multielectrode recording, sensory encoding, ganglion cells, vision
the identification of functional cell types is an essential element of a bottom-up understanding of any complex neural system. Since the time of Cajal, it has been believed that morphological types correspond in a one-to-one fashion to physiological types, and there is much evidence in support of that proposition, particularly from the retina (Berson 2008; Boycott and Wassle 1974; Dacey 1994; Roska and Werblin 2001). Current anatomical evidence suggests that mammalian retinas have at least 10–15 types of ganglion cells [for review, see Masland (2001)]. Unfortunately, this diversity has not been matched by physiological surveys. Indeed, it has been proposed more than once that the physiological properties form a continuum (Abbott and Chance 2002; Carcieri et al. 2003; Hochstein and Shapley 1976; Mechler and Ringach 2002).
It is likely that the identification of neural cell types will eventually be made on the basis of molecular markers (Gong et al. 2003; Madisen et al. 2009; Siegert et al. 2009). However, molecular markers specific to individual neuronal cell types are only slowly being identified (Huberman et al. 2008; Kim et al. 2008; Zhang et al. 2004). Here, we sought to classify mouse ganglion cells on purely physiological grounds. Classifying cells based on their responses to light remains a challenge to physiologists. The classic descriptions relied on hand- held stimuli and auditory evaluation of the responses. These methods are fast, flexible, and have the advantage that human observers can readily detect the many nonlinearities of each cell's responses. Grating stimuli (Enroth-Cugell and Robson 1966; Hochstein and Shapley 1976) provide a useful discrimination between two of the cell types present in the cat and monkey but fail for the large population of non-X, non-Y cells. Testing with random checkerboards, although elegant in conception, is largely restricted to the analysis of the size, position, polarity, and temporal dynamics of a cells receptive field (DeVries 1999; Devries and Baylor 1997; Fairhall et al. 2006; Segev et al. 2006). Nonstandard trigger features are generally not revealed, as they occur rarely during the presentation of a random checkerboard. Natural scenes should provide the ultimate test set, because they contain the stimuli for which the cells were designed, but analyzing the responses to such scenes remains an elusive goal. We sought to steer between the extremes of formal, technically “neutral” stimuli on the one hand, and natural scenes on the other. The choice was to apply stimuli that had previously been used to discriminate between cell types of the retina and represented conceptually simple differences in a cell's response properties, e.g., ON vs. OFF, transient vs. nontransient, etc.
Our goals were to confirm or refute the proposition that an unsupervised sorting of the cells would yield distinct cell types and to estimate the number of types to be expected. This work represents the largest survey to date of visual response properties of mouse retinal ganglion cells. We demonstrate that the visual response properties are classifiable into distinct groups that do not form a continuum, and we outline the functional similarities and differences within the ganglion cell populations.
MATERIALS AND METHODS
Extracellular Recording and Spike Sorting
Experiments were performed on whole mount retinas from C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME) in accordance with Massachusetts General Hospital Animal Use Committee and have been described in previous articles (Stasheff 2008; Zeck and Masland 2007). Both male and female mice (6- to 12-wk-old) were used. Briefly, mice were anesthetized by intramuscular injection of ketamine (30–100 mg/kg) and xylazine (5–10 mg/kg) and euthanized by spinal dislocation. The enucleated eye was hemisected, and the retina was peeled off the pigment epithelium under dim red light illumination. The retina was superfused with oxygenated mouse Ringer solution (124 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 1.25 mM NaH2PO2, 26 mM NaHCO3, and 22 mM glucose) at 33–35°C and mounted ganglion cell side down on a multielectrode array (Multichannel Systems, Reutlingen, Germany). The mice were not dark adapted.
In each experiment, we recorded ∼15 cells from a patch of the mouse retina, using a multielectrode array for a period of ∼6 hours. The experimental setup and the recording technique used to simultaneously record multiple retinal ganglion cells have been described in previous reports (Stasheff 2008; Zeck et al. 2005). Briefly, the activity of ganglion cells in a patch of the retina was recorded via electrodes spaced either 100 or 200 μm apart. Each electrode was connected to its own differential preamplifier (1,200 × gain, bandpass filter 10–3,000 Hz). Extracellular waveforms were recorded (DSP signal processors; Cyberkinetics, Foxborough, MA) when their amplitude exceeded a threshold set at 2–4 SD above mean noise level. The waveforms of 1.5-ms duration were stored at 33-μs time resolution. Typically, each electrode recorded waveforms from more than one cell. A supervised neural network algorithm (PowerNAP, Neuroshare) calculated the first five principal components of all recorded waveforms on one electrode. It then separated the waveforms based on their principal components using either a K-Means cluster analysis or EM-t-distribution (Shoham et al. 2003). A final adjustment to the division of the clusters was performed manually. We assumed that waveforms from one cluster originate from a single cell and stored them as a time-stamped, spike train (times of first threshold crossing) for further analysis.
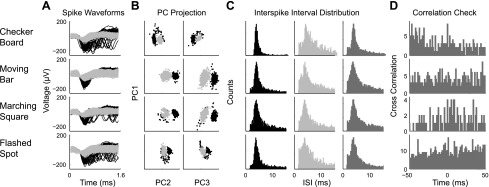
Verification that waveforms were properly sorted and assigned was performed as previously described (Segev et al. 2004; Stasheff 2008). Specifically, the appropriate assignment of individual waveforms to distinct cells was confirmed by visual inspection of the waveforms (Fig. 1, A and B) and analysis of the corresponding spike trains (Fig. 1, C and D). Interspike interval histogram distributions were computed for each spike train with a bin width of 0.2 ms (Fig. 1C). Interspike interval histograms from accepted spike trains demonstrated a refractory period of at least 1 ms (typically 2–5 ms). In addition, we calculated pairwise cross-correlograms of all spike trains on a single electrode (Fig. 1D). If this function showed a prominent break at the origin, then the two spike trains were considered to originate from the same cell; conversely, if there was no peak or a strong peak around 0, then the spikes must have originated from two separate cells. We only selected the cluster with the largest spikes on each electrode for further analysis. We determined that we can reliably find the same cell on the same electrode during different visual stimuli across recording periods of up to 6 h (Fig. 1). Spike train analysis was performed using user written routines in MATLAB.
Fig. 1.
Consistency of spike sorting across stimuli. A: waveforms of recorded spikes during the presentation of 1 of 4 stimuli: flickering checkerboard (row 1), moving bars (row 2), marching square (row 3), and flashed spot (row 4). Black lines represent those waveforms assigned to unit 1 and light grey lines to those assigned to unit 2. B: principal component (PC) projection of the waveforms demonstrates the ability to separate the waveforms based on only the first 3 principal components. In the left column the first principal component (PC1) is plotted against PC2, while in the right column PC2 is plotted against PC3. C: interspike interval (ISI) distributions of each unit alone (columns 1 and 2) test for a distinct refractory period. In column 3, the interspike interval distribution of both units combined (dark grey) test for independence of the units. D: cross-correlation functions of the spike trains of unit 1 and 2 show a flat relation, reflecting the independence of each unit.
Light Stimuli
Visual stimuli were generated using Visionworks software (Vision Research Graphics, Durham, NH) and projected with a calibrated CRT monitor (Dell-P780). The stimulus was reflected via a substage mirror and focused through a ×20 objective (LCF Plan FI NA 0.4; Olympus Optical) onto the multielectrode array. The light entered the retina through the array substrate from the ganglion-cell side.
Pseudorandom flickering checkerboard.
The receptive fields were mapped using a 16 × 16 pseudorandom flickering checkerboard stimulus; the luminance of each square was independently modulated by an m-sequence (Reid et al. 1997). The integrated light intensity at the position of the retina ranged from 0 to 2 cd/m2. The update rate of the checkerboard frames was 50 Hz, and the width of each square was 100 um2. The receptive field for each cell was calculated by reverse correlating the stimulus and spike response of the cell. The position of each cell was determined by calculating the center of mass of all checkerboard squares whose intensity at the temporal maximum of the mean effective stimulus exceeded by a factor of 3 the SD of the squares in the background (Devries and Baylor 1997).
Marching square.
A 600-μm white square was flashed on a black background at a series of locations separated, center to center, by 60 μm both in the horizontal and vertical direction (Roska and Werblin 2001) permitting the creation of a space-time map of the receptive field for each cell. The squares were presented in sequence for a duration of 2 s and an interstimulus time of 10 s. This whole sequence was repeated 10 times.
Spot size.
During each recording, the center of 15–20 cells receptive field was determined from the mean effective stimulus calculated from each cell's response to the flickering checkerboard. This analysis was performed while the marching square and moving bar stimuli were being presented to the retina. These positions were double checked for each cell after the whole experiment was completed. After each cell's receptive field was located, a series of spots was flashed, in different sizes, positioned over the center of each cell's receptive field. Both white and black spots were presented for 2 s interleaved with a homogeneous grey background for 2 s. The size of the spot presented was randomized. Each spot was then repeated for five times in a row. Total spikes occurring during the presentation of a given spot size were counted and normalized by expressing them as a fraction of the maximum number of spikes evoked by the optimal spot size.
Moving bars.
A rectangular bar 200 × 500 μm moved parallel to its long edge at 300 μm s−1 in eight directions at 45° intervals. This stimulus was presented one direction at a time with each sweep offset by 100 μm from the last in a direction perpendicular to the motion axis. This was repeated for each of the eight directions of motion five times and covered a 5- mm by 5-mm area. Each bar had a 100-μm overlap with the previously presented bar. There was an interstimulus time of 5 s.
Parameter Choice and Calculation
From the stimuli described above, 12 parameters were extracted that summarized the responses of each cell. Some were found to be uninformative, usually because they were redundant or noisy. It was important to keep the dimensions to a minimum for two reasons. First, the amount of data needed to adequately fill a multidimensional space increases exponentially with the number of dimensions. Second, the concept of distance becomes less precise as the number of dimensions grows (Beyer et al. 1999; Verleysen and Francois 2005). In particular, the distance between any data point to a particular cluster center becomes approximately the same as the dimensionality increases (Beyer et al. 1999). Weighting of parameters was avoided to reduce human bias in the classification. A set of five parameters was chosen as the basis of clustering. They were chosen in part empirically and in part from the principle that response features of known significance were most likely to be effective at discriminating among the cells. A description of the parameters and how they were calculated is presented below.
ON-OFF Bias Index (Bias Index).
The first parameter estimated whether the cell responded best to increments or decrements in light intensity or both. This parameter was calculated from the cells' responses to a flashed spot in the area-response protocol. For each cell, the spot size was selected that gave the largest response. The Bias Index was calculated by comparing the response to the spot of optimal size and sign of contrast (hence referred to as the preferred stimulus) with its response to a spot of the same size but for the nonpreferred sign of contrast. The sum of the spikes during the first 400 ms after the presentation of the stimuli was compared such that:
This index ranges from +1 for ON cells through 0 for ON-OFF cells to −1 for OFF cells.
Response Latency (Latency).
The latency of each cell was estimated by measuring the time to the peak of its response during the presentation of its preferred stimulus, i.e., the spot size that produced the most spikes, during the increasing-spot-stimulus paradigm. The response was measured as the mean firing rate measured over an integration window of 25 ms.
Transience Index (Transience).
We compared the response duration of each cell by measuring the area under its normalized 1.5-s long poststimulus time histogram (PSTH) to its preferred stimulus. The PSTH was normalized so that its peak response was 1. The area under the curve thus varied depending on the duration of the response of the cell (Fig. 2C). We report the index in seconds where 1 represents summed activity under curves equivalent to maximum firing rate for 1 of the 1.5-s PSTH duration.
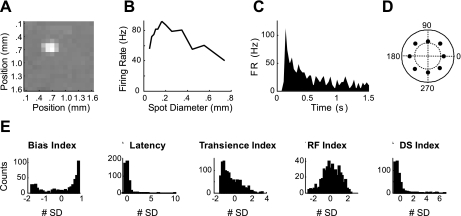
Fig. 2.
Parameter calculation and distributions. A: spike-triggered average is shown to demonstrate how the center of each cell's receptive field was determined and is defined by the center of mass of all pixels exceeding 3 SD of the mean. B: peak-firing rate is plotted vs. the presented spot size. From this plot, a Receptive-Field (RF) Index was measured that estimated the strength of each cell's surround. C: poststimulus time histogram for the stimulus producing the largest response in B; this was used to calculate the Latency and Transience Index of the response of each cell. D: a polar plot of the sum of spikes produced for each direction of motion of a moving bar. Units are average spike count during the presentation of the stimulus. Inner dotted circle represents 25 spikes, and outer solid line represents 50 spikes. Magnitude of the vector average of the response was taken as the Direction Selective (DS) Index. E: histograms of the different parameters used, plotted as the means ± SD.
Receptive Field Index (RF Index).
This index is an estimate of a cell's preference for stimuli that are larger and/or smaller than its optimal spot stimulus. It was calculated as the area under the normalized, size-response curve (Fig. 2B). The size-response curve represents the total number of spikes produced during the presentation of spots of different diameters. After the peak was normalized to 1, the area under the curve can then be calculated. The index is reported as a fraction of the total possible area under the curve. A value of 1 would mean the cell responded with its maximum response at each stimulus size. Smaller values indicate a reduction in the cell's response when presented stimuli larger and/or smaller than the optimum.
Direction Selectivity Index (DS Index).
This measures the directional tuning of each cell. It was calculated from the cells' responses to the moving bar protocol. We calculated the average firing rate during the presentation of a bar moving in eight equally separated directions (Fig. 2D). The direction selectivity of each cell was defined as the vector average of mean firing rate:
Where Nd are vectors pointing in the direction of the stimulus and having length Rd, equal to the mean firing rate recorded during the stimulus presentation. The DS Index ranges from 0, when the average firing rates are equal in all stimulus directions, to 1, when a cell responds to a single direction only.
Cluster Analysis
The objective of the cluster analysis was to organize the population of recorded ganglion cells, each with a unique set of visual response properties, into an optimal number of groups to get an estimate of the number of visual channels of the mouse retina. Deciding how many groups exist in a data set using cluster analysis is a two-step process. First, the data are clustered into a defined number of groups. Second, the best result, number of clusters, is determined using a set of validity measures. The quality of the results depend equally on the clustering algorithms ability to correctly partition the data and the ability of the validity measures to choose the correct number of clusters.
To determine which clustering method to use, we performed a comparison of 7 clustering algorithms on 27 data sets of various geometries (see Supplemental Material, Supplemental Figs. 1 and 2; Supplemental Material for this article is available online at the J Neurophysiol website). The primary result of this analysis was the superior reliability of the Fuzzy Gustafson-Kessel (Fuzzy-GK) algorithm to determine the correct partitioning of data sets as the complexity of the data sets increased with noise, number of clusters, number of dimensions and, importantly, the degree of elongation of the clusters (Supplemental Fig. 1A). Additionally, we found that the Fuzzy C-Means (Fuzzy-C), K-Means, Partitioning Around Medoids (PAM), and Affinity Propagation (AP) all performed well on most data sets but failed when challenged with elongated clusters (see Supplemental Figs. 1A and 2). The agglomerative hierarchical methods were the poorest performers having difficulty with data sets consisting of either noisy or elongated clusters. The ability of the Fuzzy-GK algorithm to correctly find an elongated cluster, even those forming long continuums (e.g., Supplemental Fig. 2), is of particular importance as it gave us confidence that we would not artificially overestimate the number of clusters by dividing a continuum into subgroups, a feature of the other clustering algorithms tested (Supplemental Fig. 2). Below, we describe four diverse clustering algorithms whose results are compared in the results.
Gustafson-Kessel algorithm.
Our chosen method to cluster the data is the Fuzzy-GK algorithm (Gustafson and Kessel 1979). The Fuzzy-GK partitioning method is an extension of the Fuzzy-C algorithm. The Fuzzy-C algorithm is initiated by assigning each point a random partial membership value to k random centroids. One then minimizes the objective function by assigning membership values to each point to each cluster based on distance, then recalculating the centroids and repeating until minimal changes occur in the objective function below a predefined value. Gustafson and Kessel extended the standard Fuzzy-C algorithm by employing an adaptive distance norm where each cluster has its own norm-inducing matrix. This allows for the detection of clusters of different geometrical shapes and orientations. The covariance matrix of each cluster is used as the optimization variables in the c-Means function, thus allowing each cluster to adapt the distance norm to the local topological structure of the data. We employed the numerically robust version of Babuska et al. (2002). The Mahalanobis distance norm takes the form:
where Ai is defined as:
where Fi is the fuzzy covariance matrix of the i-th cluster defined by:
This gives a generalized squared Mahalanobis distance norm between xk and the cluster vi, where the covariance matrix is weighted by the membership degree in the partition matrix u.
K-Means.
K-Means clustering is a simple and popular hard partitioning method (Duda et al. 2001). The algorithm assigns each data point to one of a predetermined number of clusters to minimize the with-in cluster sum of squares:
where Ai is a set of data in the i-th cluster and vi is the mean of the data in cluster i. These algorithms have a tendency to form hyperspherical clusters. In the K-Means algorithm, the centers of each cluster are the means of the data belonging to an individual cluster.
PAM algorithm.
Unlike the K-Means algorithm the PAM algorithm (Kaufman and Rousseeuw 1990) accepts as its input a dissimilarity matrix of the data set, rather than the raw data itself. It then searches for a predefined number of representative objects within the data set, which are medoids of the grouped data. The clustering has two phases. In the first phase, an initial clustering is obtained by the selection of representative objects until the predefined number has been found. Here the sum of dissimilarities to all other objects is minimized. Subsequently, at each iteration, another object is selected which decreases the sum (over all objects) of the dissimilarities to the most similar selected object. This process is continued until the predefined number of clusters in found. In the second phase, it is attempted to improve the set of representative objects. This is done by considering all pairs of objects for which object 1 has been selected as a representative object and object 2 has not. It is determined what effect is obtained by selecting object 2 as the representative object instead of object 1. The best representative is kept. This procedure is continued until no more improvements are possible. One advantage of this method is the ease of using different measures of dissimilarity. The results presented here use a dissimilarity matrix based on the Mahalanobis distance metric.
The partitioning algorithms above can be termed “flat” as they partition the data once given a set number of clusters.
AP algorithm.
This clustering routine takes as input the measure of similarity between pairs of data points (Frey and Dueck 2007). Real-valued messages are exchanged between data points until a high-quality set of exemplars and corresponding clusters gradually emerges. There are two kinds of messages exchanged between data points, and each takes into account a different kind of competition. The “responsibility” r(i,k), sent from data point i to candidate exemplar k, reflects the accumulated evidence for how well-suited point k is to serve as the exemplar for point i, taking into account other potential exemplars. The “availability” a(i,k), sent from candidate exemplar k to point i, reflects the accumulated evidence for how appropriate it would be for point i to choose point k as its exemplar, taking into account the support from other points that point k should be an exemplar. The responsibility and availability can be viewed as log-probability ratios. AP can be viewed as a version of hierarchical clustering that makes soft decisions so that it is free to hedge its bets when forming clusters. The similarities were measured using the Mahalanobis distance. We used the MATLAB-implemented programs from http://www.psi.toronto.edu/affinitypropagation/ to calculate the optimal solutions for cluster numbers between 2 and 19.
Assessment of the Clustering
To determine which partition of the data, i.e., number of clusters, was best, we assessed the clustering solutions, for cluster numbers from 2 to 19, based on internal criteria; geometric and fuzzy membership. Generally, the validation process determines which solution has a combination of the most compact clusters with the greatest separation. To decide which combination of validity indexes to use with which clustering algorithm, the performance of each validity index was assessed on the 27 artificial data sets (Supplemental Figs. 1B and 3). A new index was then created for each clustering algorithm that was simply the average of the best three or four performing indexes tested. For the Fuzzy-GK algorithm, our validity index (VIGK) was the average of four indexes: the Calinski and Harabasz (VCH), Davies-Bouldin Index (VDB), Krzanowski and Lai Index (VKL), and the Fukuyama and Sugeno Index (VFS). We found that the VIGK found the correct number of clusters in 91.3% of the artificial data sets, compared with between 38 and 77% for individual indices. For the K-Means, PAM, and AP algorithms we used the mean of three indexes: the VCH, VDB, and VKL. This compound index, VG, found the correct number of clusters for the K-Means, PAM, and AP algorithms 56.5, 69.6, and 69.6% of the time respectively. Below we describe the individual indexes.
Calinski and Harabasz Index.
The Calinski and Harabasz (VCH) is computed as:
where n and k are the total number of items and the number of clusters in the solution, respectively (Calinski and Harabasz 1974). The B and W terms are the between and pooled within cluster sum of squares and cross product matrices. This index should be maximized.
Davies-Bouldin Index.
The Davies and Bouldin Index (VDB) is based on the similarity measure of clusters, Rij (Davies and Bouldin 1979). It measures the average similarity between each cluster and its most similar one, where Rij is:
where dij is the distance between clusters i and j and Si is defined as:
The index is then defined as:
where Ri = max(Rij), i = 1 … N. The solution for the most compact and the well-separated clusters is found with the minimum of the VDB Index. VDB uses the centroids as a point of reference.
Krzanowski and Lai Index.
The index (Krzanowski and Lai 1988) is defined as:
The parameter m represents the number of features, here 5, and Wk is calculated as the within-group dispersion matrix of the clustered data:
The index should be maximized.
Fukuyama-Sugeno Index.
The Fukuyama and Sugeno Index is defined as:
where vj is the mean of cluster j, xi is data point i, uij is the fuzzy membership of data point i to cluster j and m is the fuzzy membership exponent, set here to 2 (Fukuyama and Sugeno 1989). The first term in parenthesis measures the compactness of the clusters and the second one estimates the distance between the cluster representatives. This index should be minimized.
Comparing Clustering Results
To compare the clustering results to each other or to a known partitioning solution we computed the Rand Index (Rand 1971), which is calculated as:
where a is the number of pairs elements assigned to the same cluster in both partitions, b is the number of pairs of elements that are assigned to different clusters in both partitions, c is the number of elements assigned to the same cluster in the first partition but to different partitions in the second, and d is the number of pairs of elements assigned to different sets in the first partition but to the same cluster in the second partition. Intuitively, a + b can be thought as the number of agreements between the two partitions and c + d as the number of disagreements. A value of 1 indicates that two partitioning solutions are the same and 0 that no two pairs of data were clustered the same way. To correct for chance, we calculated the adjusted Rand Index (ARI) (Hubert and Arabie 1985), which is calculated as:
Similarity Matrix
To visualize the results of different clustering routines, we generated a similarity matrix between cells. In this matrix, each element is the similarity between cells i and j. Initially, the cells were in a random order, and therefore no features could be observed in the matrix. After applying the different clustering algorithms, we reorganized the matrix such that neurons that belong to the same class were grouped together in adjacent rows/columns. If the cells belonging to a single group are functionally similar, they should have small difference values between all pairs in the group. This will appear as white colored blocks of cells along the diagonal in the reordered similarity matrix. Conversely, the regions that correspond to ganglion cells from different functional types should all have high difference values (shown by colors close to black). Therefore, if the ganglion cell population exhibits clustering into distinct functional types, the reordered matrix will have a clear block structure.
RESULTS
To determine how the encoding properties of the population of retinal ganglion cells divide into distinct channels, the visual response properties of 471 retinal ganglion cells from 40 mice were parameterized and grouped using quantitative clustering techniques. This analysis had three goals: to confirm or refute that an unsupervised sorting of the cells would yield distinct physiological cell types; to estimate the number of types to be expected; and to describe some of their basic properties.
The response properties were examined using a broad range of stimuli. Initially, the positions of 15–20 ganglion cells were mapped using a flickering checkerboard stimulus (Fig. 2A). Each ganglion cell was then presented with a set of standard stimuli. Flashed spots of different sizes targeted to each cell's receptive field center were used to determine its spatial and temporal response proprieties. Specifically, an ON-OFF Bias Index was calculated to describe the relative preference for increments and decrements of light. Second, two standard parameters associated with the time course were calculated, the Latency and Transience of the response to light and dark spots (Fig. 2C). Additionally, by comparing the response of the retinal ganglion cells to flashed spots of different sizes, a RF Index was calculated that estimated the strength of its surround (Fig. 2B). A second stimulus, a series of moving bars was used to determine if the cell was directionally selective (DS Index; Fig. 2D). Finally, to visualize the receptive field of each ganglion cell, a 600-μm white square was marched across its receptive field.
Basic Organization of Visual Channels
To ascertain if the parameterized data contained a robust structure containing distinct physiological cell types, the data were organized into groups using four different clustering algorithms that fell into three classes: two hard-partitioning algorithms, a fuzzy-partitioning algorithm, and an agglomerative algorithm. The advantages of applying diverse algorithms were twofold. First, each algorithm applied distinct criteria to assign cells to a group, suggesting that if the broad organization of the data were robust, different algorithms would group the data in a similar manner. Second, during the validation step, distinct criteria could be used to assess the quality of the clustering results and determine the optimal solution of each algorithm (Table 1; Supplemental Fig. 3). Specifically, the partial membership values could be used to assess the fuzzy-clustering algorithm, while validity indexes using the geometry of the data was used to evaluate the results of all clustering algorithms. We looked for solutions between 2 and 19 clusters. Our final judgment of each algorithm was based on the combined results from a group of validity indexes; the average of the normalized values of each validation index, where 1 indicated the best solution for a particular index and 0 the worst (see materials and methods and Supplemental Fig. 3). The maximum of this combined validity measure was assumed to be the optimal solution from each algorithm.
Table 1.
Clustering methods and validation indexes
| Cluster Routine | Description | Validation Index |
|---|---|---|
| K-Means | Hard flat partition | VCH, VKL, VDB |
| PAM | Hard flat partition | VCH, VKL, VDB |
| Fuzzy-GK | Fuzzy flat partition | VCH, VKL, VDB, VFS |
| Affinity Propagation | Fuzzy hierarchical agglomerative | VCH, VKL, VDB |
A brief description of each clustering algorithm is given along with a list of the validation algorithms applicable to determine the optimal solution of each. PAM, Partitioning Around Medoids; Fuzzy-GK, Fuzzy Gustafson-Kessel algorithm. See materials and methods for Validation Index definitions.
Three characteristics of the output from the different clustering algorithms suggest that the response properties of our data set when clustered using an unsupervised approach yield distinct physiological cell types. First, the number of clusters found by each algorithm was similar, either 12 or 13 (Fig. 3, bottom row). Second, the properties that distinguished the clusters from each other are consistent across the solutions from the different algorithms (Fig. 4). Finally, the relative number of cells assigned to the clusters with similar properties was comparable across the solutions from the different algorithms. Quantitatively, these qualitative similarities translated into an agreement between the optimal Fuzzy-GK solution and that of K-Means of 90.4%, of AP of 90.3%, and of PAM of 89.4%. A more detailed comparison of the similarities among the outputs from the different algorithms is presented below.
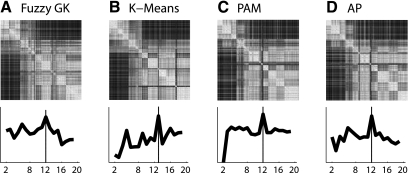
Fig. 3.
Broad structure of the data. Grouping of the different cells is displayed in a color coded plot of the sorted similarity matrices (top row) where each matrix represents the best solution of the different clustering algorithms (bottom row). Top row: each row or column represents a single ganglion cell and the color of each point reflects the similarity between individual cells. White hues indicate cells with more similarity and black with less similarity. Each similarity matrix was sorted by the value of median Bias Index value of each cluster, with the most OFF group located at the top left corner and the most ON group the bottom right. Bottom row: validation assessment deciding which clustering solution, from 2 to 19 clusters, is best. Black line is the compound validity index for each clustering algorithm (see materials and methods). A: Fuzzy-Gustav-Kessel (Fuzzy-GK). B: K-Means. C: Partition Around the Median (PAM). D: Affinity Propagation (AP).
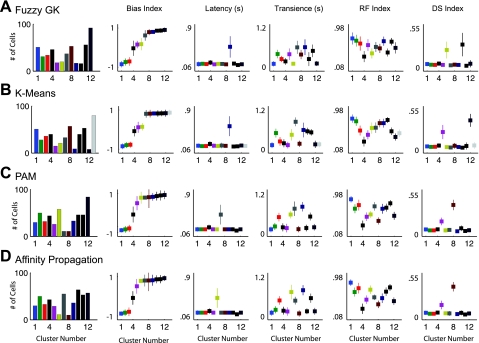
Fig. 4.
Comparison of results from each clustering algorithm. Best results of the different clustering routines (A–D) are compared by examining the relative values of cluster centers for each parameter (columns 2–6). Column 1: histograms of the number of cells making up each cluster. Columns 2–6: for the results of each clustering algorithm, the median values ± SD of the different clusters are plotted for each parameter, respectively. In A–D, the clusters are organized and color coded in ascending order according to their ON vs. OFF index value from minimum to maximum. A: 12 cluster solution of the Fuzzy-GK algorithm. B: 13 cluster solution of the K-Means algorithm. C: 12 cluster solution of the PAM algorithm. D: results of the AP algorithm at 12 clusters.
Visual inspection of the results of the different clustering algorithms revealed a broad organizational structure to the parameterized description of the retinal ganglion cell response properties. The grouping of the different cells is displayed in a color coded plot of the sorted similarity matrices (see materials and methods), where each matrix represents the best solution of each clustering algorithm (Fig. 3, top row). The matrices were sorted by the value of median Bias Index value of each cluster, with the most OFF group located at the top left corner and the most ON group the bottom right. Each row or column represents a single ganglion cell, and the color of each point reflects the similarity between the different cells. We found broad similarities among the results of the different clustering algorithms. Inspecting each similarity matrix one can see white squares formed along the diagonal. These squares represent groups of cells with similar properties compared with cells in other groups. It is clear that each algorithm groups the cells clearly according to their relative preference for ON vs. OFF stimuli. However, this projection does not allow for a complete view of the response properties that delineate the different clusters.
A more comprehensive view of the properties differentiating the different groupings of retinal ganglion cells is illustrated in Fig. 4. Here the results of the different clustering routines are compared by examining the relative values of cluster centers for each parameter (Fig. 4, columns 2–6). In Fig. 4, A–D, the clusters are organized in ascending order of the value of their Bias Index. For the results of each clustering algorithm the color of a single cluster remains constant across the columns. For example, in row 1, the results of the Fuzzy-GK algorithm, cluster 6 is yellow with a Bias Index of 0.18, Latency of 0.13, Transience of 0.14, RF Index of 0.38, and DS Index of 0.24. Additionally, histograms of the number of cells making up each cluster are shown (Fig. 4, column 1). Each row (Fig. 4, A–D) represents the best result from a single clustering algorithm.
The robustness of the partitioning solutions is evident in the similarity of the results of the four algorithms. Clear similarities among the solutions of the different algorithms can be seen by comparing the parameter values of the individual cluster centers (Fig. 4, A-D). In particular, three properties stand out that are consistent across each of the four clustering algorithms. First, each algorithm finds two groups of cells that can be described as direction selective (Fig. 4, column 6). Of these, each algorithm delineates one cluster with an “ON” Bias Index and one that contains ON-OFF cells. These two groups are likely made up of the three ON and four ON-OFF direction selective types of ganglion cells (Oyster and Barlow 1967; Sun et al. 2006; Weng et al. 2005). Second, of the clusters with an ON-OFF Bias Index one cluster always contains the group that has the smallest RF Index (cluster 4 in each case). This cluster has properties, such as a small receptive field and ON-OFF bias, that are consistent with the known properties of the local edge detector (Levick 1967; Roska and Werblin 2001; van Wyk et al. 2006; Zeck et al. 2005). Third, one cluster in each solution contains cells with a long latency to respond (Fig. 4, column 3). Additional similarities are obvious when taking a closer look at each cluster individually.
If we focus on the preference for ON vs. OFF stimuli, each algorithm found three groups of cells with a strong preference for OFF stimuli, two or three clusters with little preference, and seven to eight clusters with a strong preference for ON stimuli (Fig. 4, A–D, column 2). This consistency among the clusters extends to the other parameters. For example, among the OFF channels each algorithm detected a channel that was transient and had a large RF Index [GK = (0.13 s, 0.70, n = 51), KM = (0.15 s, 0.74, n = 51), PAM = (0.13 s, 0.79, n = 30), AP = (0.13 s, 0.79, n = 30)]; one channel that was more sustained with a large RF Index [GK = (0.41 s, 0.65, n = 31), KM = (0.50 s, 0.68, n = 28), PAM = (0.48 s, 0.67, n = 32), AP = (0.48 s, 0.68, n = 33)]; and one that was moderately transient with a moderate RF Index [GK = (0.27 s, 0.59, n = 34), KM = (0.26 s, 0.48, n = 36), PAM = (0.21 s, 0.59, n = 50), AP = (0.21 s, 0.59, n = 50)]. The similarities among the ON-OFF clusters consisted of one cluster with an ON-OFF Bias Index that was directionally selective (GK = 0.24, n = 20; KM = 0.24, n = 21; PAM = 0.16, n = 26; AP = 0.15, n = 29) and one cluster that had very small receptive field (GK = 0.31, n = 46; KM = 0.24, n = 40; PAM = 0.23, n = 44; AP = 0.23, n = 43). Among the ON channels, the similarities among the four algorithms consisted of an ON channel with long response latency (GK = 0.50 s, n = 17; KM = 0.55 s, n = 9; PAM = 0.45 s, n = 11; AP = 0.45 s, n = 11); one transient cluster with medium-sized RF Index [GK = (0.14 s, 0.50, n = 92), KM = (0.17 s, 0.42, n = 80), PAM = (0.19 s, 0.44, n = 84), AP = (0.17 s, 0.40, n = 64)]; a sustained cluster with moderate RF Index [GK = (0.61 s, 0.51, n = 37), KM = (0.82 s, 0.54, n = 33), PAM = (0.78 s, 0.59, n = 33), AP = (0.77 s, 0.46, n = 36)]; a sustained cluster with a large RF Index [GK = (0.51 s, 0.63, n = 56), KM = (0.59 s, 0.64, n = 40), PAM = (0.72 s, 0.64, n = 46), AP = (0.54 s, 0.69, n = 53)]; and as mentioned above an ON directionally selective channel (GK = 0.32, n = 16; KM = 0.40, n = 8; PAM = 0.39, n = 11; AP = 0.41, n = 10). The similarities of the solutions of these four algorithms, both the parameter values and number of cells assigned to each cluster, provide evidence that the delineated channels are robust.
Below we describe the delineated channels from the 12 cluster solution of the Fuzzy-GK algorithm in more detail. We chose to display the partitioning solution of the Fuzzy-GK algorithm as it clearly was the most reliable of the clustering methods tested (see Supplemental Fig. 1 and 2). The full partitioning solutions of the other three algorithms are shown in Supplemental Fig. 4.
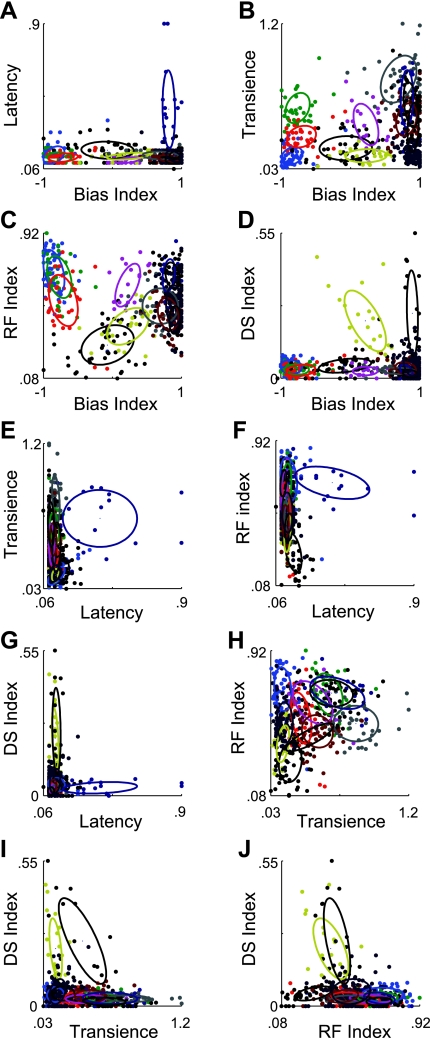
Delineating the Individual Visual Channels
The response properties of each cell have been quantified using five parameters. Scatter plots of the data illustrate how the 12 clusters, from the Fuzzy-GK solution, are distributed between the different response properties (Fig. 5, A–J; Table 2). Overlaid on the scatter plots are ellipses representing 1 SD spread of each cluster. The first property that strikes one is the amount of overlap between the clusters that exists in each of the plots. However, on closer inspection of Fig. 5, A–J, it is evident that each cluster has a separate footprint in at least one combination of parameters. For example, in Fig. 5A most of the cells group along the base of the graph, illustrating their short response latencies; however, one column of data points has been grouped together (dark blue) that share the properties of being ON cells and having a longer latency to respond than the rest (Fig. 4A, C9). A second prominent example is the two directionally selective groups (Fig. 5D). These two groups either form a voluminous cloud of cells that could be described as ON-OFF (yellow, C6) or a narrow column of cells that respond preferentially to ON stimuli (olive, C10). However, in this presentation it is not immediately obvious that each cluster occupies a unique position in the parameter space.
Fig. 5.
Twelve cluster solution of Fuzzy-GK algorithm. In A–J, the whole data set is plotted in 2 of the 5 dimensions of the parameter space. Overlaid ellipses represent the 1-SD area of each cluster. Color coding is as in Fig. 4A. The Bias, RF, and DS Indexes are expressed in normalized units, while the Latency and Transience Indexes are expressed in seconds.
Table 2.
Cluster centers and variance
| Bias Index | Latency, s | Transience, s | RF Index | DS Index | |
|---|---|---|---|---|---|
| C1 (51) | −0.85 ± 0.15 | 0.13 ± 0.05 | 0.13 ± 0.07 | 0.70 ± 0.13 | 0.04 ± 0.03 |
| C2 (31) | −0.74 ± 0.18 | 0.13 ± 0.03 | 0.41 ± 0.12 | 0.65 ± 0.12 | 0.03 ± 0.02 |
| C3 (34) | −0.70 ± 0.21 | 0.13 ± 0.03 | 0.27 ± 0.08 | 0.59 ± 0.15 | 0.03 ± 0.02 |
| C4 (46) | −0.03 ± 0.38 | 0.16 ± 0.05 | 0.20 ± 0.09 | 0.31 ± 0.12 | 0.05 ± 0.03 |
| C5 (18) | 0.16 ± 0.18 | 0.12 ± 0.02 | 0.41 ± 0.16 | 0.56 ± 0.13 | 0.03 ± 0.02 |
| C6 (20) | 0.18 ± 0.30 | 0.13 ± 0.02 | 0.14 ± 0.02 | 0.38 ± 0.11 | 0.24 ± 0.12 |
| C7 (37) | 0.62 ± 0.26 | 0.13 ± 0.02 | 0.61 ± 0.18 | 0.51 ± 0.10 | 0.03 ± 0.01 |
| C8 (53) | 0.79 ± 0.15 | 0.12 ± 0.02 | 0.40 ± 0.13 | 0.48 ± 0.09 | 0.04 ± 0.02 |
| C9 (17) | 0.80 ± 0.10 | 0.50 ± 0.02 | 0.61 ± 0.22 | 0.64 ± 0.09 | 0.03 ± 0.02 |
| C10 (16) | 0.87 ± 0.08 | 0.14 ± 0.03 | 0.27 ± 0.19 | 0.40 ± 0.08 | 0.32 ± 0.16 |
| C11 (56) | 0.87 ± 0.11 | 0.11 ± 0.02 | 0.51 ± 0.18 | 0.63 ± 0.08 | 0.02 ± 0.01 |
| C12 (92) | 0.90 ± 0.11 | 0.12 ± 0.03 | 0.14 ± 0.06 | 0.50 ± 0.12 | 0.05 ± 0.05 |
Values are means of each center ± SD. RF, receptive field; DS, direction selectivity.
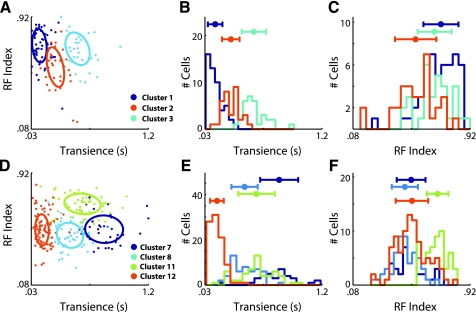
To better illustrate how some of the clusters that are less well separated are delineated, we take a closer look at the three OFF clusters (C1, C2, and C3) as well as the four ON clusters that were not directionally selective and did not have a long latency to respond (C7, C8, C11, and C12). Focusing first on the three OFF clusters, we see that they form clusters that have distinct Transience Indexes (Fig. 6, A and B). In particular, the histograms of the distribution of the cells making up each cluster forms distinct peaks along the Transience axis. Along the RF Index axis, the differences are less clear (Fig. 6C). However, the differences between the clusters increase the overall distance between the clusters when compared in the scatter plot of both the Transience and RF Index (Fig. 6A). The differences between the clusters in each dimension were found to be statistically significant (e.g., Transience C1 vs. C2, P < 0.0001; Transience C2 vs. C3, P < 0.0001; RF Index C2 vs. C3 P < 0.0002; RF Index C1 vs. C3 P < 0.0001).
Fig. 6.
Visualization of difficult to differentiate clusters. Histograms and scatter plots of the parameters that discriminate some of the difficult to differentiate clusters. A: scatter plot of the 3 OFF cells (clusters 1, 2, and 3) showing the distribution of cells across the Transience and RF Index. B: histograms of the Transience Index comparing the 3 OFF cells (clusters 1, 2, and 3). Above the plot the means ± SD of each cluster are shown. C: histograms of the RF Index comparing the 3 OFF cells (clusters 1, 2, and 3). D: scatter plot of the 4 ON cells (clusters 7, 8, 11, and 12), showing the distribution of cells across the Transience and RF Index. E: histograms of the Transience Index comparing 4 ON cells (clusters 7, 8, 11, and 12). F: histograms of the RF Index comparing the 4 ON cells (clusters 7, 8, 11, and 12).
The differences among the four ON clusters, C7, C8, C11, and C12, are a little less clear. However, the Transience Index clearly distinguishes C12 from the others (Fig. 6, D and F). Additionally, the RF Index distinguishes C11 from the others (Fig. 6, E and F). To delineate the C7 and C8, one must look carefully at the Transience Index. Here these two cluster form two distinct distributions whose 1 SD limits do not overlap. The differences between the clusters in at least one dimension were found to be statistically significant (e.g., Transience C12 vs. C7, C8, and C11, P < 0.0001; RF Index C11 vs. C7, C8, and C12, P < 0.0001; Transience C7 vs. C8, P < 0.002).
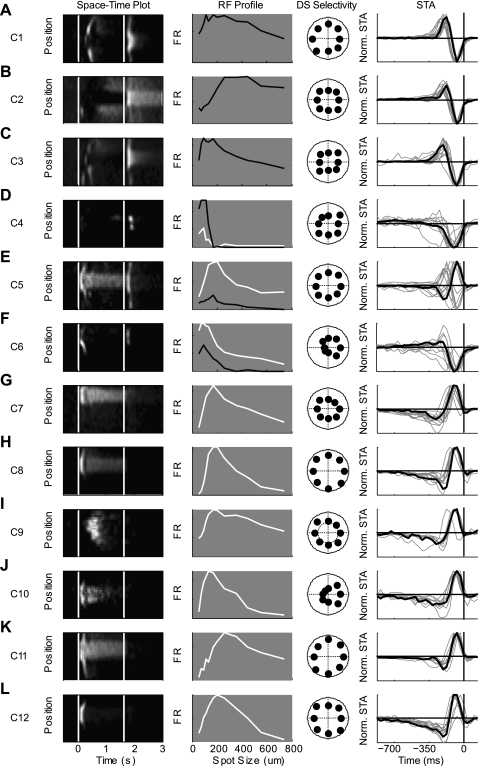
In Fig. 7, we present an example cell from the center of each of the 12 clusters. In the first column, a space-time map depicts the spatial and temporal response properties of each cell type; the second column contains a size-response curve, depicting the spatial receptive field of the neuron, a directional selective polar plot is presented in the third column and the spike triggered average, presented in the fourth column, illustrates the temporal dynamics of the cells response during a flickering checkerboard stimulus. The 12 cell types contained 6 ON cells, 3 OFF cells, and 3 ON-OFF cells. Brief descriptions of the cells follow, in ascending order of their Bias Index.
Fig. 7.
Examples of each cell group. A–L: response properties are shown for an archetypal cell from each cluster (C1–C12), the one closest to the center of each. Column 1: in the left column is a visual representation of each group's receptive field. White colors refer to high firing rates, black to low. Each row is the average of 5 responses to a flashed square for 2 s. Column 2: spatial receptive field determined by relative response during spot of increasing size. Black lines show the responses to OFF stimulus and ON stimulus are indicated with white line. Column 3: polar plots of each cell's response to moving bars in 8 different directions. Column 4: spike triggered average is shown for the example cell (black line) as well as all other neurons lying within 1 SD of the center of that cluster (grey lines). STA, spike-triggered average.
Clusters 1, 2, and 3 (C1, n = 51; C2, n = 31; C3, n = 34) contained the three clusters containing cells that unambiguously preferred decreasing steps in luminance, i.e., OFF cells. The differences between the clusters are apparent in the differences in both their Transience and RF Indexes. C1 and C3 were both transient (Transience = 0.13 and 0.27 s, respectively), while C2 was more sustained (Transience = 0.41 s). C1 and C2 had weak inhibitory surrounds (RF Index = 0.70 and 0.65), while C3 had a RF Index of 0.59, indicating a stronger inhibitory surround. These clusters likely contain the OFF transient and OFF sustained cells described by Pang et al. (2003) and van Wyk et al. (2009), as well as the approach-sensitive retinal ganglion cell of Munch et al. (2009).
Cluster 4 (C4, n = 46) contained cells that had exceptionally strong inhibitory surrounds as indicated by their small RF Index (RF Index = 0.20) and tended to respond to both increments and decrements in light intensity (Bias Index = −0.03). This was not always visible during the marching square stimulus paradigm. It was not uncommon for the space-time plot to contain two bumps at either light ON or light OFF indicating the stimuli when a contrast edge was within the receptive field of the cell. These cells resemble the local edge detectors previously described in rabbits (Roska et al. 2006; Roska and Werblin 2001; van Wyk et al. 2006; Zeck et al. 2005).
Cluster 5 (C5, n = 18) contained cells that were distinguished by their sustained response to increments in light and their transient response to decrements in light intensity (Bias Index = 0.16). The strength of the inhibitory surround was relativity weak in these cells (RF Index = 0.56), and they were not directionally selective (DS Index = 0.03).
The cells of cluster 6 (C6, n = 20) responded to both increments and decrements in light intensity (Bias Index = 0.18). In addition, they are directionally selective (DS Index = 0.24). They tended to have transient responses to changes in light intensity (Transience = 0.14 s) and strong inhibitory surrounds (RF Index = 0.38). This cluster appears to contain mainly the ON-OFF directionally selective ganglion cells, one of the most distinct and well-studied cell types in the retina (Barlow and Hill 1963; Huberman et al. 2009; Oyster and Barlow 1967; Weng et al. 2005).
The last six clusters all responded to increases in light intensity (Bias Index > 0.6). Cluster 7 (C7, n = 37) and cluster 8 (C8, n = 53) both had RF Indexes around the mean of the whole population (RF Index = 0.51 and 0.48, respectively), were not directionally selective (DS Index = 0.03, 0.04) and had relatively short latency of response. What differentiated them was their relative transience to flashes of light. While C7 was sustained, maintaining a high firing rate during the entire duration of the stimulus (Transience = 0.61 s), C8 was less so (Transience = 0.40 s), responding with a sharp rise that was followed by a much lower level of activity until the stimulus ended.
Cluster 9's (C9, n = 17) outstanding feature was its long latency (Latency = 0.50 s). It also had a weak inhibitory surround, was not directionally selective and had a sustained response to increments of light.
Cluster 10 (C10, n = 16), like C6, is directionally selective (DS Index = 0.32). Unlike C6, the cells in this cluster did not respond to decrements in light intensity but only to increases in light intensity (Bias Index = 0.87). The cells of this cluster are predominately the ON directionally selective cells, which play a strong role in controlling image stabilization (Oyster and Barlow 1967; Sun et al. 2006; Yonehara et al. 2009; Yoshida et al. 2001).
Cluster 11 (C11, n = 56), like C7, responded to increases in brightness with a sustained spiking activity lasting the whole presentation time of the stimulus. What distinguishes the two cell types is the size of their receptive fields. C11 had a much larger RF Index (0.63 vs. 0.51).
Cluster 12 (C12, n = 92) are ON transient cells with response properties resembling the ON parasol cells of the rabbit (Roska et al. 2006; Roska and Werblin 2001). They had a RF Index of 0.50 and were not directionally selective.
Variance of Responses
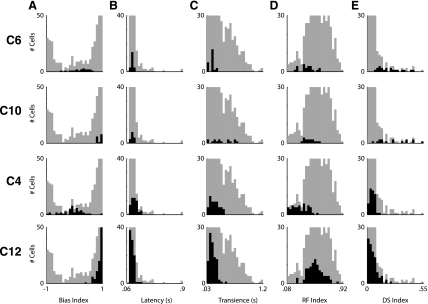
Two distinct characteristics about the distribution of the data are clear (Fig. 2E). First, the data do not form symmetric distribution about a mean, except for the RF Index. Instead, they are either heavily skewed towards one end of the parameter space or towards each extreme. How does the data of a single cluster for the individual parameters vary? An overview of the variance of data within each cluster is apparent in the SD of each cluster center organized by parameter (Fig. 4A; Table 2). For most clusters, the variance was quite small. In particular, focusing on the Bias Index, the variance of the seven clusters that had a Bias Index greater than 0.75 or less than −0.75 had smaller variances then the five clusters that did not. The variance of the 11 clusters that had a Latency <200 ms was at least five times smaller than that of C9, the single cluster with a long latency to respond. The clusters all had similar variances for the Transience and RF Indexes. With regards to the DS Index, the 10 nondirectionally selective clusters had very small variances, while the 2 DS clusters, C6 and C10, showed much greater variance.
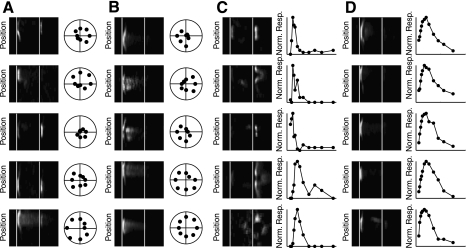
To explore the nature of the variability of data within a single cluster, we took a closer look at the receptive fields of the individual cells making up the directionally selective clusters, C6 and C10, as well as C4, the putative local edge detectors, and C12 containing ON transient-like cells (Fig. 8A). First, we look at C6, putatively containing the ON-OFF directionally selective cells. The mean response of the neurons in this cluster suggests an almost equal preference for increments and decrements of light, together with selectivity for the motion of bars in a particular direction. To highlight their similarity, one can see that each cell within 1 SD of the cluster center is directionally selective (Fig. 8A, column 2). However, there is distinct variability among the responses of the different cells to the presentation of a square of light (Fig. 8A, column 1). In particular, the differences in ON-OFF preference are notable. Where some neurons in the group responded almost exclusively to the presentation of the white square, others clearly preferred the OFF stimulus. How the data representing C6 fit into the overall distribution of the data is evident in Fig. 9, first row. Note that while the distributions for Latency, Transience, and RF Index are quite narrow, those for the Bias and DS Index are quite wide. This variety is qualitatively similar to that seen in a genetically homogeneous subgroup of ON-OFF directionally selective cells (see Fig. 2 of Huberman et al. 2009).
Fig. 8.
Illustration of within cluster variance. Five examples are shown from 4 different clusters to illustrate the variance that exists within each cluster. For each cluster, a space-time plot of the cell to flashed squares illustrates the variation in the receptive field structure of cells lying within a single cluster. Additionally, either the polar plot of the direction selectivity (A and B) or the size-response plot (C and D) is shown to illustrate a response parameter which was qualitatively significant in defining the cell type. A: cluster 6, putative ON-OFF directionally selective cells. B: cluster 10, putative ON directionally selective cells. C: cluster 4, putative local edge detector cells. D: cluster 12. ON transient cells.
Fig. 9.
Distribution of parameters from individual selected clusters. Histograms of each parameter are shown for all the data (grey) and the selected cluster (black) A: distribution of Bias Index. B: distribution of Latency. C: distribution of Transience. D: distribution of RF Index. E: distribution of DS Index. Row 1: cluster 6, putative ON-OFF directionally selective cells. Row 2: cluster 10, putative ON directionally selective cells. Row 3: cluster 4, putative local edge detector cells. Row 4: cluster 12, ON transient cells.
Focusing on C10, putatively containing the ON directionally selective cells, we find that each cell displayed is directionally selective (Fig. 8B, column 2). However, there is distinct variability to the Transience of the response of each cell (Fig. 8B, column 1). This variability is also evident in the histograms of the parameters of the C10 (Fig. 9, second row).
Of the cells making up cluster C4, it is evident that the parameter distinguishing this cluster from the rest is the size of its RF Index. The small RF Index values appear a consequence of these cells' strong inhibitory surrounds. This is evident in the examples shown in Fig. 8C as for each there is a steep negative slope to the size-response curve as each cell is presented with larger spots. In the histogram of the response properties, the distribution for most parameters, except for the ON-OFF Bias Index, is narrow.
Within cluster C12, containing ON transient cells, it is evident that each cell is an ON cell and responds in a transient manner (Fig. 8D, column 1). The cells making up this group showed a remarkable consistency on response profiles. This is evident in both space-time maps and the size response profiles (Fig. 8D). Unlike the directionally selective cells or the putative local edge detectors this group of cells showed relatively narrow distribution of response properties for each of the parameters measured (Fig. 9, fourth row).
The relatively broad distributions of responses within a single cluster have two likely sources. One is that the stimuli presented do not stimulate the cells best trigger feature in a robust manner. Thus the measured responses contain variable responses.
The second possibility is that there is real biological variability in the response profile of the cells of a single type. This is supported by the recording made from genetically marked cells (Huberman et al. 2009; Kim et al. 2008).
DISCUSSION
Here we have quantitatively clustered and described 12 distinct visual channels of the mouse retina. The experimental sample included 471 retinal cells, while the stimulus protocol and parameter space were quite broad: including temporal and spatial information about each cell's receptive field and some nonlinear response characteristics (e.g., direction selectivity). Although we relied on quantitative methodologies that made few assumptions about the structure of the data, certain aspects of the study did require some arbitrary choices to be made. These are discussed below.
Choosing Stimulus Sets
As discussed earlier (see Introduction) the choice of stimulus set was a hybrid between severely reduced stimuli used for tractability and the other extreme of truly natural scenes. As always, this choice of stimulus characteristics may have limited or biased the response repertoire of the cells. Certain nonlinear characteristics (e.g., “looming”) are not directly represented. Furthermore, the responses to even a seemingly simple series of stimuli can change as the parameter is varied. One clear example is the measure of ON vs. OFF Bias Index during spots of increasing sizes. Cells often show different responses depending on the size of the spot presented. This is even true for genetically labeled, anatomically distinct cells of the JamB and DRD4-GFP retinas (Huberman et al. 2008; Kim et al. 2008). The ganglion cells labeled in the JamB mouse show a predominately OFF response to spots <250 μm, predominately ON responses to spots >400 μm and ON-OFF responses to spots between 250 and 400 μm [see Fig. 2 in Kim et al. (2008)]. Similar variability of responses is evident in homogenous group of ON-OFF directionally selective cells labeled in the DRD4-GFP mice. In compensation for these limitations, the stimuli are easily described and reproduced. An important consideration is the future need for reproducibility. The cell types discriminated here can readily be specified in terms of the responses of the cell to each of the stimuli tested (Table 2).
In addition to the visual response properties, the intrinsic electrophysiological properties of ganglion cells have been used to discriminate between cell types (Devries and Baylor 1997; O'Brien et al. 2002; Zeck and Masland 2007). Although we found some consistency among the autocorreleograms and median interspike intervals of the spike trains during the presentation of a flickering checkerboard, we did not find these as robust of a determinant of cell type as the measured response to the different light stimuli. It does not appear that in the mouse these measures are as useful as in the rabbit.
Selection of Clustering Methods
Cluster analysis aims to identify groups of similar objects and uncover distributed patterns in large data sets. Difficulties in applying clustering routines to a particular data set occur as any inherent structure is unknown. As each clustering algorithm is essentially using a predetermined model of how data should be grouped, each routine will have its own biases. To limit the bias of our results, we performed two separate comparisons. First, we tested the performance of 7 clustering algorithms on 27 artificial data sets that tested each algorithm's ability to handle different levels of noise, number of clusters, number of dimensions, and shapes of the clusters (Supplemental Figs. 1 and 2). This analysis allowed us to determine which algorithms are the most reliable across a wide variety of data set geometries. We found that the Fuzzy-GK algorithm was the most reliable and focused our analysis on its optimal solution.
Additionally, we assessed how robust the clustering of the visual response properties were by comparing the optimal partitioning solution from different clustering methods (Fig. 3 and 4). We compared the Fuzzy-GK, K-Means, PAM, and AP algorithms due to their diverse assumptions of how to cluster data and their reliability in our clustering assessment (Supplemental Fig. 1). We found that the different algorithms partitioned the neurons in a similar manner. This suggests that the clustering of the data was robust and that the solution of the Fuzzy-GK algorithm was a good approximation of the correct groups that exist within the mouse retina.
The second stage of any clustering methodology, independent of the algorithm employed, is to decide which of the solutions is best. This is accomplished by applying a validation procedure (Bezdek and Pal 1998; Milligan and Cooper 1985; Wang and Zhang 2007). As with the clustering routines themselves, the validity measures also make different assumptions about what is the best solution. To avoid prejudice for any single validity measure, we compared the results of different indexes; on the same 27 data sets we tested the clustering algorithms (Supplemental Fig. 3). We then used a combination of the best performing indexes for each algorithm (Table 1). We found that the combination of four indexes applicable to the Fuzzy-GK algorithm allowed us to find the correct number of clustering in the artificial data sets 91.3% of the time. Other then the analysis presented in Supplemental Fig. 3, we favor the results of the fuzzy partitioning algorithms as distinct geometric and fuzzy-set arguments, or a combination, can be used to determine the best solution (Wang and Zhang 2007).
Functional Classifications of Mouse Ganglion Cells
The ultimate goal of this study, and others like it, is a description of the fundamental language of vision: the words or phrases by which the visual world is communicated to the brain. We do not pretend to have achieved a definitive statement of them here, but we can begin to see the rough shape that such a “dictionary” would have. Our classification of the output of the mouse retina into 12 distinct channels is in rough agreement with the number of distinct anatomically defined ganglion cells (Badea and Nathans 2004; Coombs et al. 2006; Kong et al. 2005; Sun et al. 2002) and the basic physiological response properties previously described (Carcieri et al. 2003; Kim et al. 2008; Pang et al. 2003; Weng et al. 2005; Zeck et al. 2005).
The total population of ganglion cells could accommodate a dozen subtypes and still achieve coverage of the retina by each. Electron microscopic counts of axons in the optic nerve of C57BL6 mice gave a total of 44,860 ganglion cell axons. Assuming for simplicity that these ganglion cells are distributed uniformly across a 17-mm2 retina (Jeon et al. 1998), the density of ganglion cells on the retinal surface is 2,639 cells/mm2. How many cell types could this number of ganglion cells accommodate? For comparison, two mouse lines with single ganglion cell types labeled, include the JamB cells and the posterior ON-OFF directionally selective cell; for these cases, both the dendritic field size and density of the single ganglion cell types are known (Huberman et al. 2009; Kim et al. 2008). The JamB cells have a spatial density of ∼200 cells/mm2 and one can estimate the dendritic field area at ∼25,000 um2 [see Figs. 1, C, G, and H in Kim et al. (2008)]. This yields a coverage factor (cell density × dendritic area) of five. The reported density of the posterior motion sensitive ON-OFF directionally selective cells is ∼250 cells/mm2 with an estimated dendritic field area of ∼30,000 um, giving a coverage factor of 7.5. If these values were representative for all retinal ganglion cells in the mouse, the number of retinal ganglion cell types would range from ∼10 (assuming other cells to be like the ON-OFF directionally selective cells) to 13 for the JamB cells (2,639 total ganglion cells/mm2 divided by 250 or 200 cells/mm2, respectively). This value would appear to be in excellent agreement with our results and previous anatomical ones.
However, the agreement may well be fortuitous, such that the true number of types is even larger. The JamB and ON-OFF directionally selective cells are two of the smaller mouse ganglion cells so far described (Badea and Nathans 2004; Coombs et al. 2006; Kong et al. 2005; Sun et al. 2002; Volgyi et al. 2009). Since larger cells can cover the retinal surface using fewer cells, e.g., ∼20 cells/mm2 for cells of CB2-GFP:b2nAChR−− mice (Huberman et al. 2008), larger cells would allow for more cell types than the 12 predicted here. If we assume that the densities of individual cell types are evenly spaced between the smallest and largest ganglion cells, we obtain an average density of ∼135 cells/mm2 across the different cell types. This would suggest a total number of cell types of ∼20. Which of these values is correct will depend on the sizes of the dendritic arbors of the cells and on the extent of their overlaps, but published surveys of ganglion cell morphology make it seem clear that the true value falls in the window between 12 and 20 cell types. The estimate derived here is at the low end of this range, partly for reasons that are obvious: as noted above, there are parameter dimensions we failed to explore. The most notable of these is color. Mice, like many small terrestrial mammals, express a dual gradient in opsins, with M opsin concentrated superiorly and the S opsin concentrated inferiorly. A set of ganglion cells has indeed been shown to be selective for different wavelengths of light in guinea pig (Yin et al. 2009). In addition, luminance levels were not explored and cells have been shown to partition functionally along luminance sensitivity curves (Volgyi et al. 2004). Thus it is likely that our clustering results group cells that could, with a wider choice of stimuli, display distinct response properties. Thus we reject the null hypothesis that the physiological properties of the cells form a continuum: the cells can be sorted by unsupervised methods into a large number of distinct clusters. However, it remains for future studies to determine their definitive number and to match them with morphologically defined types.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kim Fetchel, David Balya, Tamas Szikra, and Janine Hall for carefully reading the manuscript.
Present address for R. Masland: Howe Laboratory of Ophthalmology, Department of Ophthalmology, Mass. Eye and Ear Infirmary, Boston, Massachusetts.
REFERENCES
- Abbott LF, Chance FS. Rethinking the taxonomy of visual neurons. Nat Neurosci 5: 391–392, 2002 [DOI] [PubMed] [Google Scholar]
- Babuska R, Veen PJvd, Kaymak U. Improved covariance estimation for Gustafson-Kessel clustering. In: IEEE International Conference on Fuzzy Systems, 2002, p. 1081–1085 [Google Scholar]
- Badea TC, Nathans J. Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol 480: 331–351, 2004 [DOI] [PubMed] [Google Scholar]
- Barlow HB, Hill RM. Selective sensitivity to direction of movement in ganglion cells of the rabbit retina. Science 139: 412–414, 1963 [DOI] [PubMed] [Google Scholar]
- Berson DM. Retinal ganglion cell types and their central projections. In: The Senses: a Comprehensive Reference, edited by Bushnell MC, Smith DV, Beauchamp GK, Firestein SJ, Dallos P, Oertel D, Masland RH, Albright T, Kaas JH, Gardner EP, Basbaum AI. San Diego, CA: Academic GW, 2008, p. 491–520 [Google Scholar]
- Berson DM. Strange vision: ganglion cells as circadian photoreceptors. Trends Neurosci 26: 314–320, 2003 [DOI] [PubMed] [Google Scholar]
- Beyer KS, Goldstein J, Ramakrishnan R, Shaft U. When is “nearest neighbor” meaningful? Lecture Notes Comp Sci 1540: 217–235, 1999 [Google Scholar]
- Bezdek JC, Pal NR. Some new indexes of cluster validity. IEEE Trans Syst Man Cybern B Cybern 28: 301–315, 1998 [DOI] [PubMed] [Google Scholar]
- Boycott BB, Wassle H. The morphological types of ganglion cells of the domestic cat's retina. J Physiol 240: 397–419, 1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carcieri SM, Jacobs AL, Nirenberg S. Classification of retinal ganglion cells: a statistical approach. J Neurophysiol 90: 1704–1713, 2003 [DOI] [PubMed] [Google Scholar]
- Coombs J, van der List D, Wang GY, Chalupa LM. Morphological properties of mouse retinal ganglion cells. Neuroscience 140: 123–136, 2006 [DOI] [PubMed] [Google Scholar]
- Dacey DM. Physiology, morphology and spatial densities of identified ganglion cell types in primate retina. Ciba Found Symp 184: ; discussion 28–34, 63–70, 199412–28 [DOI] [PubMed] [Google Scholar]
- Davies DL, Bouldin DW. A cluster separation measure. IEEE Trans Pattern Anal Machine Intell 1: 95–104, 1979 [PubMed] [Google Scholar]
- DeVries SH. Correlated firing in rabbit retinal ganglion cells. J Neurophysiol 81: 908–920, 1999 [DOI] [PubMed] [Google Scholar]
- Devries SH, Baylor DA. Mosaic arrangement of ganglion cell receptive fields in rabbit retina. J Neurophysiol 78: 2048–2060, 1997 [DOI] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Storck DG. Pattern Classification. New York: Wiley Interscience, 2001 [Google Scholar]
- Enroth-Cugell C, Robson JG. The contrast sensitivity of retinal ganglion cells of the cat. J Physiol 187: 517–552, 1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhall AL, Burlingame CA, Narasimhan R, Harris RA, Puchalla JL, Berry MJ., II Selectivity for multiple stimulus features in retinal ganglion cells. J Neurophysiol 96: 2724–2738, 2006 [DOI] [PubMed] [Google Scholar]
- Frey BJ, Dueck D. Clustering by passing messages between data points. Science 315: 972–976, 2007 [DOI] [PubMed] [Google Scholar]
- Fu Y, Liao HW, Do MT, Yau KW. Non-image-forming ocular photoreception in vertebrates. Curr Opin Neurobiol 15: 415–422, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425: 917–925, 2003 [DOI] [PubMed] [Google Scholar]
- Gustafson EE, Kessel WC. Fuzzy clustering with a fuzzy covariance matrix. In: IEEE CDC. San Diego, CA: 1979, p. 761–766 [Google Scholar]
- Hochstein S, Shapley RM. Quantitative analysis of retinal ganglion cell classifications. J Physiol 262: 237–264, 1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Manu M, Koch SM, Susman MW, Lutz AB, Ullian EM, Baccus SA, Barres BA. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron 59: 425–438, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman AD, Wei W, Elstrott J, Stafford BK, Feller MB, Barres BA. Genetic identification of an on-off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron 62: 327–334, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L, Arabie P. Comparing partitions. J Classification 2: 193–218, 1985 [Google Scholar]
- Jeon CJ, Strettoi E, Masland RH. The major cell populations of the mouse retina. J Neurosci 18: 8936–8946, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IJ, Zhang Y, Yamagata M, Meister M, Sanes JR. Molecular identification of a retinal cell type that responds to upward motion. Nature 452: 478–482, 2008 [DOI] [PubMed] [Google Scholar]
- Kong JH, Fish DR, Rockhill RL, Masland RH. Diversity of ganglion cells in the mouse retina: unsupervised morphological classification and its limits. J Comp Neurol 489: 293–310, 2005 [DOI] [PubMed] [Google Scholar]
- Krzanowski WJ, Lai YT. A criterion for determining the number of groups in a data set using sum-of-squares clustering. Biometrics 44: 23–34, 1988 [Google Scholar]
- Levick WR. Receptive fields and trigger features of ganglion cells in the visual streak of the rabbits retina. J Physiol 188: 285–307, 1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masland RH. Neuronal diversity in the retina. Curr Opin Neurobiol 11: 431–436, 2001 [DOI] [PubMed] [Google Scholar]
- Mechler F, Ringach DL. On the classification of simple and complex cells. Vision Res 42: 1017–1033, 2002 [DOI] [PubMed] [Google Scholar]
- Milligan GW, Cooper MC. An examination of procedures for determining the number of clusters in a data set. Psychometrika 50: 159–179, 1985 [Google Scholar]
- Munch TA, da Silveira RA, Siegert S, Viney TJ, Awatramani GB, Roska B. Approach sensitivity in the retina processed by a multifunctional neural circuit. Nat Neurosci 2009 [DOI] [PubMed] [Google Scholar]
- O'Brien BJ, Isayama T, Richardson R, Berson DM. Intrinsic physiological properties of cat retinal ganglion cells. J Physiol 538: 787–802, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyster CW, Barlow HB. Direction-selective units in rabbit retina: distribution of preferred directions. Science 155: 841–842, 1967 [DOI] [PubMed] [Google Scholar]
- Pang JJ, Gao F, Wu SM. Light-evoked excitatory and inhibitory synaptic inputs to ON and OFF alpha ganglion cells in the mouse retina. J Neurosci 23: 6063–6073, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand W. Objective criteria for evaluation of clustering methods. J Am Stat Assoc 66: 846–850, 1971 [Google Scholar]
- Reid RC, Victor JD, Shapley RM. The use of m-sequences in the analysis of visual neurons: linear receptive field properties. Vis Neurosci 14: 1015–1027, 1997 [DOI] [PubMed] [Google Scholar]
- Roska B, Molnar A, Werblin FS. Parallel processing in retinal ganglion cells: how integration of space-time patterns of excitation and inhibition form the spiking output. J Neurophysiol 95: 3810–3822, 2006 [DOI] [PubMed] [Google Scholar]
- Roska B, Werblin F. Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature 410: 583–587, 2001 [DOI] [PubMed] [Google Scholar]
- Segev R, Goodhouse J, Puchalla J, Berry MJ., II Recording spikes from a large fraction of the ganglion cells in a retinal patch. Nat Neurosci 7: 1154–1161, 2004 [DOI] [PubMed] [Google Scholar]
- Segev R, Puchalla J, Berry MJ., II Functional organization of ganglion cells in the salamander retina. J Neurophysiol 95: 2277–2292, 2006 [DOI] [PubMed] [Google Scholar]
- Shoham S, Fellows MR, Normann RA. Robust, automatic spike sorting using mixtures of multivariate t-distributions. J Neurosci Methods 127: 111–122, 2003 [DOI] [PubMed] [Google Scholar]
- Siegert S, Scherf BG, Del Punta K, Didkovsky N, Heintz N, Roska B. Genetic address book for retinal cell types. Nat Neurosci 12: 1197–1204, 2009 [DOI] [PubMed] [Google Scholar]
- Stasheff SF. Emergence of sustained spontaneous hyperactivity and temporary preservation of OFF responses in ganglion cells of the retinal degeneration (rd1) mouse. J Neurophysiol 99: 1408–1421, 2008 [DOI] [PubMed] [Google Scholar]
- Sun W, Deng Q, Levick WR, He S. ON direction-selective ganglion cells in the mouse retina. J Physiol 576: 197–202, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li N, He S. Large-scale morphological survey of mouse retinal ganglion cells. J Comp Neurol 451: 115–126, 2002 [DOI] [PubMed] [Google Scholar]
- van Wyk M, Taylor WR, Vaney DI. Local edge detectors: a substrate for fine spatial vision at low temporal frequencies in rabbit retina. J Neurosci 26: 13250–13263, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wyk M, Wassle H, Taylor WR. Receptive field properties of ON- and OFF-ganglion cells in the mouse retina. Vis Neurosci 26: 297–308, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verleysen M, Francois D. The curse of dimensionality in data mining and time series prediction. Lecture Notes Comp Sci 3512: 758–770, 2005 [Google Scholar]
- Volgyi B, Chheda S, Bloomfield SA. Tracer coupling patterns of the ganglion cell subtypes in the mouse retina. J Comp Neurol 512: 664–687, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgyi B, Deans MR, Paul DL, Bloomfield SA. Convergence and segregation of the multiple rod pathways in mammalian retina. J Neurosci 24: 11182–11192, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang Z. ON fuzzy validity indices. Fuzzy Sets Syst 158: 2095–2117, 2007 [Google Scholar]
- Weng S, Sun W, He S. Identification of ON-OFF direction-selective ganglion cells in the mouse retina. J Physiol 562: 915–923, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L, Smith RG, Sterling P, Brainard DH. Physiology and morphology of color-opponent ganglion cells in a retina expressing a dual gradient of S and M opsins. J Neurosci 29: 2706–2724, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonehara K, Ishikane H, Sakuta H, Shintani T, Nakamura-Yonehara K, Kamiji NL, Usui S, Noda M. Identification of retinal ganglion cells and their projections involved in central transmission of information about upward and downward image motion. PLos One 4: e4320, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Watanabe D, Ishikane H, Tachibana M, Pastan I, Nakanishi S. A key role of starburst amacrine cells in originating retinal directional selectivity and optokinetic eye movement. Neuron 30: 771–780, 2001 [DOI] [PubMed] [Google Scholar]
- Zeck GM, Masland RH. Spike train signatures of retinal ganglion cell types. Eur J Neurosci 26: 367–380, 2007 [DOI] [PubMed] [Google Scholar]
- Zeck GM, Xiao Q, Masland RH. The spatial filtering properties of local edge detectors and brisk-sustained retinal ganglion cells. Eur J Neurosci 22: 2016–2026, 2005 [DOI] [PubMed] [Google Scholar]
- Zhang DQ, Stone JF, Zhou T, Ohta H, McMahon DG. Characterization of genetically labeled catecholamine neurons in the mouse retina. Neuroreport 15: 1761–1765, 2004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.