Abstract
Sos-1, a guanine nucleotide exchange factor (GEF), eps8 and Abi1, two signaling proteins, and the lipid kinase phosphoinositide 3-kinase (PI3-K), assemble in a multimolecular complex required for Rac activation leading to actin cytoskeletal remodeling. Consistently, eps8 –/– fibroblasts fail to form membrane ruffles in response to growth factor stimulation. Surprisingly, eps8 null mice are healthy, fertile, and display no overt phenotype, suggesting the existence of functional redundancy within this pathway. Here, we describe the identification and characterization of a family of eps8-related proteins, comprising three novel gene products, named eps8L1, eps8L2, and eps8L3. Eps8Ls display collinear topology and 27–42% identity to eps8. Similarly to eps8, eps8Ls interact with Abi1 and Sos-1; however, only eps8L1 and eps8L2 activate the Rac-GEF activity of Sos-1, and bind to actin in vivo. Consistently, eps8L1 and eps8L2, but not eps8L3, localize to PDGF-induced, F-actin–rich ruffles and restore receptor tyrosine kinase (RTK)-mediated actin remodeling when expressed in eps8 –/– fibroblasts. Thus, the eps8Ls define a novel family of proteins responsible for functional redundancy in the RTK-activated signaling pathway leading to actin remodeling. Finally, the patterns of expression of eps8 and eps8L2 in mice are remarkably overlapping, thus providing a likely explanation for the lack of overt phenotype in eps8 null mice.
INTRODUCTION
Small GTPases act as binary switches that cycle between inactive GDP-bound and active GTP-bound states (Campbell et al., 1998; Bar-Sagi and Hall, 2000). Among small GTPases, Rac, which belongs to the Rho-family of GTPases, is essential in the pathway originating from receptor tyrosine kinases (RTKs) and leading to the reorganization of the actin cytoskeleton (Van Aelst and D'Souza-Schorey, 1997; Hall, 1998; Kaibuchi et al., 1999).
The activation of small GTPases depends on guanine nucleotide exchange factor (GEFs; Cherfils and Chardin, 1999; Schmidt and Hall, 2002). The large number of GEFs, far exceeding that of GTPases, suggests that a single GTPase is under the control of multiple activators (Zohn et al., 1998; Schmidt and Hall, 2002). GEFs are themselves regulated at multiple levels. The catalytic domain present in GEFs for RhoGTPases is the Dbl homology domain (DH), which is almost invariably preceded by a pleckstrin homology (PH) domain, suggesting an integrated function for the DH-PH tandem (Whitehead et al., 1997). Accordingly, the DH-PH module frequently represents the minimal catalytic unit (Crompton et al., 2000; Hoffman and Cerione, 2002). However, in other cases, the DH-PH might function as a self-inhibitory device, as suggested by findings that isolated PH domains bind to and inhibit the activity of some DH domain (Nimnual et al., 1998; Das et al., 2000; Paduch et al., 2001).
Additional regulatory complexity is implied by the observation that GEFs can assemble into multimolecular complexes (Cherfils and Chardin, 1999; Schmidt and Hall, 2002). An example is provided by the complex formed by eps8, Abi1, p85 (the regulatory subunit of phosphatidylinositol 3 kinase [PI3-K]), and the GEF Sos-1 (Fazioli et al., 1993; Biesova et al., 1997; Scita et al., 1999; Di Fiore and Scita, 2002; Innocenti et al., 2002, 2003; Nimnual and Bar-Sagi, 2002). Sos-1 belongs to a class of exchange factors endowed with dual catalytic ability, on Ras and Rac GTPases (Nimnual and Bar-Sagi, 2002; Schmidt and Hall, 2002). The catalytic specificity of Sos-1 is dictated by its engagement into different protein complexes (Innocenti et al., 2002). Thus, when coupled to the adaptor molecule Grb2, Sos-1 is recruited to the plasma membrane by activated RTK and catalyzes nucleotide exchange on Ras. Conversely, Sos-1, when in complex with eps8, Abi1, and p85, displays Rac-GEF activity (Innocenti et al., 2002).
Eps8 is critical for the activation of the eps8-Abi1-p85-Sos-1 complex. Consistently, disruption of the complex, by genetic removal of eps8 abrogates Rac activation and Rac-dependent actin remodeling induced by various stimuli (Scita et al., 1999). Within the complex, Abi1 acts as a scaffold on which eps8, p85, and Sos-1 assemble (Scita et al., 1999). This topological arrangement is thought to facilitate a low affinity interaction between eps8 and Sos-1, which is sufficient for the catalytic activation of Sos-1 (Scita et al., 2001). Thus, two features are critical to the function of eps8: the association with Abi1 via its SH3 domain, and the activating interaction with Sos-1. Furthermore, eps8 binds directly to F-actin and directs in vivo the localization of Abi1 and Sos-1 to F-actin–rich structures (Scita et al., 2001). Thus, the formation of an eps8-based macromolecular complex is essential for both Sos-1 catalytic activation and localization of the complex to sites where its activity is required.
Despite the essential role of eps8 in signaling, eps8 null mice are healthy, fertile, and devoid of any obvious phenotypic alteration (Scita et al., 1999). This is in contrast with the early embryonic lethal phenotype exhibited by null mice for other proteins involved in the same signaling pathway, such as Rac1, and Abi1 (Sugihara et al., 1998, and A.M. Pendergast, personal communication). An obvious explanation for the lack of phenotype of the eps8 –/– mice is the existence of redundant functions in vivo. Here, we describe the identification and characterization of a family of eps8-related proteins (eps8Ls, standing for eps8-like) and provide biochemical and biological evidence that members of this family act redundantly in the pathway leading to Rac activation. Thus, the eps8Ls constitute a novel family of proteins, which links growth factor stimulation to actin dynamics and provide a safety device to ensure propagation of signals leading to the activation of Rac and Rac-dependent pathways.
MATERIALS AND METHODS
Cloning of EPS8-like Genes
A UNI-ZapXR Human Placenta library (CLONTECH, Palo Alto, CA), a pCEV-29–based library from M426 human fibroblasts (a kind gift of T. Miki), and a λ phage (λ NM1149) library derived from HeLa cells (a kind gift of C. Schneider) were used to obtain the entire open reading frames (ORFs) of human EPS8L1, EPS8L2, and EPS8L3, respectively. The ORFs of the EPS8Ls were individually cloned into pBSK+ vectors and subcloned into pCDNA3.1 flag-containing eucaryotic expression vectors. The mouse sequences were obtained by sequencing of EST clones obtained from the IMAGE consortium and by RT-PCR–based cloning from E16.5 dpc mouse head RNA. The human and mouse sequences were deposited in GenBank (AY074928, AY074931, AY074929, AY074930, AY074932).
In Situ Hybridization
Timed matings were set up between adult C57BL6 males and females. The day of plug was considered 0.5 d postconception (dpc). Embryos were dissected at the indicated stages, processed, and paraffin-embedded. In situ hybridization was carried out on 5-μm sagittal sections as previously described (Rugarli et al., 1993). Sense and antisense 35S-labeled riboprobes were transcribed from 600-base pair mouse cDNA fragments, corresponding to positions 1480–2100 of eps8L1, 1560–2100 of eps8L2, and 1630–2250 of eps8L3, using T3 and T7 RNA polymerase (numbering is according to the GenBank-deposited sequences).
Northern Blot and Semiquantitative RT-PCR
Total RNA was extracted from adult mouse tissues using Triazol (GIBCO BRL, Rockville, MD) according to the manufacturer's instructions. Northern blotting was performed using 10 μg of total RNA according to standard procedures. For RT-PCR, oligo dT-primed first-strand synthesis was performed using Superscript II (GIBCO BRL) on 1 μg of total RNA. PCR was performed with specific primers located in the 5′ coding region of the genes. Cycle number was determined for each gene individually. The sequences of the primers and the exact PCR conditions for each primer set are available on request.
Antibodies and Biochemical Procedures
Monoclonal antibodies to flag (M2, Sigma, St. Louis, MO), actin (Sigma), and eps8 (Transduction Laboratories, Lexington, KY) were obtained commercially. The polyclonal anti-Abi1 has been previously described (Biesova et al., 1997). Anti-myc (9E10) was produced in synthetic medium. Standard procedures of protein analysis (Western blotting, immunoprecipitations, and coimmunoprecipitations) were performed as previously described (Fazioli et al., 1993).
In vitro GEF activity toward Rac was performed as described (Scita et al., 1999). Data are the mean ± SE of at least three independent experiments performed in triplicates. Results are expressed as the percentage of [3H]GDP released after 20 min relative to time 0, after subtracting the background counts released in control reactions (obtained in the presence of immunoprecipitates performed with control IgGs).
Actin cosedimentation assays were performed as described (Scita et al., 2001). Briefly, monomeric rabbit G-acting (1 mg/ml) was induced to polymerize at 37°C in F-actin buffer (5 mM Tris/HCl, pH 7.8, 1 mM ATP, 0.5 mM DDT, 0.2 mM CaCl2, 0.2 mM MgCl2, and 100 mM KCl). Recombinant and purified GST-fusion proteins were subsequently incubated with 10 μM F-actin for 45 min at 37°C, flowed by ultracentrifugation. Equal amount of starting materials, supernatants, and pellets were solubilized in loading buffer, boiled, and resolved on SDS-PAGE.
Transfection Procedures and Indirect Immunofluorescence
Cells seeded on gelatin were transfected with the indicated expression vectors using the LIPOfectamine reagent (GIBCO BRL), according to the manufacturer's instructions. Alternatively, nuclei of quiescent fibroblasts were injected with 100 ng/ml the appropriate expression vectors. Three to 6 h later, cells were processed for immunostaining. At least 100 microinjected cells were analyzed in each experiment. For indirect immunofluorescence cells were fixed in 4% paraformaldeyde for 10 min, permeabilized in 0.1% Triton X-100 and 2% BSA for 30 min, and then incubated with the primary antibody for 45 min, followed by the secondary antibody for 30 min. F-actin was detected by staining with rhodamine-conjugated phalloidin (Sigma), at a concentration of 6.7 U/ml (ca. 0.22 μm). Where indicated, cells were treated with PDGF (10 ng/ml) for 10 min.
RESULTS
A Family of eps8-related Proteins Regulates the RacGEF Activity of Sos-1
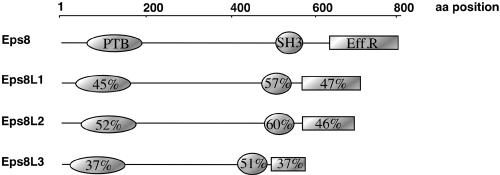
An in silico screening of the expressed sequence tags (EST) database led to the identification of three human genes displaying high homology to the human EPS8 gene. These genes were named EPS8-Like1, 2, and 3 (EPS8L1, 2, and 3), respectively (Mongiovi et al., 1999; Tocchetti et al., 2003). Full-length clones of EPS8Ls were obtained by screening of human cDNA libraries. The predicted proteins displayed collinear topology and an overall amino acid identity, among themselves, ranging from 27 to 42%. Eps8Ls share a common modular organization, consisting of three domains: an N-terminal region (37–52% identity with eps8), whose predicted fold resembles a phosphotyrosine binding domain (PTB; Tocchetti et al., 2003; Calderwood, 2003); a central SH3 domain (51–60% identity with eps8); and a C-terminal region (37–47% identity with eps8) similar to the “effector region” of eps8 (Scita et al., 2001) (Figure 1).
Figure 1.
The eps8 family. Schematic representation of human eps8 and of the three related proteins eps8L1, eps8L2, and eps8L3. The PTB and SH3 domains were identified using the SMART module. The effector region (Eff.R., amino acids 649–822 in eps8) was previously functionally identified in eps8 (Scita et al., 2001). The percentages of identity between eps8 and individual eps8Ls, in the three domains, are shown, as calculated by pairwise alignments using a BLOSUM62 matrix.
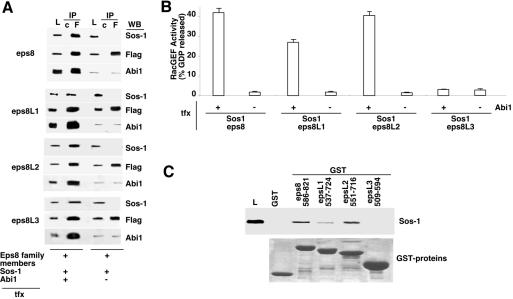
The conserved modular organization within the family suggested overlapping biochemical and functional properties among its members. Eps8 is essential for the activation of the RacGEF activity of the eps8-Abi1-p85-Sos-1 complex. Thus, we initially tested the ability of eps8Ls to bind to Abi1 in vivo. As shown in Figure 2, all eps8 family members, expressed as flag-tagged proteins, coimmunoprecipitated with endogenous Abi1.
Figure 2.
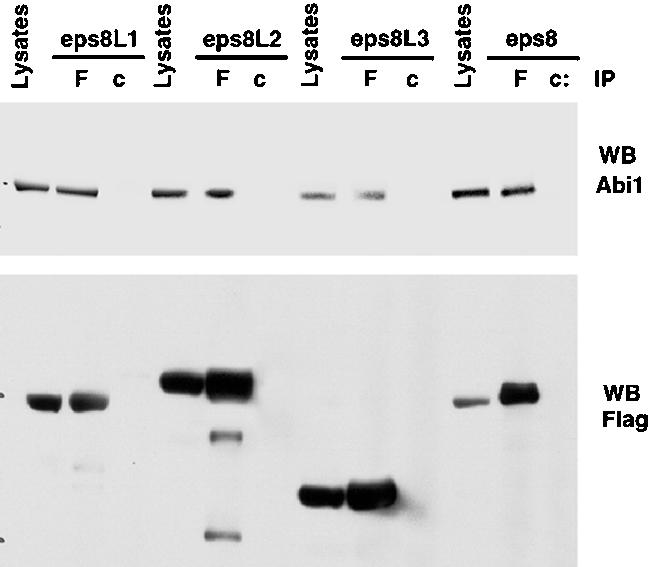
Eps8Ls coprecipitate with Abi1. 293T cells were transfected with flag-tagged eps8Ls (as indicated at the top, Tfx), followed by immunoprecipitation (IP) with an antiflag (F) or control IgGs (c). Lysates and immunoprecipitates were immunoblotted (WB) with the indicated antibodies.
We next assessed whether RacGEF activity could be recovered in eps8Ls immunoprecipitates, under conditions in which eps8Ls-Abi1-Sos-1 complexes are formed. To do this, we engineered a model system in 293T cells, in which flag-tagged eps8 family members were individually transfected together with Sos-1 and either with or without an Abi1 expression vector (Figure 3, A and B). We have previously demonstrated that an endogenous complex, containing eps8, Abi1, and Sos-1, can be recovered by immunoprecipitation with anti-eps8 antibodies from untransfected cells (Innocenti et al., 2002). However, in order to perform enzymatic assays for Rac-GEF activity, recovery of sizable amounts of the complex can be obtained only under conditions of overexpression of the three components (Scita et al., 1999). In addition, the low amounts of endogenous Abi1 (which scaffolds together eps8 and Sos-1) in 293T cells makes the detection of an eps8/Abi1/Sos-1 complex rather difficult, even in the presence of overexpressed eps8 and Sos-1 (Figure 3A, top left panel). However, this can be turned into an experimental advantage, because the simultaneous overexpression of Abi1 allows for the detection of the complex in an Abi1-dependent manner. Indeed, under conditions of triple transfection, all of the eps8 family members could be found in association with Abi1 and Sos-1 (Figure 3A). In addition, the formation of the complex was Abi1 dependent, as shown by lack of coimmunoprecipitation between eps8 family members and Sos-1, in the absence of overexpressed Abi1 (Figure 3A).
Figure 3.
Rac-GEF activity of complexes containing various eps8 family members. (A) 293T cells were cotransfected with the constructs (Flag-tagged eps8 family members, Sos-1 and Abi1) indicated at the bottom. The individual eps8 family member transfected in each set is indicated on the left. Total cellular lysates were immunoprecipitated with an anti-Flag antibody (F) or control IgGs (c). Lysates (L) and immunoprecipitates (IP) were immunoblotted with the indicated antibodies (WB). (B) RacGEF activity was measured on aliquots of the IPs shown in A. For each set, the combination of transfected expression vectors (eps8 family members, Sos-1, with or without Abi1) is indicated at the bottom. Data are expressed as described in MATERIALS AND METHODS. (C) Immobilized, C-terminal fragments of various eps8 family members (the amino acid boundaries are indicated), or GST alone, were incubated with total cellular lysates from Sos-1–expressing 293T cells. Bound proteins were resolved by SDS-PAGE and immunoblotted with anti-Sos-1 (top panel). A Comassie staining, to verify equal loading of GST fusions, is also shown (bottom panel, GST-proteins).
We then tested Rac-GEF activity in antiflag immunoprecipitates from the triple transfectants (Figure 3B). Eps8-, eps8L1-, and eps8L2-, but not eps8L3-containing immunocomplexes displayed RacGEF activity (Figure 3B). Of note, the presence of the Rac-GEF activity, in the immunocomplexes, was dependent on the simultaneous transfection with Abi1, as predicted (Figure 3B).
The lack of Rac-GEF activity in eps8L3-containing complexes indicated that some additional molecular characteristics, shared by eps8, eps8L1, and eps8L2, are not present in eps8L3. We have previously demonstrated that the C-terminal region of eps8 (effector region) can contract a direct low-affinity interaction with Sos-1 (Scita et al., 2001). We further showed that this interaction is necessary and sufficient to cause the catalytic activation of Sos-1 as a Rac-GEF, both in vitro and in vivo (Scita et al., 2001). Using purified proteins, we showed in vitro that holo-eps8 cannot interact with Sos-1; however, the interaction can be unmasked if the C-terminal fragment of eps8 is used (Scita et al., 2001). Although this probably reflects the need for some conformational change in vivo, which affects eps8 in the eps8/Abi1/Sos-1 complex and which is mimicked by the isolation of the effector region from the context of the holoprotein, it provided an assay to investigate biochemical differences within the eps8 family. Thus, we tested the ability of the C-termini of eps8L1, eps8L2, and eps8L3 to associate to Sos-1 in vitro. Native Sos-1 could be recovered onto immobilized GST-C-terminal fragments of eps8, eps8L2, and eps8L1 (in the latter case, at lower stochiometry). No association could be detected with eps8L3 (Figure 3C). Thus, eps8L1 and eps8L2, but not eps8L3 share critical biochemical properties with eps8.
Differential Interaction of eps8 Family Members with Dynamic F-actin–rich Structures In Vivo
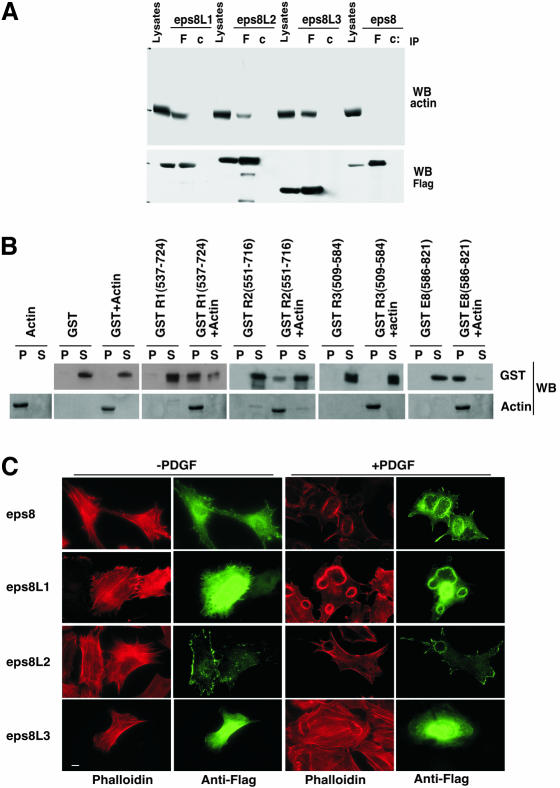
An important and biologically relevant feature of eps8 is its ability to interact with actin. This interaction is essential for the intracellular localization of eps8 and is thought to localize eps8-based complexes to sites where actin remodeling is required (Scita et al., 2001). Similarly to eps8, eps8L1 and eps8L2, but not eps8L3 coprecipitated with actin (Figure 4A). The C-terminal region of eps8, encompassing the last 173 amino acids, mediates a direct interaction with F-actin (Scita et al., 2001). Because this region is also highly conserved among the member of the eps8-related genes (Figure 1), we next tested the ability of purified C-terminal fragments of the various Eps8Ls to associate directly with polymerized actin in cosedimentation assays. Consistent with the immunoprecitation results, eps8, eps8L1, and L2, but not eps8L3 could directly associate to F-actin, albeit with different stoichiometry, pointing to the existence of a novel, conserved F-actin–binding surface present in the C-terminus of eps8Ls. This is further supported by the observation that eps8L3, whose C-terminal region is significantly shorter and the least conserved among the eps8Ls, failed to interact with F-actin. The ability of eps8Ls to associate to filamentous actin may contribute to their intracellular localization at site where F-actin is enriched, such as membrane ruffles. To this end, the cellular distribution of the eps8Ls was assessed. Flag-tagged eps8Ls were expressed in mouse fibroblasts. Eps8L1 and eps8L2, similarly to eps8, were found enriched in PDGF-induced, F-actin–rich ruffles (Figure 4B). In contrast, eps8L3 was inefficiently recruited to ruffles, in agreement with its lack of interaction with actin (Figure 4, A and B). It has to be pointed out that the direct binding to F-actin may not be sufficient to direct the localization of eps8Ls at membrane protrusions. Additional structural features, first and foremost the SH3 domains, may participate in dictating a proper cellular distribution. Consistent with this notion, all eps8Ls are capable of associating to Abi1, which was shown to localize specifically at the leading edge of protruding membranes (Stradal et al., 2001), possibly contributing to the localization of eps8Ls.
Figure 4.
Interaction between actin and eps8 family members. (A) 293T cells were transfected with flag-tagged eps8 family members (as indicated at the top, Tfx), followed by immunoprecipitation (IP) with an antiflag (F) or control IgGs (c). Lysates and immunoprecipitates were immunoblotted (WB) with the indicated antibodies. The flag (bottom) panel is the same as in Figure 2, bottom, because the two experiments were performed together. (B) Actin cosedimentation assay. The indicated C-terminal fragments of eps8 and eps8 related proteins (eps8L1 [537–724]; Eps8L2 [551–716]; eps8L3 [509–594]; eps8 [586–821]) fused to GST (1 μM) were mixed in the presence (+ actin) or absence of 10 μM of purified F-actin, as described in MATERIALS AND METHODS, and subjected to ultracentrifugation. GST alone was used as a negative control. Each lane shows the supernatant (S) and the pellet (P) obtained after ultracentrifugation. The lanes (actin) on the left show the supernatant and the pellet of F-actin alone. Detection was by immunoblotting (WB) with the indicated abs. (C) MEFs, microinjected with flag-tagged eps8 family members (indicated on the left), were serum-starved and stimulated with 10 ng/ml PDGF (+PDGF) or mock-treated (–PDGF). Cells were fixed and stained with an antiflag antibody (Anti-Flag, green) or rhodaminated phalloidin to visualize F-actin (Phalloidin, red).
Eps8L1 and eps8L2, but not eps8L3, Reconstitute the PDGF-induced Ruffling Response in eps8 –/– Fibroblasts
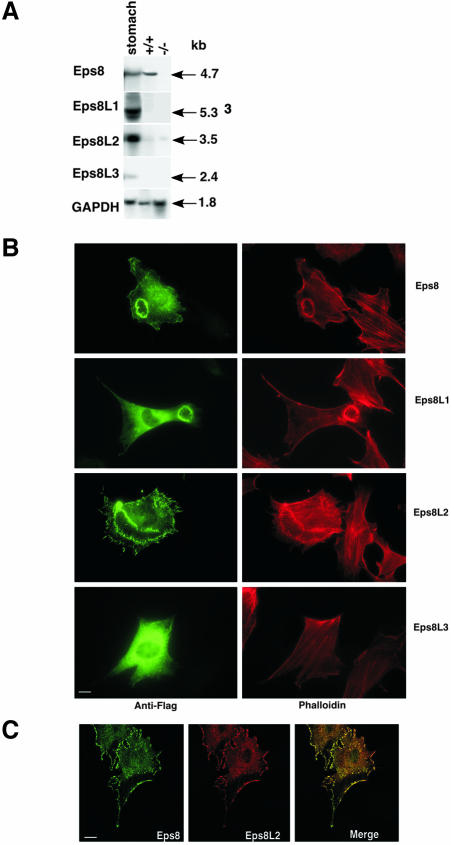
The ability to form an active complex with Abi1 and Sos-1 and to associate with F-actin–rich structures suggested that eps8L1 and eps8L2 might compensate for loss of eps8 function in vivo. To verify this prediction, we expressed the various eps8Ls in mouse embryo fibroblasts (MEFs) derived form eps8 knockout mice and analyzed RTK-mediated ruffling. Eps8 –/– MEFs do not express detectable levels of mRNA for eps8L genes (Figure 5A) and are unable to form membrane ruffles in response to growth factor stimulation (Scita et al., 1999). Ectopic expression of eps8, eps8L1, or eps8L2, but not of eps8L3, restored ruffling formation, in response to growth factor stimulation, in eps8 null MEFs (Figure 5B). Notably, eps8L2 was as efficient as eps8 (see legend to Figure 5). This is consistent with the observation that eps8 and eps8L2 display the highest degree of amino acid identity, within the family, and quantitatively comparable biochemical properties (eps8L2 and eps8 bind to and activate Sos-1 with similar efficiency; see Figure 3). Furthermore, they show a very similar intracellular distribution as witnessed by the large extent of colocalization between the two proteins by confocal analysis (Figure 5C). Thus, the members of the eps8 family display overlapping functional properties, which can be ranked by their ability to elicit Rac-GEF activity and to substitute for eps8 (eps8 = eps8L2 > eps8L1 ≫ eps8L3), and generate functional redundancy in pathway connecting RTK to Rac.
Figure 5.
Eps8L1 and eps8L2, but not eps8L3 reconstitute ruffling in response to PDGF in eps8 –/– MEFs. (A) Northern blot analysis of total RNAs from wt (+/+) and eps8 null (–/–) MEFs. An adult stomach RNA, in which the expression of all family members is visible, is also shown as a positive control. Probes specific for the various eps8Ls detected a single transcript of the indicated size (Kb). GAPDH was used to control for loading. (B) eps8 –/– MEFs were microinjected with flag-tagged eps8 family members (indicated on the right), serum-starved, and stimulated with 10 ng/ml PDGF. Cells were fixed and stained with antiflag (Anti-Flag, green) and rhodaminated phalloidin (Phalloidin, red). Quantitation (n = 3, at least 100 cells counted/experiment) was performed by determining the percentage of cells expressing eps8 family members that underwent ruffling: eps8, 78 ± 3; eps8L1, 42 ± 2; eps8L2, 71 ± 4; eps8L3, 4 ± 1; and untransfected cells, 5 ± 1. (C) eps8 –/– MEFs, cotransfected with myc-eps8 and flag-eps8L2, were fixed and stained with the appropriate antitag antibodies (eps8, antimyc in green; eps8L2, antiflag in red). Colocalization of eps8 and eps8L2 is evidenced by the appearance of yellow color in the merge. Central confocal sections are shown. Bar, 10 μM
Differential Patterns of Expression of eps8Ls in Developing and Adult Mice
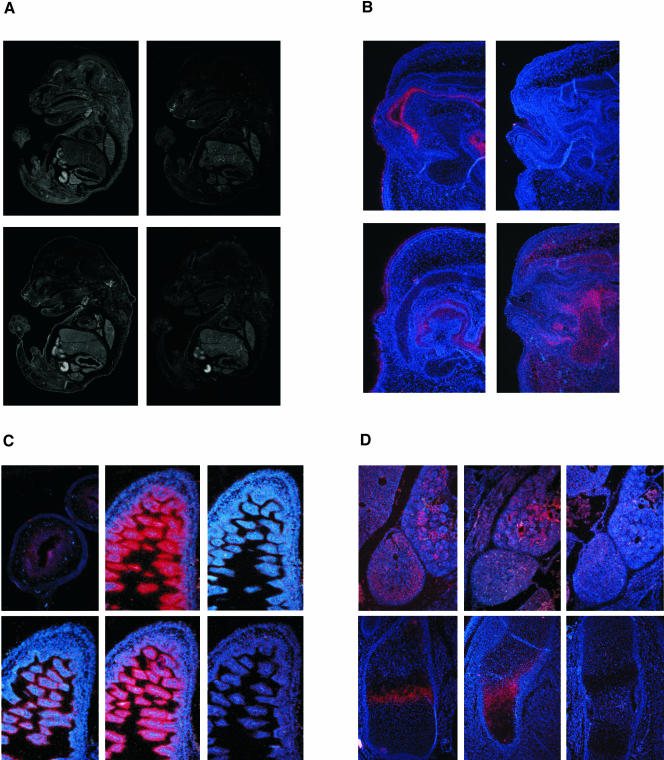
The overlapping functions of eps8L-family members provide tentative hypothesis to explain the lack of phenotype in eps8 null mice. To gain insight into this issue, we analyzed the pattern of expression of eps8-family members by in situ hybridization, in the developing mouse. The expression of eps8, but not the other eps8Ls, was detected as early as 10.5 d postconception (dpc; our unpublished results, and Avantaggiato et al., 1995). However, at 16.5 dpc expression of all four genes could be observed. Eps8 and eps8L2 displayed a widespread and largely overlapping pattern of expression (Figure 6, A–D, and Table 1). Eps8L1 and eps8L3 in contrast showed restricted expression (Table 1 and Figure 6). Eps8L1 was detectable almost exclusively in the naris externis (Figure 6, A and B). In this location, the expression of eps8L1 overlapped with eps8L2, but not with eps8, whose expression, instead, was restricted to the cartilage that gives rise to the nasal bones (Figure 6B). Weak expression of eps8L1 was also observed in the skin (Figure 6B). The expression of eps8L3 was confined to the inner mucosal layer of the intestine, where it was first detectable at 14.5 dpc (Figure 6C), rising to relatively high levels by 16.5 dpc (Figure 6C). Expression of eps8L3 was higher in the hindgut than in the midand foregut (Figure 6A). In this location, eps8L3 was coexpressed with eps8 and eps8L2, whereas eps8L1 was completely absent (Figure 6C).
Figure 6.
Eps8Ls displays a distinct expression pattern in the developing mouse embryo. (A) Expression of the eps8Ls in 16.5 dpc mouse embryos. Low-magnification photomicrographs demonstrate nearly ubiquitous expression of eps8 and widespread expression of eps8L2 in 16.5 dpc mouse embryos. Eps8L1 and eps8L3 in contrast show a very restricted expression, limited to the naris externis and the developing gut, respectively. (B) Expression of eps8Ls in the nasal anlage. The eps8L1 antisense probe (a), but not the sense control (b) displays a specific signal on the naris externis. Expression of eps8L2 was localized in the naris externis, in the surrounding nasal cartilage and in the skin (c). Eps8 expression was limited to the nasal cartilage (d). (C) Expression of eps8Ls in the hindgut. An antisense probe for eps8L3 detects expression in the embryonic gut starting from 14.5 dpc (a), increasing to very high levels at 16.5 dpc (b). No signal is obtained with the corresponding sense probe (c). Eps8 (d) and eps8L2 (e), but not eps8L1 (f) were detected in the inner mucosal layer of the gut. (D) Examples of overlapping expression of eps8 and eps8L2. An antisense probe for eps8 detects expression in the kidney and adrenal gland (a), and in the cartilage primordium of the developing femur (d). Eps8L2 displayed a similar expression pattern (b, kidney and adrenal gland; e, cartilage primordium of the scapula). No signal was detected with the sense probe for eps8L2 (c and f). The red color represents the in situ hybridization signal and the blue color shows the nuclei stained with the Hoechst 33258 dye.
Table 1.
Expression of eps8 family members in the 16.5 dpc mouse embryo
| eps8 | eps8L1 | eps8L2 | eps8L3 | |
|---|---|---|---|---|
| Brain | + | - | - | - |
| Dorsal root ganglia | - | - | - | - |
| External naris | - | + | + | - |
| Heart | - | - | - | - |
| Lung | ± | - | ± | - |
| Liver | - | - | ± | - |
| Kidney | + | - | + | - |
| Adrenal gland | + | - | + | - |
| Salivary gland | + | - | + | - |
| Esophagus | ± | - | ± | - |
| Stomach | ± | - | + | - |
| Gut | ± | - | + | + |
| Bladder | ± | - | ± | - |
| Muscle | - | - | - | - |
| Cartilage | + | - | + | - |
| Skin | ± | ± | + | - |
Expression levels were assessed by in situ hybridization and are expressed in a semiquantitative fashion: (-) negative, (±) weakly positive, (+) positive.
Northern blot analysis and RT-PCR were used to assess the expression of eps8Ls in adult mice (Table 2). Consistently with data obtained in embryos, eps8 and eps8L2 showed a relative broad pattern of expression. In contrast, eps8L1 and eps8L3 displayed a restricted expression pattern also in adult tissues (Table 2). Of note, both in developing and adult mice, eps8 was the sole family member that displayed detectable expression in the brain (Tables 1 and 2).
Table 2.
Expression of eps8 family members in mouse adult tissues
| Tissue | Methoda | eps8 | eps8L1 | eps8L2 | eps8L3 |
|---|---|---|---|---|---|
| Testis | N, P | + | ± | ± | ++ |
| Ovary | P | + | ± | + | ± |
| Placenta | P | + | ++ | ++ | ++ |
| Liver | N, P | - | - | ± | - |
| Skin | P | + | + | ++ | ± |
| Adrenal gland | P | + | ± | + | + |
| Salivary gland | P | + | ± | + | ± |
| Mammary gland | P | + | + | ± | - |
| Lymph node | P | ± | ± | ± | - |
| Thymus | P | - | ± | ± | ± |
| Bone marrow | P | + | + | - | - |
| Brain | N | + | - | - | - |
| Kidney | N | ++ | ± | ++ | - |
| Heart | N | ± | ± | - | - |
| Intestine | N | ++ | - | ++ | ++ |
| Stomach | N | + | ++ | ++ | ± |
Expression levels were measured by either Northern blot analysis (N) or semiquantitative PCR (P). Results refer to intragene comparison (after normalization for control GAPDH) and are expressed in a semiquantitative fashion: (-) negative, (±) weakly positive, (+) positive, (++) strongly positive.
DISCUSSION
In this study, we describe a new family of proteins, the eps8 family, whose founding member, eps8, was previously shown to be a key component of a multimolecular complex endowed with RacGEF activity (Scita et al., 1999; Innocenti et al., 2003). Our results show that members of this family link growth factor stimulation to actin organization, generating functional redundancy in the pathways that regulate actin cytoskeletal remodeling.
Members of the eps8 family share a modular organization consisting of a putative PTB domain, a central SH3 domain and a C-terminal effector region. The SH3 domains of eps8Ls display unique binding preferences for peptides containing a proline-X-X-aspartate-tyrosine (pXXDY) consensus and constitute a phylogenetically distinct subfamily within the SH3 domain family (Mongiovi et al., 1999). In keeping with this, all eps8Ls can associate in vivo, with comparable efficiencies, to Abi1, a known interactor of the SH3 of eps8 that is required for the assembly of the multimeric Rac-GEF complex. However, in the case of eps8L3, this is not sufficient to form an active RacGEF or to substitute for eps8 in eps8 null MEFs, suggesting that additional structural or functional features are required. The C-terminal effector region fulfills this requirement. This region, in eps8, is essential for interaction with Sos-1 and its activation as a RacGEF. In addition, it mediates a direct interaction with F-actin, determining the intracellular localization of the protein at sites where actin is highly dynamic (Scita et al., 2001). Consistently, eps8L1 and eps8L2, which display the highest homology to the eps8 in the C-terminal effector region, are capable of restoring ruffling response to RTK stimulation in eps8 null fibroblasts. Moreover, they can associate to actin in vivo and accumulate in PDGF-induced ruffles, thus further supporting the notion that relocalization of eps8Ls-based complexes is a prerequisite for their function in vivo. The observation that eps8L3, instead, does not contain a functional effector domain raises the question of its physiological role. We note that eps8L3 displays a punctuate, cytoplasmic staining reminiscent of proteins typically associated with vesicular organelles. Thus, it is tempting to speculate that eps8L3 may be involved in intracellular trafficking, a possibility currently under investigation.
Eps8L1 and eps8L2, similarly to eps8, mediate RTK-dependent actin remodeling, most likely by entering into a RacGEF complex with Abi1 and Sos-1. The existence of a family of proteins capable of forming multisubunit RacGEFs underscores additional complexity in the regulation of Rac. Multiple mechanisms have, indeed, been implicated in the activation of Rac by active plasma membrane receptors (Di Fiore and Scita, 2002). These rely mainly on the existence of a variety of Rac-specific GEFs (Schmidt and Hall, 2002), which are all comparably efficient in mediating Rac activation. Physiologically, however, the intracellular and the tissue distribution of the various RacGEFs is thought contribute to their specificity. The tissue distribution of the eps8 family of proteins supports this hypothesis.
Finally, the lack of overt phenotypic alterations in mice lacking eps8 can be rationalized in light of the observation that, at the mRNA level, the pattern of expression of eps8 overlaps with that of eps8L2, which, in turn, exhibits complete functional redundancy with eps8, at the protein level. It is of note that in organisms where eps8 gene duplication has not yet occurred or where only two members have evolved (Caenorhabditis elegans and in Drosophila, respectively) the genetic removal of eps8 leads to a number of developmental defects (A. Croce and P.P. Di Fiore, unpublished observations, and K. Moffat, personal communication). In mammalian systems, altered phenotypes (impaired Rac activation and actin remodeling in response to growth factor stimulation), caused by genetic removal of eps8, could be unmasked only in mouse embryo fibroblasts, which express exclusively eps8 (Scita et al., 2001, and this study). This suggests that possible alterations in eps8 null mice might be confined to those tissues or organs that do not show redundant expression of other family members, such as the brain. The lack of gross morphological defects in this organ (Scita et al., 2001, and our unpublished results), however, points to the possibility that more subtle phenotypes, such as behavioral deficits, might be linked to eps8 removal. We are currently investigating this possibility.
Acknowledgments
This work was supported by grants from AIRC (Associazione Italiana Ricerca sul Cancro) to G.S. and P.P.D.F.; from Human Science Frontier Program to G.S.; from the Italian Ministry of Health (grant R.F. 02/18h) to G.S., and from Telethon Foundation, the CNR (Target project Biotechnology), and the EC (V Framework) to P.P.D.F.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–06–0427. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-06-0427.
Abbreviations used: RTK, receptor tyrosine kinase, dpc, days postconception; MEF, mouse embryo fibroblast; PI3K, phosphoinositides 3 kinase; GEF, guanine nucleotide exchange factor.
References
- Avantaggiato, V., Torino, A., Wong, W.T., Di Fiore, P.P., and Simeone, A. (1995). Expression of the receptor tyrosine kinase substrate genes eps8 and eps15 during mouse development. Oncogene 11, 1191–1198 [PubMed] [Google Scholar]
- Bar-Sagi, D., and Hall, A. (2000). Ras and Rho GTPases: a family reunion. Cell 103, 227–238 [DOI] [PubMed] [Google Scholar]
- Biesova, Z., Piccoli, C., and Wong, W.T. (1997). Isolation and characterization of e3B1, an eps8 binding protein that regulates cell growth. Oncogene 14, 233–241 [DOI] [PubMed] [Google Scholar]
- Campbell, S.L., Khosravi-Far, R., Rossman, K.L., Clark, G.J., and Der, C.J. (1998). Increasing complexity of Ras signaling. Oncogene 17, 1395–1413. [DOI] [PubMed] [Google Scholar]
- Cherfils, J., and Chardin, P. (1999). GEFs: structural basis for their activation of small GTP-binding proteins. Trends Biochem. Sci. 24, 306–311 [DOI] [PubMed] [Google Scholar]
- Crompton, A.M., Foley, L.H., Wood, A., Roscoe, W., Stokoe, D., McCormick, F., Symons, M., and Bollag, G. (2000). Regulation of Tiam1 nucleotide exchange activity by pleckstrin domain binding ligands. J. Biol. Chem. 275, 25751–25759 [DOI] [PubMed] [Google Scholar]
- Das, B., Shu, X., Day, G.J., Han, J., Krishna, U.M., Falck, J.R., and Broek, D. (2000). Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275, 15074–15081 [DOI] [PubMed] [Google Scholar]
- Di Fiore, P.P., and Scita, G. (2002). Eps8 in the midst of GTPases. Int. J. Biochem. Cell Biol. 34, 1178–1183 [DOI] [PubMed] [Google Scholar]
- Fazioli, F., Minichiello, L., Matoska, V., Castagnino, P., Miki, T., Wong, W.T., and Di Fiore, P.P. (1993). Eps8, a substrate for the epidermal growth factor receptor kinase, enhances EGF-dependent mitogenic signals. EMBO J. 12, 3799–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509–514 [DOI] [PubMed] [Google Scholar]
- Hoffman, G.R., and Cerione, R.A. (2002). Signaling to the Rho GTPases: networking with the DH domain. FEBS Lett. 513, 85–91 [DOI] [PubMed] [Google Scholar]
- Innocenti, M., Frittoli, E., Ponzanelli, I., Falck, J.R., Brachmann, S.M., Di Fiore, P.P., and Scita, G. (2003). Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 160, 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innocenti, M., Tenca, P., Frittoli, E., Faretta, M., Tocchetti, A., Di Fiore, P.P., and Scita, G. (2002). Mechanisms through which Sos-1 coordinates the activation of Ras and Rac. J. Cell Biol. 156, 125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibuchi, K., Kuroda, S., and Amano, M. (1999). Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 68, 459–486 [DOI] [PubMed] [Google Scholar]
- Mongiovi, A.M., Romano, P.R., Panni, S., Mendoza, M., Wong, W.T., Musacchio, A., Cesareni, G., and Di Fiore, P.P. (1999). A novel peptide-SH3 interaction. EMBO J. 18, 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimnual, A., and Bar-Sagi, D. (2002). The two hats of SOS. Sci STKE 2002, PE36. [DOI] [PubMed] [Google Scholar]
- Nimnual, A.S., Yatsula, B.A., and Bar-Sagi, D. (1998). Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279, 560–563 [DOI] [PubMed] [Google Scholar]
- Paduch, M., Jelen, F., and Otlewski, J. (2001). Structure of small G proteins and their regulators. Acta Biochim. Pol. 48, 829–850 [PubMed] [Google Scholar]
- Rugarli, E.I., Lutz, B., Kuratani, S.C., Wawersik, S., Borsani, G., Ballabio, A., and Eichele, G. (1993). Expression pattern of the Kallmann syndrome gene in the olfactory system suggests a role in neuronal targeting. Nat. Genet. 4, 19–26 [DOI] [PubMed] [Google Scholar]
- Schmidt, A., and Hall, A. (2002). Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 16, 1587–1609 [DOI] [PubMed] [Google Scholar]
- Scita, G., Nordstrom, J., Carbone, R., Tenca, P., Giardina, G., Gutkind, S., Bjarnegard, M., Betsholtz, C., and Di Fiore, P.P. (1999). EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401, 290–293 [DOI] [PubMed] [Google Scholar]
- Scita, G. et al. (2001). An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J. Cell Biol. 154, 1031–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stradal, T., Courtney, K.D., Rottner, K., Hahne, P., Small, J.V., and Pendergast, A.M. (2001). The Abl interactor proteins localize to sites of actin polymerization at the tips of lamellipodia and filopodia. Curr. Biol. 11, 891–895 [DOI] [PubMed] [Google Scholar]
- Sugihara, K. et al. (1998). Rac1 is required for the formation fo three germ layers during gastrulation. Oncogene 17, 3427–3433 [DOI] [PubMed] [Google Scholar]
- Tocchetti, A., Confalonieri, S., Scita, G., Di Fiore, P.P., and Betsholtz, C. (2003). In silico analysis of the EPS8 gene family: genomic organization, expression profile and protein structure. Genomics 2, 234–244. (in press). [DOI] [PubMed] [Google Scholar]
- Van Aelst, L., and D'Souza-Schorey, C. (1997). Rho GTPases and signaling networks. Genes Dev. 11, 2295–2322 [DOI] [PubMed] [Google Scholar]
- Whitehead, I.P., Campbell, S., Rossman, K.L., and Der, C.J. (1997). Dbl family proteins. Biochim. Biophys. Acta 1332, F1–F23. [DOI] [PubMed] [Google Scholar]
- Zohn, I.M., Campbell, S.L., Khosravi-Far, R., Rossman, K.L., and Der, C.J. (1998). Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene 17, 1415–1438. [DOI] [PubMed] [Google Scholar]