Summary
Raf kinase inhibitors can induce ERK cascade signaling by promoting dimerization of Raf family members in the presence of oncogenic or normally activated Ras [1–3]. This interaction is mediated by a dimer interface region in the Raf kinase domain that is conserved in members of the ERK cascade scaffold family, Kinase-suppressor of Ras (KSR) [4, 5]. In this study, we find that most Raf inhibitors also induce the binding of KSR1 to wild-type and oncogenic B-Raf proteins, including V600E B-Raf, but promote little complex formation between KSR1 and C-Raf. The inhibitor-induced KSR1/B-Raf interaction requires direct binding of the drug to B-Raf and is dependent on conserved dimer interface residues in each protein, but, unexpectedly, is not dependent on binding of B-Raf to activated Ras. Inhibitor-induced KSR/B-Raf complex formation can occur in the cytosol and is observed in normal mouse fibroblasts, as well as a variety of human cancer cell lines. Strikingly, we find that KSR1 competes with C-Raf for inhibitor-induced binding to B-Raf and, as a result, alters the effect of the inhibitors on ERK cascade signaling.
Keywords: KSR1, B-Raf, Raf inhibitors, ERK cascade
Results and Discussion
Raf Inhibitors Induce KSR1/B-Raf binding
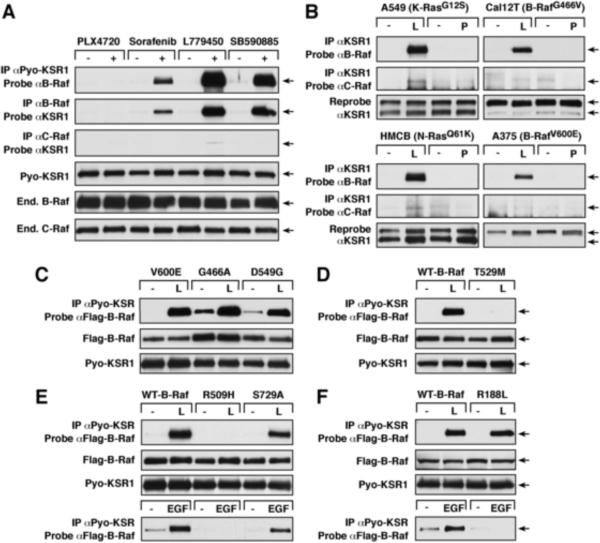
ATP-competitive Raf inhibitors can promote dimerization of the Raf kinases in the presence of oncogenic or activated wild-type (WT) Ras [1–3]. Because the KSR1 scaffold also interacts with Raf in response to Ras activation [6, 7] and contains residues homologous to those in the Raf dimer interface region [4, 5] that are critical for inhibitor-induced Raf dimerization [1], we examined whether the Raf inhibitors might also promote KSR1/Raf binding. KSR−/− mouse embryonic fibroblasts (MEFs; [8]) that stably express Pyo-tagged WT-KSR1 (WT-KSR1 MEFs) were treated with the Raf inhibitors PLX4720, Sorafenib, L779450, or SB590885 for 1 hr, following which complex formation between KSR1 and the endogenous Raf proteins was examined. As shown in Fig. 1A, Sorafenib, L779450, and SB590885 all induced robust association of KSR1 with B-Raf, but little or no interaction with C-Raf. Inhibitor-induced KSR1/B-Raf binding was dose-dependent, increasing with greater drug concentration (Fig. S1A). Notably, PLX4720, did not promote binding of KSR1 to either Raf protein. Further investigation revealed that only when C-Raf was highly over-expressed (~25-fold above endogenous levels) could some interaction between KSR1 and C-Raf be detected following L779450 treatment; however, even with high C-Raf over-expression, binding was still not observed in PLX4720-treated cells (Fig. S1B). Unlike the other inhibitors tested, PLX4720 binding causes a shift in the B-Raf α-helix [1, 9], which may perturb the KSR1/B-Raf interaction and account for the absence of KSR1/B-Raf complexes in PLX4720-treated cells.
Figure 1.
Raf Inhibitors Induce KSR1/B-Raf Binding
(A) Cycling WT-KSR1 MEFs were treated with the indicated drugs (10 μM for 1 hr). Pyo-KSR1 or endogenous B-Raf or C-Raf complexes were isolated and examined by immunoblot analysis as indicated. (B) A549, Cal12T, A375 and HMCB cancer lines were treated with L779450 or PLX7420 (10 μM for 1 hr). Endogenous KSR1 complexes were examined for the presence of endogenous B-Raf or C-Raf. (C–F) Cycling WT-KSR1 MEFs stably expressing the indicated Flag-B-Raf proteins were treated with L779450 (L, 10 μM for 1 hr), following which Pyo-KSR1/Flag-B-Raf binding was assessed. In parallel experiments (E and F), Pyo-KSR1 complexes were isolated from serum-starved cells treated with EGF (100 ng/ml for 5 min). Of note, Raf inhibitor-induced KSR1/B-Raf binding was increased in comparison to EGF-mediated binding and required shorter exposure times for detection. Protein expression levels are also shown.
Because the Raf inhibitors are normally used in the context of oncogenic signaling, a panel of melanoma and non-small cell lung carcinoma lines were screened for KSR1 expression, and four representative lines were evaluated that possessed detectable KSR1 levels and oncogenic mutations in either Raf or Ras (Fig. S1C). In these experiments, treatment with L779450, but not PLX4720, induced strong KSR1/B-Raf binding in A549 and HMCB cells that possess oncogenic Ras proteins, in the Cal12T line that contains an impaired activity B-Raf mutant, and, surprisingly, in A375 melanoma cells that are homozygous for V600E-B-Raf (Fig. 1B and S1D). This later finding is in contrast to Raf inhibitor-induced C-Raf/B-Raf dimerization, which involves binding of C-Raf to WT B-Raf or impaired activity B-Raf mutants, but not to the high activity V600E-B-Raf (Fig. S1D, [1–3]). Of note, little or no inhibitor-induced binding between KSR1 and C-Raf was observed in these lines (Fig. 1B).
To further evaluate the ability of oncogenic B-Raf proteins to interact with KSR1 upon inhibitor treatment, WT-KSR1 MEFs stably expressing Flag-tagged V600E (high activity)-, G466A (moderate activity)-, or D594G (impaired activity)-B-Raf [10] were treated with L779450 and examined for binding of KSR1 to the Flag-B-Raf proteins. As shown in Fig. 1C, basal association of KSR1 was higher for the moderate and impaired activity B-Raf mutants; however, inhibitor-induced binding was equivalent for V600E, G466A, and D594G-B-Raf (Fig. 1C), indicating that the activity level of B-Raf has no impact on the drug-mediated interaction with KSR1.
Inhibitor-induced KSR1/B-Raf Complex Formation Requires Binding of the Inhibitor to B-Raf and an Intact B-Raf Dimer Interface, but is Not Ras-dependent
To investigate the requirements in B-Raf for inhibitor-induced binding to KSR1, WT-KSR1 MEFs stably expressing various Flag-tagged B-Raf proteins were treated with L779450 and examined. In co-immunoprecipitation assays, the T529M-B-Raf gatekeeper mutant [11] failed to interact with KSR1 (Fig. 1D), demonstrating that as with inhibitor-induced C-Raf/B-Raf dimerization [1–3], drug-mediated KSR1/B-Raf complex formation requires direct binding of the inhibitor to B-Raf. Mutation of a conserved arginine residue (R509H) in the B-Raf dimer interface region [5] also disrupted the interaction between KSR1 and B-Raf (Fig. 1E), consistent with previous reports that an intact B-Raf dimer interface is required for inhibitor-induced C-Raf/B-Raf dimerization [1] and for growth factor-mediated KSR1/B-Raf binding (Fig. 1E; [5]). However, S729A-B-Raf, which contains a mutation in the C-terminal 14-3-3 binding site, was competent to bind KSR1 in response to either L779450 or EGF treatment (Fig. 1E). Surprisingly, mutation of the Ras binding site in B-Raf (R188; [12]) had no effect on L779450-induced KSR1/B-Raf binding (Fig. 1F). Thus, in contrast to both inhibitor-induced C-Raf/B-Raf dimerization [2] and EGF-mediated KSR1/B-Raf binding (Fig. 1F), the inhibitor-induced KSR1/B-Raf interaction is not Ras-dependent (requirements summarized in Table S1).
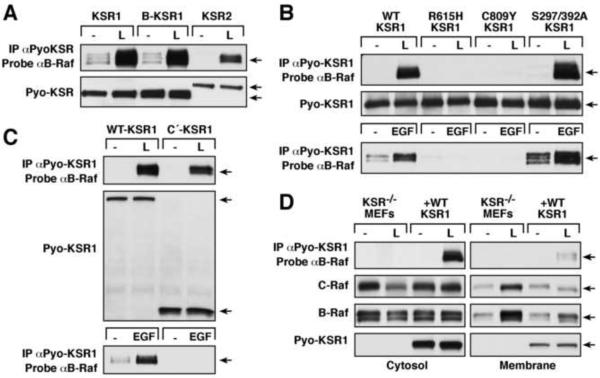
All KSR Proteins Competent to Bind B-Raf Can Form Inhibitor-induced Complexes
To investigate the requirements in KSR for inhibitor-induced binding to B-Raf, KSR−/− MEFs stably expressing various Pyo-tagged WT and mutant KSR proteins were treated with L779450 and analyzed for KSR/B-Raf complex formation. As shown in Fig. 2A, the mammalian KSR proteins KSR1 [4], KSR2 [13], and B-KSR1 (a brain specific splice variant of KSR1 [14]) were all competent to bind B-Raf in response to L779540 treatment. Interestingly, inhibitor treatment promoted little KSR1/KSR2 heterodimerization (Fig. S2A) and no detectable KSR1/KSR1 homodimerization (Fig. S2B) in MEFs. Further evidence that the Raf inhibitors may bind only weakly to the KSR proteins, mutation of the predicted gatekeeper site in KSR1 (T636M) reduced, but did not eliminate, complex formation with B-Raf in L779450-treated cells (Fig. S2C). In contrast, mutation of the conserved arginine residue in the KSR1 dimer interface (R615H; [5]) abolished B-Raf binding following L779450 treatment, as well as EGF stimulation (Fig. 2B). Inhibitor-induced KSR1/B-Raf complex formation was also perturbed by the C809Y mutation (Fig. 2B), which disrupts the KSR1/MEK interaction [14, 15] and growth factor-mediated KSR1/B-Raf binding [7]. However, S297A/S392A-KSR1, a mutant that is defective in 14-3-3 binding [16] and exhibits enhanced growth factor-mediated binding to B-Raf (Fig. 2B), demonstrated an increased interaction with B-Raf following L779450 treatment (Fig. 2B).
Figure 2.
Requirements in KSR1 for Inhibitor-induced B-Raf/KSR1 Binding
(A–C) Cycling KSR−/− MEFs expressing the indicated KSR proteins were treated with L779450 (L, 10 μM for 1 hr), following which Pyo-KSR/B-Raf binding was assessed. In parallel experiments (B and C), Pyo-KSR1 complexes were isolated from serum-starved cells treated with EGF (100 ng/ml for 5 min). (D) Cycling KSR−/− and WT-KSR1 MEFs were treated with L779450 (10 μM for 1 hr) prior to cell fractionation. Membrane and cytosolic fractions were examined for KSR1/B-Raf complex formation and for Pyo-KSR1, B-Raf, and C-Raf levels.
Surprisingly, a truncated KSR1 protein containing only the C-terminal region of KSR1 (C′-KSR1, amino acids 530–873) formed complexes with B-Raf in L779450-treated cells (Fig. 2C), despite the fact that this mutant lacks the N-terminal, membrane-targeting C1 domain [17] and cannot localize to the plasma membrane or interact with B-Raf following EGF treatment (Fig. 2C; [7]). This finding together with the observation that binding of B-Raf to KSR1 in response to L779450 treatment is Ras-independent indicates that the inhibitor-induced KSR1/B-Raf interaction can occur in the cytosol. In further support of this conclusion, a KSR1 protein containing mutations that disrupt the structure of the membrane-targeting C1 domain (C1m-KSR1; [17]) also formed inhibitor-induced complexes with B-Raf (Fig. S2C) and KSR1/B-Raf complexes were abundantly detected in the cytosolic compartment of WT-KSR1 MEFs that were fractionated after inhibitor treatment (Fig. 2D).
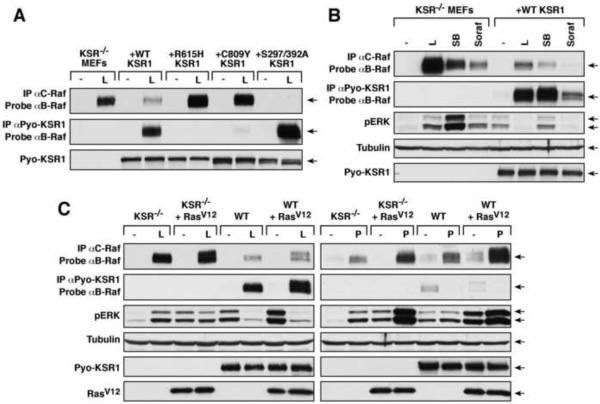
KSR1 Competes with C-Raf for Inhibitor-induced Binding to B-Raf
The finding that inhibitor-induced complex formation between KSR1/B-Raf can occur in the cytosol raises the interesting possibility that KSR1 might compete with C-Raf for inhibitor-induced binding to B-Raf, which has been shown to be Ras-dependent and occur at the plasma membrane [1, 2]. To begin to investigate this possibility, KSR−/− MEFs and those expressing WT-KSR1 or various KSR1 mutants were treated with L779450 and analyzed for complex formation between the endogenous C-Raf/B-Raf proteins and for association of KSR1 to endogenous B-Raf. As shown in Fig 3A, L779450 strongly induced C-Raf/B-Raf dimerization in KSR−/− MEFs; however, this interaction was significantly reduced in WT-KSR1 MEFs, where induction of KSR1/B-Raf binding was observed. L779450-induced B-Raf/C-Raf dimerization was not reduced in MEFs expressing the R615H- and C809Y-KSR1 mutants unable to bind B-Raf; whereas B-Raf/C-Raf dimerization was abolished in cells expressing S297A/S392A-KSR1 that exhibits enhanced inhibitor-induced B-Raf binding (Fig. 3A). The presence of WT-KSR1 also reduced complex formation between C-Raf and the oncogenic G466A- and D594G-B-Raf proteins in L779450-treated cells (Fig. S3). Moreover, in cell fractionation experiments, B-Raf and C-Raf were strongly recruited into the membrane fraction of L779450-treated KSR−/− MEFs, while in L779450-treated WT-KSR1 MEFs that contain high levels of cytosolic KSR1/B-Raf complexes, little to no membrane recruitment was observed (Fig. 2D). In further support that KSR1 can compete with C-Raf for inhibitor-induced binding to B-Raf, C-Raf/B-Raf dimerization was consistently reduced in WT-KSR1 MEFs versus KSR−/− MEFs when cells were treated with any of the Raf inhibitors capable of promoting KSR1/B-Raf binding (Fig. 3B and 3C), and C-Raf/B-Raf dimerization was still reduced in L779450-treated WT-KSR1 MEFs even when RasV12 was expressed (Fig. 3C). In contrast, equivalent C-Raf/B-Raf dimerization was observed in KSR−/− and WT-KSR1 MEFs treated with PLX4720, which does not promote KSR1/B-Raf binding, and PLX4720-induced C-Raf/B-Raf dimerization was significantly increased in both cells lines when RasV12 was expressed (Fig. 3C).
Figure 3.
KSR1 Competes with C-Raf for Inhibitor-induced Binding to B-Raf
(A–C) Cycling KSR−/− MEFs and those expressing the indicated KSR1 proteins were treated with L779450 (L), SB590885 (SB), Sorafenib (Soraf) or PLX4720 (P) (10 μM for 1 hr). Pyo-KSR1 or endogenous C-Raf complexes were isolated and examined for endogenous B-Raf. Lysates were analyzed for pERK and tubulin (loading control) levels. Cells stably expressing RasV12 were also examined in (C).
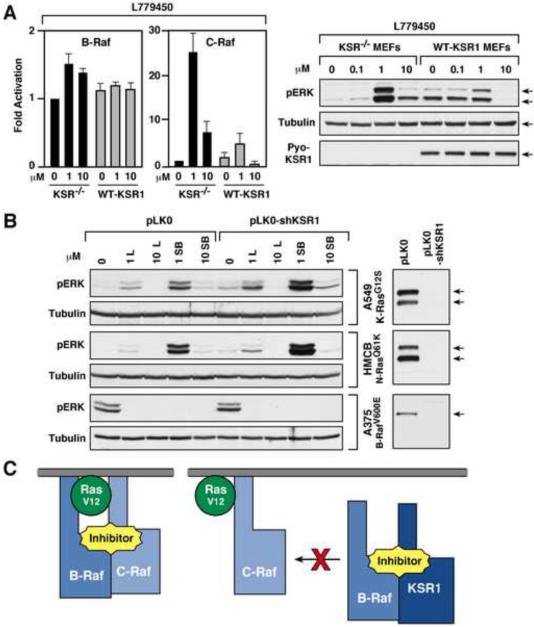
KSR1 Can Alter the Effect of Raf Inhibitors on ERK Cascade Signaling
Given the above results that KSR1 can compete with C-Raf for inhibitor-induced binding to B-Raf, we next examined the effect of KSR1 on inhibitor-induced ERK cascade signaling. Strikingly, we found that activated pERK levels differed significantly in KSR−/− verses WT-KSR1 MEFs when treated with inhibitors that promote KSR/B-Raf binding. As shown in Fig. 3B, although basal ERK activation was higher in WT-KSR1 MEFs, pERK levels were either unchanged (SB590885) or inhibited in WT-KSR1 cells (L779450 and Sorafenib) treated with inhibitors that promote KSR1/B-Raf binding. Conversely, these inhibitors all stimulated ERK activation in KSR−/− MEFs, with SB590885 inducing significantly higher pERK levels than seen in untreated or SB590885-treated WT-KSR1 MEFs (Fig. 3B). Oncogenic RasV12 increased the basal pERK levels in both KSR−/− and WT-KSR1 MEF; however, treatment with L779450 was still able to suppress ERK activation in cells expressing KSR1, but had no inhibitory effect in KSR−/− cells (Fig. 3C). Interestingly, pERK levels were equivalent and not suppressed in both KSR−/− and WT-KSR1 MEFs treated with PLX4720, which does not promote KSR1/B-Raf binding.
When Raf catalytic activity was monitored (Fig. 4A), we observed a slight change in B-Raf activity in L779450-treated KSR−/− MEFs (1.2–1.5 fold), but no change in WT-KSR1 MEFs. Notably, C-Raf activity increased ~25 fold with 1 μM L779450 treatment in KSR−/− cells but only 2–3 fold in WT-KSR1 MEFs. Moreover, 10 μM L799450 still activated C-Raf 8–10 fold in KSR−/− MEFs, but inhibited Raf activity in WT-KSR1 cells (Fig. 4A). ERK activation in the KSR−/− and WT-KSR1 MEFs was also found to correlate with the effect of L779450 dosage on C-Raf activity (Fig. 4A). As expected, the presence of KSR1 had no effect on either B-Raf or C-Raf activity in cells treated with PLX4720 (Fig. S4).
Figure 4.
Effects of KSR1 on Inhibitor-induced ERK Cascade Activation
(A) KSR−/− and WT-KSR1 MEFs were treated with L779450 (10 μM for 1 hr). The catalytic activity of endogenous B-Raf and C-Raf proteins was measured in immune-complex kinase assays using kinase-inactive MEK as a substrate. Lysates were analyzed for pERK and tubulin (loading control) levels. (B) A549, HMCB, and A375 cells expressing either the pLKO.1 vector or pLKO.1-KSR1 shRNA were treated as indicated for 1hr. Lysates were analyzed for pERK and tubulin levels. Depletion of KSR1 is also shown. (C) Inhibitor-induced Raf dimerization occurs at the plasma membrane and is Ras-dependent (Left). Inhibitor-induced KSR1/B-Raf complexes can form in the cytosol, thus inhibiting the formation of C-Raf/B-Raf dimers at the membrane (Right).
To determine if KSR1 can influence the effect of inhibitor treatment in cancer cell lines, a lentivirus expressing a shKSR1 RNA was used to generate A549, HMCB and A375 lines depleted of KSR1 (Fig. 4B). ERK activation was then compared in these and the vector control lines following drug treatment. Consistent with studies showing that L779450 and SB590885 can effectively inhibit V600E-B-Raf activity [2, 3], depletion of KSR1 had no effect on the ability of these drugs to block ERK activation in A375 cells homozygous for V600E-B-Raf. As previously reported [3], high inhibitor treatment also blocked ERK activation in the HMCB and A549 lines that contain oncogenic Ras proteins (Fig. 4B). However, treatment with lower doses (1μM) of L779450 or SB590885 promoted ERK activation in the HMCB and A549 lines, and pERK levels were increased 2–5 fold in cells depleted of KSR1.
Concluding comments
Determining effective therapeutic strategies for treating human cancer is a significant challenge for the scientific and medical communities. Recent studies have shown that treatment with Raf kinase inhibitors can paradoxically induce ERK cascade signaling by promoting dimerization of Raf family members in the presence of activated Ras. Our findings reveal that select Raf inhibitors can also promote KSR1/B-Raf binding. Like inhibitor-induced C-Raf/B-Raf dimerization, drug-induced KSR1/B-Raf binding requires an intact KSR1/B-Raf dimer interface and binding of the inhibitor to the ATP binding pocket of B-Raf. However, drug-mediated KSR1/B-Raf complex formation differs from inhibitor-induced C-Raf/B-Raf dimerization and normal growth factor-mediated KSR1/B-Raf binding in that it is Ras-independent can occur in the cytoplasm. Strikingly, and as a result of this difference, we find that KSR1 competes with C-Raf for inhibitor-induced binding to B-Raf (Fig. 4C) and attenuates the activating effect of the inhibitors on ERK cascade signaling. Together, these findings suggest that KSR1 expression levels may impact the therapeutic effect of select Raf inhibitors.
Experimental Procedures
DNA Constructs and Generation of Stable Cell Lines
pBabe retroviral constructs encoding Pyo-KSR1 and Flag-B-Raf have been described [18, 19] and constructs encoding HA- H-RasG12V, Pyo-KSR2, or Pyo-BKSR1 were generated by subcloning. Point mutations were introduced by site directed mutagenesis and confirmed by DNA sequencing. Stable cell lines were generated by retroviral infection of KSR−/− MEFs, which are null for both KSR1 and KSR2 [8].
Cell Lysis, Co-immunoprecipitation Assays, Immune Complex Kinase Assays
Cells were lysed in 1% Nonident P-40 buffer [20 mM Tris (pH 8.0), 137 mM NaCl, 10% glycerol, 1% Nonident P40, 0.15 unit/mL aprotinin, 1 mM phenylmethylsulfonyl fluoride (PMSF), 20 μM leupeptin, and 0.5 mM sodium vanadate and lysates were clarified by centrifugation. Protein concentrations were determined and equivalent amounts of protein lysate were incubated with the appropriate antibody and Protein G Sepharose beads for 3 hr at 4°C. Immune complexes were washed and analyzed either by immunoblotting or in immune complex kinase assays as described in Ritt et al [19].
Cell Fractionation
Cell fractionation of KSR−/− or WT-KSR1 MEFs was performed as described previously [20]. The purity of the membrane and cytoplasmic fractions were monitored by the presence of tubulin (cytoplasmic) and H-Ras (membrane).
Supplementary Material
Acknowledgements
We thank members of the Laboratory of Cell and Developmental Signaling for helpful discussions. This project has been funded by Federal funds from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information Supplemental Information includes Supplemental Experimental Procedures, four figures, and one table.
References
- 1.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, Ludlam MJ, Stokoe D, Gloor SL, Vigers G, Morales T, Aliagas I, Liu B, Sideris S, Hoeflich KP, Jaiswal BS, Seshagiri S, Koeppen H, Belvin M, Friedman LS, Malek S. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431–435. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 2.Heidorn SJ, Milagre C, Whittaker S, Nourry A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer CJ, Pritchard C, Marais R. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 5.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 6.Ritt DA, Zhou M, Conrads TP, Veenstra TD, Copeland TD, Morrison DK. CK2 Is a component of the KSR1 scaffold complex that contributes to Raf kinase activation. Curr Biol. 2007;17:179–184. doi: 10.1016/j.cub.2006.11.061. [DOI] [PubMed] [Google Scholar]
- 7.McKay MM, Ritt DA, Morrison DK. Signaling dynamics of the KSR1 scaffold complex. Proc Natl Acad Sci U S A. 2009;106:11022–11027. doi: 10.1073/pnas.0901590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kortum RL, Lewis RE. The molecular scaffold KSR1 regulates the proliferative and oncogenic potential of cells. Mol Cell Biol. 2004;24:4407–4416. doi: 10.1128/MCB.24.10.4407-4416.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, Bremer R, Gillette S, Kong J, Haass NK, Sproesser K, Li L, Smalley KS, Fong D, Zhu YL, Marimuthu A, Nguyen H, Lam B, Liu J, Cheung I, Rice J, Suzuki Y, Luu C, Settachatgul C, Shellooe R, Cantwell J, Kim SH, Schlessinger J, Zhang KY, West BL, Powell B, Habets G, Zhang C, Ibrahim PN, Hirth P, Artis DR, Herlyn M, Bollag G. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc Natl Acad Sci U S A. 2008;105:3041–3046. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, Marais R. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 11.Whittaker S, Kirk R, Hayward R, Zambon A, Viros A, Cantarino N, Affolter A, Nourry A, Niculescu-Duvaz D, Springer C, Marais R. Gatekeeper mutations mediate resistance to BRAF-targeted therapies. Sci Transl Med. 2010;2:35ra41. doi: 10.1126/scitranslmed.3000758. [DOI] [PubMed] [Google Scholar]
- 12.Marais R, Light Y, Paterson HF, Mason CS, Marshall CJ. Differential regulation of Raf-1, A-Raf, and B-Raf by oncogenic ras and tyrosine kinases. J Biol Chem. 1997;272:4378–4383. doi: 10.1074/jbc.272.7.4378. [DOI] [PubMed] [Google Scholar]
- 13.Dougherty MK, Ritt DA, Zhou M, Specht SI, Monson DM, Veenstra TD, Morrison DK. KSR2 is a calcineurin substrate that promotes ERK cascade activation in response to calcium signals. Mol Cell. 2009;34:652–662. doi: 10.1016/j.molcel.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller J, Cacace AM, Lyons WE, McGill CB, Morrison DK. Identification of B-KSR1, a novel brain-specific isoform of KSR1 that functions in neuronal signaling. Mol Cell Biol. 2000;20:5529–5539. doi: 10.1128/mcb.20.15.5529-5539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart S, Sundaram M, Zhang Y, Lee J, Han M, Guan KL. Kinase suppressor of Ras forms a multiprotein signaling complex and modulates MEK localization. Mol Cell Biol. 1999;19:5523–5534. doi: 10.1128/mcb.19.8.5523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cacace AM, Michaud NR, Therrien M, Mathes K, Copeland T, Rubin GM, Morrison DK. Identification of constitutive and ras-inducible phosphorylation sites of KSR: implications for 14-3-3 binding, mitogen-activated protein kinase binding, and KSR overexpression. Mol Cell Biol. 1999;19:229–240. doi: 10.1128/mcb.19.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou M, Horita DA, Waugh DS, Byrd RA, Morrison DK. Solution structure and functional analysis of the cysteine-rich C1 domain of kinase suppressor of Ras (KSR) J Mol Biol. 2002;315:435–446. doi: 10.1006/jmbi.2001.5263. [DOI] [PubMed] [Google Scholar]
- 18.McKay MM, Morrison DK. Caspase-dependent cleavage disrupts the ERK cascade scaffolding function of KSR1. J Biol Chem. 2007;282:26225–26234. doi: 10.1074/jbc.M702692200. [DOI] [PubMed] [Google Scholar]
- 19.Ritt DA, Monson DM, Specht SI, Morrison DK. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol Cell Biol. 2010;30:806–819. doi: 10.1128/MCB.00569-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stokoe D, McCormick F. Activation of c-Raf-1 by Ras and Src through different mechanisms: activation in vivo and in vitro. Embo J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.