Abstract
The primary pathway of metabolism of dietary alcohol is via its oxidation in liver by alcohol dehydrogenases (ADH). Differences in the ADH enzyme activity or levels of enzyme present could affect the risk for alcoholism. Regulatory variations have been shown to affect the promoter activity and thereby affect the risk for alcoholism. In this study the functional effects of the two SNPs (rs1159918 and rs1229982) in the proximal promoter region of ADH1B that were associated with alcoholism were explored. We examined the effects of five naturally occurring haplotypes on the promoter activity. We observed that a C to A change at rs1229982 increased promoter activity 1.4-fold.
Keywords: alcohol dehydrogenase 1B, alcoholism, polymorphism, SNP, promoter, gene expression, haplotype
1. Introduction
Alcoholism is a complex disease affecting millions in the world. It is influenced by both genetic and environmental factors. Evidence that greater than 50% of the differences in risk can be attributed to genetic factors has been obtained from family, twin and adoption studies [1–10].
Alcohol dehydrogenases metabolize alcohol to acetaldehyde in the first step of alcohol metabolism. The rate at which ethanol is oxidized influences the concentration of ethanol and acetaldehyde. This is the hypothesized mechanism by which ADH variations affect the risk for alcoholism. There are seven alcohol dehydrogenases in humans: ADH1A, ADH1B, ADH1C, ADH4, ADH5, ADH6, and ADH7 [11]. Class I ADHs (ADH1A, ADH1B and ADH1C) contribute to 70% of ethanol metabolism in the liver [12]. Coding variations in ADH1B have a strong effect on the risk for alcoholism; a single nucleotide polymorphism (SNP) that leads to an arginine to histidine change in the coding sequence (rs1229984; R48H) increases the catalytic rate of the enzyme and was shown to be protective against alcoholism [13–20].
The rate of alcohol metabolism can also be affected by regulatory variations that influence the amount of enzyme present. Several studies have reported regulatory variations of the ADHs. A SNP at position -136 (relative to the +1 translational start site) in the promoter of the ADH4 gene affects the promoter activity in hepatoma cells, with the A allele having 2-fold higher activity than the C allele [21]. This variation has been associated with the risk for alcoholism in a Brazilian population [22]. Polymorphisms that affect expression levels were also identified in a regulatory element 3 kb upstream of ADH1C promoter [23]. Non-coding variations in and near other ADH genes have been associated with alcohol consumption and the risk for alcoholism [1, 24–27].
Two SNPs in the ADH1B promoter region, rs1229982 and rs1159918, are associated with alcohol dependence defined by meeting both DSM-IIIR criteria for alcohol dependence plus criteria for Feighner definite alcoholism [25]. We tested the effect of the two polymorphisms and others in the ADH1B promoter region on promoter activity.
2. Materials and Methods
2.1 Cloning of test fragments
The region extending to 1484 bp upstream of the start of the ADH1B coding sequence (+1 CDS) was amplified by PCR using high fidelity platinum Pfx polymerase (Invitrogen, Carlsbad, CA), using HE3003 (CGAGATCTGTCTTCTCTGCCCACCAGC) and HE3116 (GTGGTACCCTGGGGCTATCTTCTTTCCG) as primers. PCR conditions were: 94C for 5 min, (94C for 15 s, 62C for 20 s, 68C for 90 s) × 10 cycles; (94C for 15 s, 60C for 20 s, 68C for 90 s) × 30 cycles, 68C for 7 min. DNA from five different individuals was used as template. Fragments were cloned into KpnI and BglII sites in the pXP2 luciferase reporter vector [28]. Clones were sequenced by BigDye terminator v3.1 cycle sequencing kit (Applied Biosystems, Santa Clara, CA); five different haplotypes were obtained.
2.2 Transient transfections and Reporter gene assays
HepG2 human hepatoma cells (HB-8065; ATCC, Manassas, VA) were cultured in MEM (ATCC) with 10% FBS (Invitrogen, Carlsbad, CA), 4 mM glutamine (Thermo Scientific Hyclone, Waltham, MA) and 1X Penicillin and Streptomycin (Thermo Scientific Hyclone) on cell bind surface plates (Corning Inc, Corning, NY) at 37C. For transient transfection assays 8 × 105 cells were seeded per well in 6-well cell binding surface plates. 24 h after seeding, cells were transfected in serum free media with 2 μg of test DNA, along with 140 ng of CMV-galactosidase plasmid (Clontech, Mountain View, CA) and 1.2 μg of pUC19 DNA, using 3 μl Fugene HD (Roche, Indianapolis, IN). Complete medium was added 6 h after addition of DNA and cells were cultured for another 24 h. Cells were harvested 30 h after addition of DNA by scraping into ice-cold 1X PBS, pelleted by centrifugation and suspended in 100 μl of 1X Reporter lysis buffer (Promega, Madison, WI). Cell extracts were prepared by three repeated cycles of freeze-thawing followed by centrifugation at 16,000 × g for 1 min. Luciferase assays were carried out using 20 μl of the extract and the Luciferase assay system (Promega, Madison, WI). β-galactosidase assays were carried out using 5 μl of the extract and the Galacto-Light System (Tropix, Bedford, MA). Activities were measured on a Lmax Plate Luminometer (Molecular Devices Sunnyvale, CA).
All test constructs were transfected at least in triplicate in each individual experiment, with experiments repeated four times, on different days. Promoter activity was defined as luciferase activity normalized to β-galactosidase activity, to correct for the transfection efficiency. A t-test assuming unequal variances was carried out in Microsoft Excel, considering each transfection as an independent data point.
3. Results and Discussion
Two SNPs in the non-coding region of ADH1B have been associated with alcohol dependence [25]; both rs1229982 and rs1159918 are located in the proximal promoter region. Allele and genotype frequencies of rs1229982 and rs1159918 are shown in Table 1. The Single Nucleotide Polymorphism database (dbSNP; www.ncbi.nlm.nih.gov/projects/SNP) reports a total of 12 SNPs in the region between −1484 bp and −10 bp (with respect to the coding sequence start site; Figure 1).
Table 1. Allele and genotype frequencies for two SNPs in the ADH1B proximal promoter region.
Allele and genotype frequencies in different populations for the two SNPs that were associated with alcoholism were obtained from HapMap (release 27, Phase II and III; hapmap.ncbi.nlm.nih.gov). Populations are Utah residents with ancestry from northern and western Europe (CEU), Han Chinese in Beijing, China (CHB), Japanese in Tokyo, Japan (JPT), Yoruba in Ibadan, Nigeria (YRI).
| rs1229982 | rs1159918 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Allele | Genotype | Allele | Genotype | |||||||
| HapMap Population | A | C | A/A | C/A | C/C | T | G | T/T | T/G | G/G |
| CEU | 0.2 | 0.8 | 0.0 | 0.3 | 0.7 | 0.3 | 0.7 | 0.1 | 0.4 | 0.5 |
| CHB | 0.0 | 1.0 | 0.0 | 0.1 | 0.9 | 0.2 | 0.8 | 0.0 | 0.3 | 0.6 |
| JPT | 0.0 | 1.0 | 0.0 | 0.1 | 0.9 | 0.2 | 0.8 | 0.0 | 0.3 | 0.7 |
| YRI | 0.6 | 0.4 | 0.3 | 0.5 | 0.2 | 0.9 | 0.1 | 0.8 | 0.2 | 0.0 |
Figure 1. Variations in the ADH1B proximal promoter region.
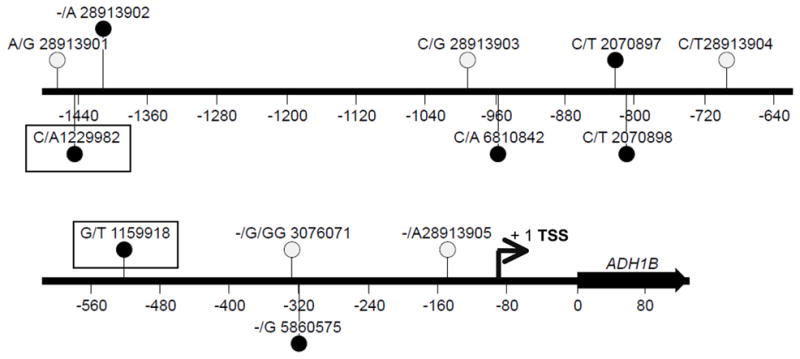
Twelve variations in the proximal promoter are shown as vertical lines with the SNP ID (rs number) and the two alleles to the left of the rs number. Filled circles represent those variations that were tested in the transfection assays. Variations associated with the risk for alcoholism are outlined by rectangular boxes. For some variations (shown as open circles), the same allele was present in all the haplotypes tested. Numbering is relative to the ADH1B translational start site (+1). The transcriptional start site (TSS) is indicated by an arrow.
The effects of different alleles of rs1229982 and rs1159918, along with the other natural variations, were tested by transient transfection assays. The region was amplified from DNA of different individuals to generate the natural haplotypes, and cloned into the pXP2 reporter plasmid. Five different haplotypes (Table 2) were obtained; all five had the same allele for SNPs rs28913901, rs28913903, rs28913904, rs3076071 and rs28913905 (which have low minor allele frequencies); they differed in the remaining seven positions (Figure 1).
Table 2. Tested haplotypes of the ADH1B proximal promoter.
Five haplotypes with seven SNPs that were obtained from cloning the proximal promoter region are shown; they were identical at all other positions. Alleles of the two SNPs that were associated with alcoholism are shown in bold.
| Haplotype | rs1229982 | rs28913902 | rs6810842 | rs2070897 | rs2070898 | rs1159918 | rs5860575 |
|---|---|---|---|---|---|---|---|
| 1 | A | - | A | C | C | T | - |
| 2 | A | T | C | T | C | G | - |
| 3 | C | T | C | T | T | G | - |
| 4 | C | T | C | C | C | T | - |
| 5 | C | T | A | C | C | T | G |
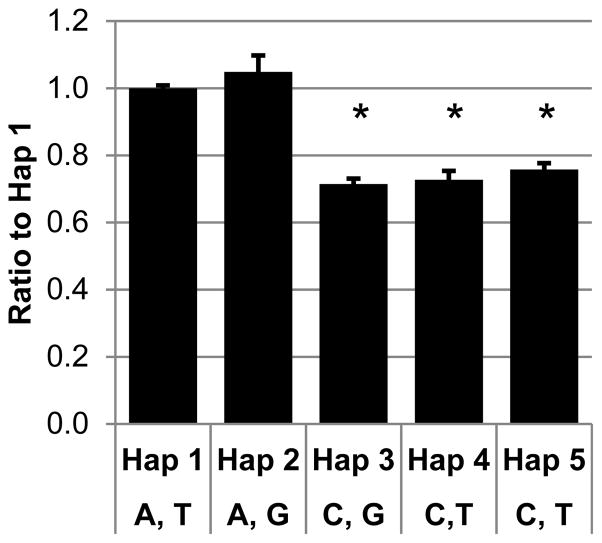
Five haplotypes were tested in HepG2 cells by transient transfections. The activity of each was normalized to that of haplotype 1, which had the sequence closest to the reference sequence in GenBank (accession number NT_016354.17). A significant decrease in the promoter activity relative to haplotype 1 was observed in haplotypes 3, 4 and 5 (p-value ≤5 × 10−7), whereas no difference in activity was seen with haplotype 2 (p-value = 0.39; Figure 2). Haplotypes 1 and 2 share an A at rs1229982 (Table 2), suggesting that rs1229982 might be responsible for the decrease in promoter activity.
Figure 2. Variations in the ADH1B promoter affect activity.
Five different haplotypes of the ADH1B proximal promoter region were tested in transient transfections in HepG2 cells (n = 12). The promoter activity of each haplotype was normalized to the promoter activity of Haplotype 1. Alleles of rs1229984, and rs1159918, respectively are shown on the x-axis. The error bars indicate standard errors of the mean. * p-value 7le;7 × 10−4.
Data from the different haplotypes were therefore grouped based on the allele of each SNP (e.g., the average activities of all haplotypes with the rs1229982 A allele were compared to those of all haplotypes with C at that position) for analysis. A significant difference in expression was observed for only one SNP, rs1229982 (Table 3). Promoters with the A allele at rs1229982 have, on average, 39% more activity than those with C.
Table 3. Analysis of the haplotypes grouped by the allele at each SNP.
The promoter activities (β-galactosidase normalized luciferase activity) of different haplotypes were grouped by the alleles of four SNPs; the other three SNPs could not be compared as there was only one representative haplotype for one of the alleles. The ratio of the activity of the two groups was determined and the significance of this ratio was calculated by t-test.
| Allele 1 | Allele 2 | Ratio of allele 1/allele 2 | p-value | |
|---|---|---|---|---|
| rs1229982 | A | C | 1.39 | 6.2×10−6 |
| rs6810842 | C | A | 0.95 | 0.46 |
| rs2070897 | C | T | 0.94 | 0.42 |
| rs1159918 | T | G | 0.94 | 0.42 |
The ADH1B proximal promoter region extending to 270 bp upstream of the translational start site has binding sites for multiple transcription factors [29–30]. A putative CCAAT enhancer binding protein (C/EBP) family of transcription factor was predicted by PROMO [31] at rs1229982, which might explain the effect on promoter activity.
The A allele of rs1229982 is found at a low frequency (0.2) in European Americans (CEU); it is at high frequency in the Nigeria (YRI) population (60%) but is completely lost in the Asian populations (CHB, JPT). In the European-American population [25], the A allele was observed to be overtransmitted to alcoholics. This is somewhat surprising, given that higher ADH activity coding alleles are protective [13–20].
Because we did not detect a functional effect for rs1159918, its association with alcoholism could either be due to functional effects in other tissues or through linkage disequilibrium (LD) with another SNP that is functional. In the CEU population, rs1159918 is reported to be in high LD with rs6810842 (r2 = 0.949), but rs6810842 also doesn’t appear to have an effect on promoter activity (Table 3); the two SNPs are in different phase in 1 of the 5 haplotypes we tested. There are other SNPs in the ADH cluster which are in LD (r2 >0.25) with rs1159918 but have not been tested for function.
In summary, we have identified a functional effect of the variation rs1229982 in the ADH1B proximal promoter; the A allele at this position had higher promoter activity, and was overtransmitted to alcoholics. These results support our hypothesis that regulatory variations could alter the levels of ADH enzymes and thus affect the risk for alcoholism.
Acknowledgments
We thank Jun Wang for technical assistance. This research was supported by grant R37AA006460 from the National Institute on Alcohol Abuse and Alcoholism, NIH.
Footnotes
Conflict of interest statement: The authors declare that there are no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Birley AJ, et al. Genetic time-series analysis identifies a major QTL for in vivo alcohol metabolism not predicted by in vitro studies of structural protein polymorphism at the ADH1B or ADH1C loci. Behav Genet. 2005;35:509–524. doi: 10.1007/s10519-005-3851-6. [DOI] [PubMed] [Google Scholar]
- 2.Goodwin DW, et al. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch Gen Psychiatry. 1973;28:238–243. doi: 10.1001/archpsyc.1973.01750320068011. [DOI] [PubMed] [Google Scholar]
- 3.Goodwin DW, et al. Drinking problems in adopted and nonadopted sons of alcoholics. Arch Gen Psychiatry. 1974;31:164–169. doi: 10.1001/archpsyc.1974.01760140022003. [DOI] [PubMed] [Google Scholar]
- 4.Kendler KS, et al. Temperance board registration for alcohol abuse in a national sample of Swedish male twins, born 1902 to 1949. Arch Gen Psychiatry. 1997;54:178–184. doi: 10.1001/archpsyc.1997.01830140090015. [DOI] [PubMed] [Google Scholar]
- 5.Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGue M. A behavioral-genetic perspective on children of alcoholics. Alcohol Health Res World. 1997;21:210–217. [PMC free article] [PubMed] [Google Scholar]
- 7.McGue M. The Behavioral Genetics of Alcoholism. Current Directions in Psychological Science. 1999;8:109–115. [Google Scholar]
- 8.Nurnberger JI, Jr, et al. A family study of alcohol dependence: coaggregation of multiple disorders in relatives of alcohol-dependent probands. Arch Gen Psychiatry. 2004;61:1246–1256. doi: 10.1001/archpsyc.61.12.1246. [DOI] [PubMed] [Google Scholar]
- 9.Prescott CA, Aggen SH, Kendler KS. Sex differences in the sources of genetic liability to alcohol abuse and dependence in a population-based sample of U.S. twins. Alcohol Clin Exp Res. 1999;23:1136–1144. doi: 10.1111/j.1530-0277.1999.tb04270.x. [DOI] [PubMed] [Google Scholar]
- 10.Prescott CA, Kendler KS. Genetic and environmental contributions to alcohol abuse and dependence in a population-based sample of male twins. Am J Psychiatry. 1999;156:34–40. doi: 10.1176/ajp.156.1.34. [DOI] [PubMed] [Google Scholar]
- 11.Edenberg HJ. Regulation of the mammalian alcohol dehydrogenase genes. Progress in Nucleic Acid Research and Molecular Biology. 2000;64:295–341. doi: 10.1016/s0079-6603(00)64008-4. [DOI] [PubMed] [Google Scholar]
- 12.Hurley TD, Edenberg HJ, Li T-K. Pharmacogenomics of Alcoholism. In: Julio Licinio M-LW, editor. Pharmacogenomics: The Search for Individualized Therapies. Culinary and Hospitality Industry Publications Services; 2002. pp. 417–441. [Google Scholar]
- 13.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- 14.Xu YL, et al. Genotyping of human alcohol dehydrogenases at the ADH2 and ADH3 loci following DNA sequence amplification. Genomics. 1988;2:209–214. doi: 10.1016/0888-7543(88)90004-3. [DOI] [PubMed] [Google Scholar]
- 15.Thomasson HR, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–681. [PMC free article] [PubMed] [Google Scholar]
- 16.Whitfield JB. Meta-analysis of the effects of alcohol dehydrogenase genotype on alcohol dependence and alcoholic liver disease. Alcohol Alcohol. 1997;32:613–619. doi: 10.1093/oxfordjournals.alcalc.a008303. [DOI] [PubMed] [Google Scholar]
- 17.Borras E, et al. Genetic polymorphism of alcohol dehydrogenase in europeans: the ADH2*2 allele decreases the risk for alcoholism and is associated with ADH3*1. Hepatology. 2000;31:984–989. doi: 10.1053/he.2000.5978. [DOI] [PubMed] [Google Scholar]
- 18.Whitfield JB. Alcohol dehydrogenase and alcohol dependence: variation in genotype-associated risk between populations. Am J Hum Genet. 2002;71:1247–1250. doi: 10.1086/344287. author reply 1250–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luczak SE, Glatt SJ, Wall TL. Meta-analyses of ALDH2 and ADH1B with alcohol dependence in Asians. Psychol Bull. 2006;132:607–621. doi: 10.1037/0033-2909.132.4.607. [DOI] [PubMed] [Google Scholar]
- 20.Chen CC, et al. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edenberg HJ, Jerome RE, Li M. Polymorphism of the human alcohol dehydrogenase 4 (ADH4) promoter affects gene expression. Pharmacogenetics. 1999;9:25–30. doi: 10.1097/00008571-199902000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Guindalini C, et al. Association of genetic variants in alcohol dehydrogenase 4 with alcohol dependence in Brazilian patients. Am J Psychiatry. 2005;162:1005–1007. doi: 10.1176/appi.ajp.162.5.1005. [DOI] [PubMed] [Google Scholar]
- 23.Chen HJ, Tian H, Edenberg HJ. Natural haplotypes in the regulatory sequences affect human alcohol dehydrogenase 1C (ADH1C) gene expression. Hum Mutat. 2005;25:150–155. doi: 10.1002/humu.20127. [DOI] [PubMed] [Google Scholar]
- 24.Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- 25.Edenberg HJ, et al. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Human Molecular Genetics. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- 26.Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15:755–768. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- 28.Nordeen SK. Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques. 1988;6:454–458. [PubMed] [Google Scholar]
- 29.Brown CJ, Zhang L, Edenberg HJ. Tissue-specific differences in the expression of the human ADH2 alcohol dehydrogenase gene and in binding of factors to cis-acting elements in its promoter. DNA Cell Biol. 1994;13:235–247. doi: 10.1089/dna.1994.13.235. [DOI] [PubMed] [Google Scholar]
- 30.Brown CJ, Zhang L, Edenberg HJ. Gene expression in a young multigene family: tissue-specific differences in the expression of the human alcohol dehydrogenase genes ADH1, ADH2, and ADH3. DNA Cell Biol. 1996;15:187–196. doi: 10.1089/dna.1996.15.187. [DOI] [PubMed] [Google Scholar]
- 31.Farre D, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]