Abstract
The specification of an appropriate number of cardiomyocytes from the lateral plate mesoderm requires a careful balance of both positive and negative regulatory signals. To identify new regulators of cardiac specification, we performed a phenotype-driven ENU mutagenesis forward genetic screen in zebrafish. In our genetic screen we identified a zebrafish ctr9 mutant with a dramatic reduction in myocardial cell number as well as later defects in primitive heart tube elongation and atrioventricular boundary patterning. Ctr9, together with Paf1, Cdc73, Rtf1 and Leo1, constitute the RNA polymerase II associated protein complex, PAF1. We demonstrate that the PAF1 complex (PAF1C) is structurally conserved among zebrafish and other metazoans and that loss of any one of the components of the PAF1C results in abnormal development of the atrioventricular boundary of the heart. However, Ctr9, Cdc73, Paf1 and Rtf1, but not Leo1, are required for the specification of an appropriate number of cardiomyocytes and elongation of the heart tube. Interestingly, loss of Rtf1 function produced the most severe defects, resulting in a nearly complete absence of cardiac precursors. Based on gene expression analyses and transplantation studies, we found that the PAF1C regulates the developmental potential of the lateral plate mesoderm and is required cell autonomously for the specification of cardiac precursors. Our findings demonstrate critical but differential requirements for PAF1C components in zebrafish cardiac specification and heart morphogenesis.
Keywords: cardiac specification, heart tube, PAF1 complex, PAF1C, zebrafish
Introduction
The specification of an appropriate number of cardiac precursors is essential for the development of a functional heart. In vertebrates, cardiac progenitor cells are specified from the lateral plate mesoderm (LPM) to form a bilaterally symmetric heart field. These precursor cells then migrate medially and fuse at the midline. In zebrafish, the fusion of the heart field generates a cone-like structure, which develops into a pulsatile primitive heart tube via a process of cellular migration and rearrangement.
The size of the zebrafish heart field is regulated by both positive and negative signals originating in the LPM and surrounding tissues (Chen and Fishman, 2000; Yelon, 2001; Yelon et al., 2002; Zaffran and Frasch, 2002). Inductive signals such as gata4/5/6, bmp2b, fgf8, hand2 and nkx2.5 in the LPM push mesodermal cells toward a cardiac fate (Chen and Fishman, 1996; Holtzinger and Evans, 2007; Marques et al., 2008; Peterkin et al., 2007; Reifers et al., 2000; Reiter et al., 1999; Reiter et al., 2001b; Trinh et al., 2005; Yelon et al., 2000), while RA signaling negatively regulates cardiac progenitor production by activating the expression of hoxb5b in the limb fields (Waxman et al., 2008). Expression of scl and etsrp in the LPM anterior to the heart field establish the anterior boundary of cardiac progenitors (Schoenebeck et al., 2007), while unknown signals from the notochord establish the posterior heart field boundary (Goldstein and Fishman, 1998). Precise regulation of the expression of all the factors involved in specification of the heart field is vitally important for the development of a functional heart.
The PAF1 complex (PAF1C) is an RNA Polymerase II-interacting protein complex required for the development of the heart and other organs (Akanuma et al., 2007; Nguyen et al., 2010). The PAF1C was originally identified in yeast, and consists of five core proteins: Paf1, Ctr9, Rtf1, Cdc73, and Leo1. Loss of the PAF1C components Paf1 and Ctr9 in yeast prevents transcriptional activation of the G1 cyclin CLN2, resulting in defective cell cycle control and cell growth (Koch et al., 1999). Yeast Paf1, Ctr9, and Rtf1 are also essential for the Set1-mediated methylation of histone H3 lysine-4 and Paf1 and Rtf1 are additionally required for Dot1- mediated histone H3 lysine-79 methylation (Krogan et al., 2003). Furthermore, Paf1 is necessary for proper poly(A) site utilization of a subset of yeast genes (Penheiter et al., 2005).
In multicellular organisms, the PAF1C is essential for numerous signaling pathways and developmental processes. In Arabidopsis, PAF1C proteins are important regulators of flowering-time. Loss of the Arabidopsis homologs of Paf1, Ctr9, and Rtf1 results in an early flowering phenotype because of a reduction in histone H3 lysine-4 trimethylation at the promoter of genes required to repress flowering (He et al., 2004; Oh et al., 2004). The Rtf1 subunit of the PAF1C also regulates the Notch signaling pathway in Drosophila and zebrafish (Akanuma et al., 2007; Tenney et al., 2006). Loss of Rtf1 in zebrafish causes a reduction in the expression level of Notch-regulated genes in the somites, resulting in aberrant somitogenesis (Akanuma et al., 2007) and loss of Leo1 in zebrafish produces defects in atrioventricular boundary and neural crest development (Nguyen et al., 2010). Interestingly, mutation in zebrafish cdc73 is able to suppress the defects in erythroid gene transcription elongation of Transcription Intermediary Factor 1gamma mutants, resulting in a rescue of blood development (Bai et al., 2010). Lastly, Cdc73 has also been shown to positively regulate the Wnt and Hedgehog signaling pathways in Drosophila and mammalian cells via direct interactions with beta-catenin and Gli proteins (Mosimann et al., 2006, 2009). However, the function of each component of the PAF1C has not yet been evaluated systematically in any animal model.
From our ENU mutagenesis screen for genes affecting cardiovascular development, we identified a mutant, LA961, with a severe reduction in cardiac cell number. Positional cloning of this mutant revealed a mutation in the zebrafish homolog of ctr9, a member of the PAF1C. Key differences are present in the cardiac phenotypes of ctr9 and leo1 mutants, suggesting distinct requirements for the individual proteins of the PAF1C during cardiogenesis. By morpholino knockdown, we found that Paf1, Cdc73 and Rtf1, like Ctr9, are important for establishing an appropriate number of cardiomyocytes and for elongation and patterning of the primitive heart tube. PAF1C components are not essential for the formation of LPM, but instead regulate the specification of LPM derivatives. Furthermore, by transplanting Rtf1-deficient cells into wild type embryos, we found that the PAF1C plays a cell-intrinsic role in the specification of cardiomyocytes.
Materials and Methods
Zebrafish and ENU mutagenesis
Zebrafish colonies were cared for and bred under standard conditions. Developmental stages of zebrafish embryos were determined using standard morphological features of fish raised at 28.5°C (Westerfield, 2000). ENU mutagenesis of Tg(kdrl:GFP)LA116 male zebrafish was performed as previously described (Nguyen et al., 2010).
Positional cloning
The LA961 mutation was identified in the AB background and heterozygous carriers of this mutation were crossed to the polymorphic WIK strain for mapping. Embryo lysis and PCR-based bulk segregant analysis were performed as previously described (Nguyen et al., 2010). Using the Massachusetts General Hospital microsatellite map, we found that LA961 was linked to markers Z1239 and Z41496 on chromosome 7. The markers generated during fine mapping include 1162-5 and 395-1 and their primer sequences are as follows (from 5’ to 3’):
1162-5-F: ACTCCTGTGGGATGTGTGTG;
1162-5-R: CAAATAAATTGTATGGAACCCAAT;
395-1-F: TCAGTTTTGACATTCCTGCATC;
395-1-R: GCTGTGGTTTCATGTTCGTC.
To identify the ctr9LA961 mutation, total RNA was isolated from 1 day post fertilization (dpf) LA961 mutants using RNA Wiz (Ambion, Austin, TX) and cDNA was synthesized using the Superscript II Kit (Invitrogen). cDNA fragments were amplified with Phusion polymerase (NEB) and cloned into pCR-Blunt II-TOPO (Invitrogen) for sequencing.
Cardiomyocyte cell counting
Tg(myl7:EGFP) transgenic (Huang et al., 2003) wild type and ctr9LA961 mutant embryos were dissociated in Ca2+-free Ringers solution and flat mounted. Dissociated cardiac cells were photographed and counted using a Zeiss Axiovert 200M equipped with a Zeiss Axiocam camera.
Constructs and morpholinos
The ctr9WT, ctr9LA961, and leo1 full length cDNAs were amplified from 1 dpf wild-type embryo cDNA using Phusion polymerase (NEB) and cloned into pCS2+myc or pCS2+3XFLAG for tagging with the Myc and FLAG epitopes. Full length cDNA for cdc73, paf1, and rtf1 was amplified with KOD polymerase (Novagen) and cloned into pCS2+3XFLAG. Plasmids were cut with NotI and SP6 RNA polymerase was used to generate mRNA for injection.
Morpholino oligonucleotides targeting the translation initiation sites of ctr9, cdc73, paf1, and rtf1 (ctr9MO, cdc73MO, paf1MO, and rtf1MO) were purchased from Gene Tools, LLC and injected at dosages of 0.75 ng (for ctr9MO), 2 ng (for cdc73MO), 1.5 ng (for paf1MO) and 0.25 ng (for rtf1MO). Cdc73MO and paf1MO were coinjected with a p53 morpholino (4 ng and 6.5 ng, respectively) to prevent non-specific cell death. The sequences of the morpholinos used are as follows (from 5’ to 3’):
ctr9MO: GATTTCAATGGATCCCCGAGACATG;
cdc73MO: GAAGAACACTCAACACGTCCGCCAT;
paf1MO: CCTGCGTCTGTATGGTAGGAGCCAT;
rtf1MO: CTTTCCGTTTCTTTACATTCACCAT (Akanuma et al., 2007);
p53 morpholino: GCGCCATTGCTTTGCAAGAATTG.
Morpholino efficacy was tested by Western blotting to detect levels of PAF1 complex proteins in 1 day old morpholino injected embryo lysates. Embryos were lysed in Rubinfield’s Lysis buffer (20mM Tris pH 8.0, 140mM NaCl, 1% Triton X-100, 10% glycerol, 1mM EGTA, 1.5mM MgCl2, 1mM Na3VO4, 50mM NaF, 1mM DTT) with protease inhibitors (Roche), and one embryo equivalent was loaded per lane on a 8% polyacrylamide gel. Electrophoresed proteins were transferred to a nitrocellulose membrane. Antibodies against PAF1 complex proteins include anti-Ctr9 (1:2,500, SAB1100738, Sigma), anti-Paf1 (1:10,000, A300-172A, Bethyl Laboratories, Inc.), anti-Cdc73 (1:5,000, C3620, Sigma), and anti-Rtf1 (1:2,000, ab70690, Abcam). Relative protein levels were determined by densitometry measurements in Adobe Photoshop CS3 using β-actin (1:5,000, A1978, Sigma) as a loading control.
Whole mount in situ hybridization
Embryos for in situ hybridization were raised in embryo medium supplemented with 0.2 mM 1-phenyl-2-thiourea to maintain optical transparency (Westerfield, 2000). Whole-mount in situ hybridization was performed as described previously (Chen and Fishman, 1996). The antisense RNA probes used in this study include cmlc2 (myl7), nkx2.5, vmhc, amhc (myh6), bmp4, notch1b, tbx2b, tbx20, gata4, gata5, hand2, and ctr9.
Antibody staining
Embryos injected with 200 pg of RNA encoding Myc-tagged zebrafish ctr9, FLAG-tagged zebrafish cdc73, FLAG-tagged zebrafish paf1, or FLAG-tagged zebrafish rtf1 were fixed in 4% PFA in PBS at 75% epiboly. The fixed embryos were incubated in primary antibody (1:50 mouse anti-FLAG M2, F1804, Sigma; 1:50 rabbit anti-Myc, 06–549, Upstate) in blocking solution (10% goat serum in PBDT) for 2 hours at room temperature followed by detection with fluorescent secondary antibodies (1:200 anti-mouse IgG1-R, sc-2084, Santa Cruz Biotechnology; 1:200 anti-rabbit IgG-Cy5, 81-6116, Invitrogen). Nuclei were stained with DAPI (Invitrogen) and embryos were embedded in 1% low-melt agarose and imaged on a Zeiss LSM510 confocal microscope equipped with a 63X water objective.
Coimmunoprecipitations
For coimmunoprecipitation analysis, 60 embryos injected with tagged constructs were lysed using Rubinfeld’s Lysis Buffer (20 mM Tris pH 8.0, 140 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EGTA, 1.5 mM MgCl2, 1 mM Na3VO4, 50 mM NaF, 1 mM DTT) with protease inhibitors (Roche). Binding of antibody (mouse anti-Myc 9E10, 05–419, Upstate) was performed overnight on a nutator at 4°C. Protein G sepharose beads (17-0618-01, GE Healthcare) were used to pull down bound proteins. Immunoprecipitated proteins were separated on a 7% polyacrylamide gel and transferred to a nitrocellulose membrane for Western analysis. Mouse anti-FLAG M2 (1:10,000, F1804, Sigma) and rabbit anti-mouse IgG-HRP (1:20,000, 61–6020, Invitrogen) antibodies were used to detect Flag-tagged proteins.
Transplantations
For experiments requiring MO treatment Tg(myl7:EGFP) donor embryos or Tg(myl7:mCherry) recipient embryos were injected with 0.25 ng rtf1MO and 0.5 ng p53MO. Donor cells were drawn into a glass needle by back pressure and 20–30 cells were transplanted into high stage recipient embryos at the margin in two locations 90° from each other. On 1 or 2 dpf recipient embryos were scored for donor cell contribution to the heart. For imaging, 2 dpf hearts were gently removed from the embryos and mounted in 1% low melt agarose. Images were taken using a Zeiss LSM510 confocal microscope.
Results
Identification of a novel zebrafish cardiac mutant, LA961
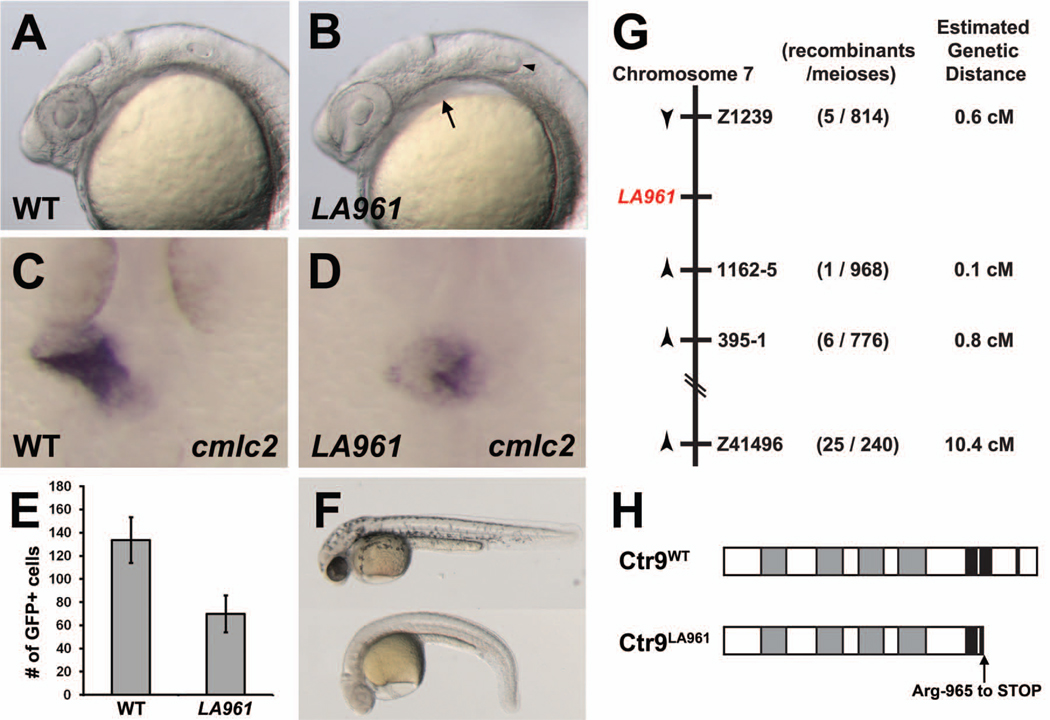
In a screen for zebrafish cardiovascular mutants, we identified a novel mutant, LA961, with severe defects in the morphogenesis of the primitive heart tube. In wild type zebrafish embryos, a beating heart tube is visible on the left side of the body by 24 hours post fertilization (hpf). At 24 hpf, LA961 mutants lack a visible heart tube. Additionally, LA961 mutants exhibit a slight pericardial edema and an absence of otoliths (Figure 1A,B). By 35 hpf, LA961 mutants develop a severely reduced and dysmorphic heart, a complete lack of pigment cells, and a downwardly curved body (Figure 1F), indicating that the LA961 gene plays a role in multiple developmental processes.
Figure 1. Mutation in zebrafish ctr9 perturbs primitive heart tube elongation and causes a reduction in cardiomyocyte cell number.
(A,B) Lateral views of 24 hpf wild type (A) and LA961 mutant (B) embryos. LA961 mutants can be distinguished from wild type embryos based on missing otoliths (arrowhead) and pericardial edema (arrow). (C,D) Wild type and LA961 mutant embryos analyzed for cmlc2 expression at 24 hpf. Wild type embryos have an elongated primitive heart tube (C), whereas LA961 mutants have a clump of cardiomyocytes at the midline (D). (E) Graph of number of GFP-positive cardiomyocytes in wild type (WT) and LA961 mutant embryos. Error bars indicate standard deviations. (F) Lateral view of wild type (top) and LA961 mutant (bottom) embryos at 35 hpf. (G) Diagram of LA961 mapping. Estimated genetic distance is in centimorgans (cM). (H) Diagram of Ctr9WT and Ctr9LA961 proteins. Grey regions are predicted tetratricopeptide repeat domains and black regions are predicted nuclear localization domains.
We used the cardiomyocyte marker cmlc2 (also known as my17) to further examine the heart phenotype in LA961 and found that these mutants have cmlc2-expressing cells; however these cardiomyocytes fail to elongate into a tube by 24 hpf and are instead clustered at the midline (Figure 1C,D). The number of cells expressing cmlc2 also appeared lower in LA961 mutants. We directly counted the number of cardiomyocytes in LA961 at 24 hpf by crossing this mutation into a Tg(myl7:EGFP) genetic background and recording the number of cells expressing green fluorescent protein (GFP). We found an average of 134 GFP-positive cells in wild type embryos at 24 hpf (n= 7), whereas LA961 mutants only possessed an average of 70 GFP-positive cells at the same developmental stage (n= 6; p = 0.00006; Figure 1E). This dramatic reduction in myl7:EGFP-positive cells indicates that the LA961 gene is critical for generating an appropriate number of cardiomyocytes in addition to its role in the morphogenesis of the primitive heart tube.
The LA961 locus encodes Ctr9
We mapped the LA961 locus to an 11 centimorgan (cM) region on chromosome 7. Using custom simple sequence length polymorphism (SSLP) markers, we refined the LA961 interval and found one marker, 1162-5, located 0.1 cM from the LA961 mutant locus (Figure 1G). Sequencing of the genes near this marker identified a nonsense mutation in the ctr9 gene. Overexpression of wild type ctr9 rescued the LA961 mutant phenotype (out of 40 injected embryos generated by crossing LA961 heterozygotes, none exhibited an LA961 mutant phenotype) and wild type embryos injected with 0.75 ng of a morpholino oligonucleotide targeting the translation start site of ctr9 (ctr9MO) exhibited phenotypes identical to those seen in ctr9 mutants (n= 80) (Supplemental Figure 1).
Zebrafish Ctr9 contains 3 putative nuclear localization signal (NLS) domains in its C-terminal tail. The ctr9LA961 mutation eliminates the third and the majority of the second of these NLSs (Figure 1H). To test if this mutation results in defective localization of the Ctr9 protein, we tagged wild type and mutant Ctr9 protein with a Myc epitope and overexpressed these constructs in zebrafish embryos. Interestingly, we found that both Ctr9WT and Ctr9LA961 localize to the nucleus (Supplemental Figure 2A,B), indicating that the final 2 NLSs are not required for the cellular localization of Ctr9. However, examination of endogenous ctr9 mRNA levels in ctr9LA961 mutants by in situ hybridization revealed a dramatic reduction in expression (Supplemental Figure 2C,D), indicating that ctr9LA961 transcripts are likely unstable and are degraded by cellular nonsense mediated decay mechanisms. Together these findings suggest that loss of function of Ctr9 is causative to the cardiac and pigment defects observed in LA961 and that the LA961 mutation may be hypomorphic.
The PAF1C is evolutionarily conserved in zebrafish
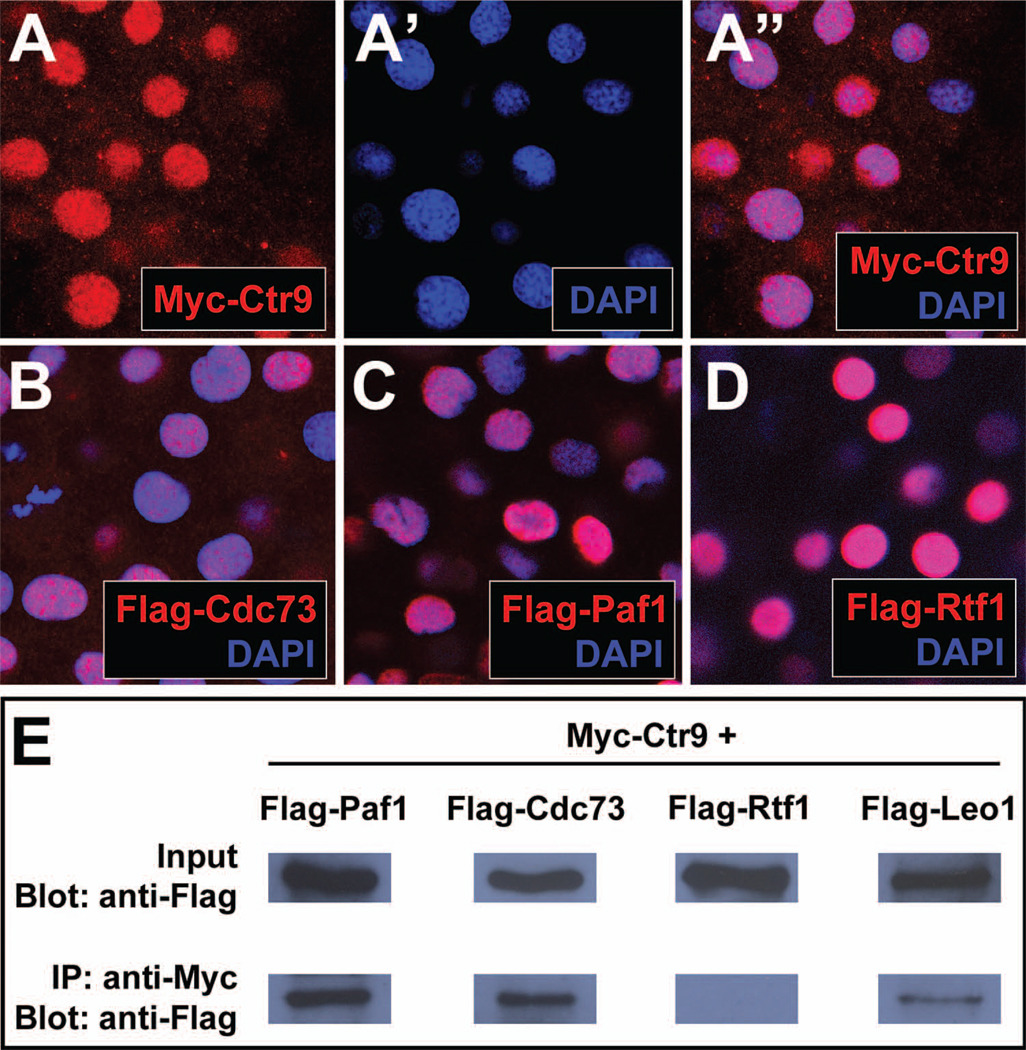
Ctr9 is a member of the multi-protein PAF1C (also consisting of Paf1, Rtf1, Cdc73, and Leo1), which was initially identified in yeast. PAF1C proteins have been conserved through evolution, and are present and interact with one another in Drosophila and humans (Adelman et al., 2006; Kim et al., 2010). We questioned whether the PAF1C was structurally conserved in the zebrafish as well. Members of the PAF1C are known to localize to the nucleus and interact with RNA polymerase II (Mueller and Jaehning, 2002; Shi et al., 1997). We have previously shown that zebrafish Leo1 exhibits a nuclear localization (Nguyen et al., 2010). Here we further tested whether the other zebrafish PAF1 component homologs were present in the same subcellular compartment. We found that Ctr9, Cdc73, Paf1, and Rtf1 all localized specifically to the nucleus (Figure 2A–D). This finding is consistent with the localization of the PAF1C in yeast, Drosophila, and humans (Adelman et al., 2006; Kim et al., 2010; Lin et al., 2007a; Lin et al., 2007b; Porter et al., 2005), and suggests that all five members of the putative zebrafish PAF1C can potentially physically interact.
Figure 2. Zebrafish PAF1C components localize to the nucleus and interact with each other in vivo.
(A-A”) Panel (A”) shows the colocalization of Myc-tagged Ctr9 protein (red, A) and nuclear DAPI staining (blue, A’). (B) Flag-tagged Cdc73 (red) and DAPI staining (blue) colocalize in the nucleus. (C) Flag-tagged Paf1 (red) and DAPI staining (blue) colocalize in the nucleus. (D) Flag-tagged Rtf1 (red) and DAPI staining (blue) colocalize in the nucleus. (E) Flag-Paf1, Flag-Cdc73, and Flag-Leo1 coimmunoprecipitate with Myc-Ctr9. Input row shows 1 embryo equivalent of total protein lysate. IP row shows immunoprecipitated protein from 20 embryo equivalents of total protein lysate.
To determine if the zebrafish PAF1C homologs physically interacted, we co-expressed different tagged PAF1 components in zebrafish embryos and performed a coimmunoprecipitation assay. We found that Paf1, Cdc73, and Leo1 efficiently coimmunoprecipitated with Ctr9, but that Rtf1 did not, despite significant levels of expressed protein (Figure 2E). This finding is consistent with previous studies showing that while Rtf1 coimmunoprecipitates with other PAF1 components in yeast, its interaction with other PAF1C proteins is comparatively weaker in animals (Adelman et al., 2006; Mueller and Jaehning, 2002; Rozenblatt-Rosen et al., 2005; Yart et al., 2005). This implies that the PAF1C in zebrafish is similar to that of other animals, consisting of at a minimum Ctr9, Paf1, Cdc73, and Leo1, and that the interaction of Rtf1 with this complex may be too weak to detect in a standard coimmunoprecipitation assay.
The PAF1C is required for multiple steps of cardiac formation in zebrafish
In addition to the ctr9 mutation, we also recently identified a mutation in the zebrafish leo1 homolog. Interestingly, the specification of cardiomyocytes and heart tube elongation are normal in leo1 mutants (Nguyen et al., 2010), indicating that loss of different members of the PAF1C can produce different effects on the process of cardiac development.
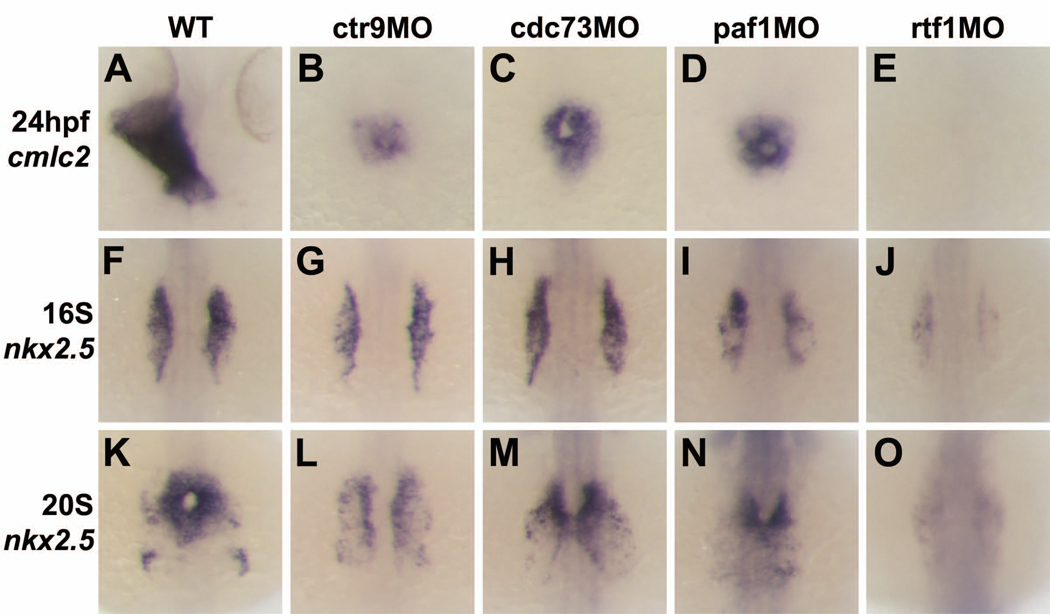
Because of the disparate roles played by Ctr9 and Leo1 during early cardiac development, we investigated the functions of the zebrafish homologs of the remaining three PAF1C components, Cdc73, Paf1, and Rtf1. We generated morpholino oligonucleotides targeting the translation start site of each gene and confirmed their efficacy by Western blotting (Supplemental Figure 3). Using these morpholinos, we found that knockdown of cdc73, paf1, and rtf1 resulted in a reduction in cardiac tissue like that observed in ctr9 mutants and morphants (Figure 3A–E). The loss of cardiac tissue was most severe in rtf1 morphants, with most of these embryos completely lacking cardiomyocytes (Figure 3E). The reduction in cardiac cell number in ctr9, cdc73, paf1, and rtf1 morphants suggests that the PAF1C plays a critical role in the production of an appropriate number of cardiomyocytes.
Figure 3. Loss of Ctr9, Cdc73, Paf1, and Rtf1 cause defects in myocardial migration, cardiomyocyte cell number, and primitive heart tube elongation.
(A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas ctr9 (B), cdc73 (C), and paf1 (D) morphants have a small clump of cardiomyocytes situated at the midline. Rtf1 morphants have no detectable cmlc2 expression at 24 hpf (E). (F–O) Dorsal views of nkx2.5 expression in 16S (F–J) and 20S (K–O) stage wild type and morphant embryos. In wild type embryos, myocardial progenitors arise in the LPM, migrate towards the midline, and fuse to form a cardiac cone (F,K). In ctr9 morphants (G,L), nkx2.5 expression is reduced and myocardial migration and fusion are delayed. Cdc73 morphants (H,M) also display delays in the migration and fusion of the myocardial progenitors, but have relatively normal nkx2.5 expression levels. Paf1 morphants (I,N) and rtf1 morphants (J,O), like ctr9 morphants, display reduced nkx2.5 expression and delayed migration and fusion of the myocardial progenitors. Expression of nkx2.5 in rtf1 morphants is barely detectable.
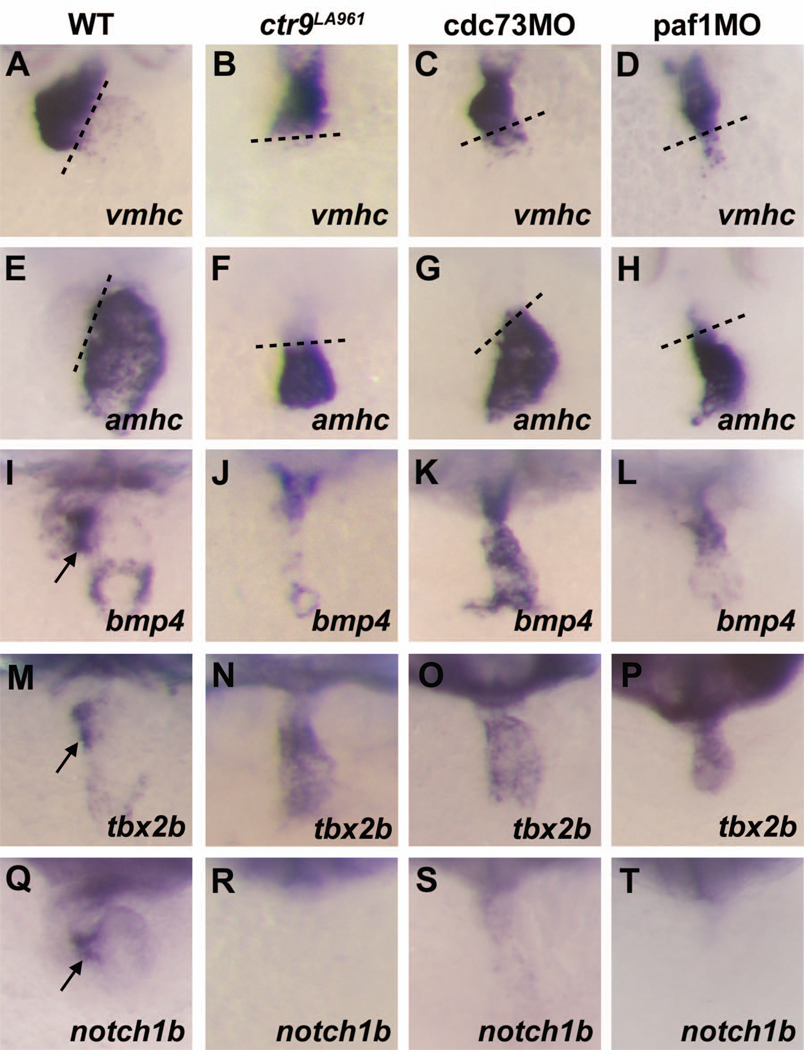
Although early cardiac development proceeds normally in leo1 mutants, our lab has previously identified a role for leo1 in the differentiation of the atrioventricular (AV) boundary. Loss of leo1 results in a failure to initiate expression of an AV boundary genetic program including bmp4, versican, notch1b, and wnt2bb (Nguyen et al., 2010). We examined the differentiation of the AV boundary in embryos deficient in the other PAF1C components to determine if the PAF1C as a whole plays a role in this developmental process. By 2 days post fertilization (dpf), rtf1 morphants have no detectable cardiac tissue (data not shown) and ctr9-, cdc73-, and paf1-deficient embryos have small, dysmorphic hearts. Like wild type embryos, ctr9-, cdc73-, and paf1-deficient embryos generate a morphologically distinct ventricle and atrium separated by an AV canal and express vmhc and amhc in a chamber-specific manner (Figure 4A–H). However, the expression patterns of markers for the AV boundary are highly abnormal in these embryos. In 2-day-old wild type embryos, bmp4 and tbx2b are strongly expressed in the myocardial cells at the AV boundary and sinus venosus (Figure 4I, M). In ctr9-, cdc73-, and paf1-deficient embryos, expression of bmp4 is not concentrated at the AV boundary (Figure 4J–L) and tbx2b is expressed throughout the heart (Figure 4N–P), suggesting that the differentiation of the AV boundary is aberrant in these embryos. In addition, we found that the cardiac expression of notch1b, which is strongly localized to the endocardial cells of the AV boundary in wild type embryos, was absent in the hearts of ctr9-, cdc73-, and paf1-deficient embryos (Figure 4Q–T).
Figure 4. Ctr9, Cdc73, and Paf1 are required for AV boundary specification.
(A–D) Cardiac expression of vmhc in 48 hpf embryos. In wild type (A), ctr9LA961 mutant (B), cdc73 morphant (C), and paf1 morphant (D) embryos, vmhc expression is detected primarily in the ventricle. (E–H) Cardiac expression of amhc in 48 hpf embryos. In wild type (E), ctr9LA961 mutant (F), cdc73 morphant (G), and paf1 morphant (H) embryos, amhc expression is detected primarily in the atrium. (I–L) Cardiac expression of bmp4 in 48 hpf embryos. In wild type embryos (I), bmp4 expression is detected in the myocardium of the ventricle, sinus venosus, and AV boundary (arrow). Ctr9LA961 mutant (J), cdc73 morphant (K), and paf1 morphant (L) embryos display ventricular expression of bmp4, but have no discernable AV boundary expression. (M–P) Cardiac expression of tbx2b in 48 hpf embryos. In wild type embryos, tbx2b is expressed by the myocardium of the AV boundary (arrow, M). Ctr9LA961 mutant (N), cdc73 morphant (O), and paf1 morphant (P) embryos instead aberrantly express tbx2b throughout the heart. (Q–T) Cardiac expression of notch1b in 48 hpf embryos. Notch1b is expressed by the AV boundary endocardium in wild type embryos (arrow, Q), but is not expressed in the hearts of ctr9LA961 mutants (R), cdc73 morphants (S), and paf1 morphants (T).
Taken together, our data show that all PAF1C components, except for Leo1, are required for establishing appropriate numbers of cardiomyocytes. This reduction in cardiac cell number has the potential to induce secondary defects in later aspects of cardiac development, like the AV boundary gene expression defects present in ctr9-, cdc73-, and paf1-deficient embryos. However, the AV boundary defects in these morphants are very similar to those observed in leo1 mutants (Nguyen et al., 2010), which have a normal number of cardiac cells, suggesting that all members of the zebrafish PAF1C may play an essential and specific role in the differentiation of the AV boundary myocardium and endocardium.
Loss of PAF1C activity causes defective production and migration of cardiac progenitors
To better understand the role of the PAF1C in early cardiac development, we examined the specification of myocardial precursor cells in the LPM during somitogenesis. We analyzed the expression of nkx2.5, a transcription factor involved in regulating the initiation of the cardiogenic differentiation program (Chen and Fishman, 1996), which marks a subset of myocardial progenitors (Schoenebeck et al., 2007).
In wild type embryos, the myocardial precursors are situated in two bilateral groups at the 16S stage (Figure 3F). By the 20S stage, the myocardial cells in wild type embryos have completed their migration to the midline and fused to form the cardiac cone (Figure 3K). In ctr9 and paf1 morphants, the expression of nkx2.5 at the 16S stage is slightly reduced compared to wild type embryos while the expression of nkx2.5 in cdc73 morphants is relatively normal (Figure 3F–I). Embryos lacking activity of ctr9, paf1, and cdc73 have reduced or slightly reduced nkx2.5 expression at the 20S stage, and the myocardial precursors in these embryos have failed to fully migrate to the midline and fuse to form a cardiac cone (Figure 3L–N). Despite this delay in migration and fusion, cardiomyocytes in ctr9-, cdc73-, and paf1-deficient embryos do reach the midline by 24 hpf (Figure 3B–D). Remarkably, we found that expression of nkx2.5 is barely detectable in rtf1 morphants throughout somitogenesis (Figure 3J,O). These results suggest that the components of the PAF1C, and especially Rtf1, are required for the initiation of the cardiogenic program and the timely migration of myocardial precursors to the midline.
The PAF1C regulates the specification of cardiomyocytes from the LPM
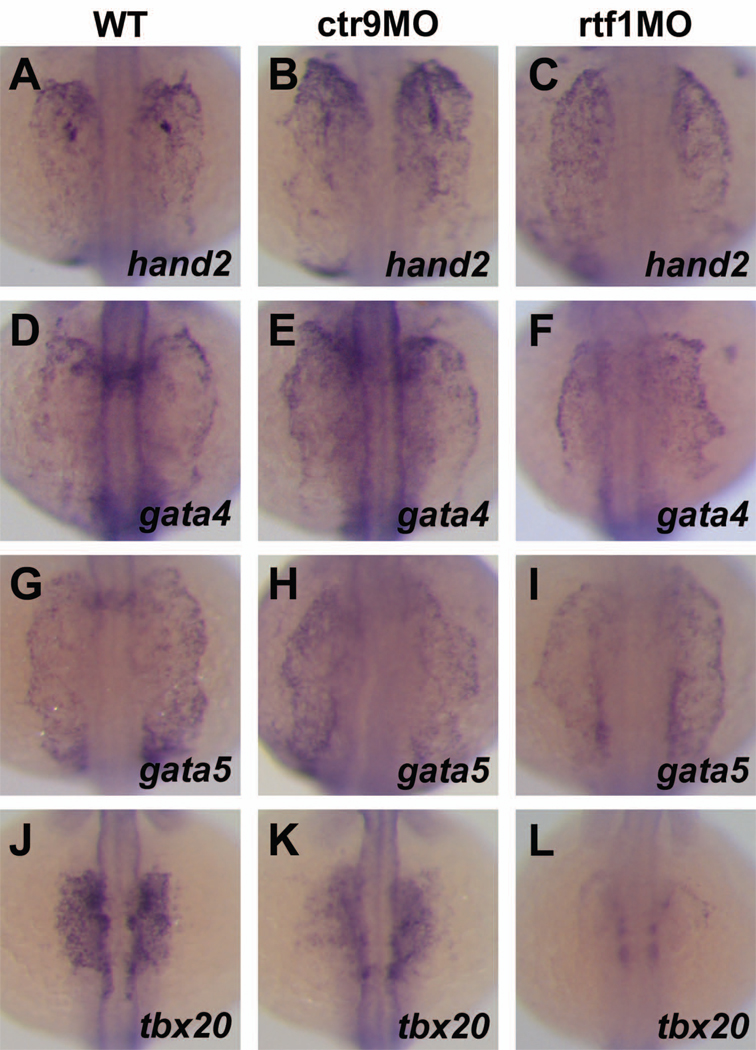
Because of the profound reduction in cardiac tissue and myocardial precursors seen in PAF1C morphant embryos, we wondered if the PAF1C was required for the formation or gene expression of the anterior lateral plate mesoderm (ALPM), the embryonic origin of cardiomyocytes. To determine if the ALPM was present in embryos lacking activity of PAF1C members, we analyzed the expression patterns of several ALPM markers. We examined the expression of hand2, which is expressed in both anterior and posterior compartments of the LPM, and gata4 and gata5, which are expressed in the ALPM and portions of the endoderm (Angelo et al., 2000; Reiter et al., 1999; Reiter et al., 2001a; Serbedzija et al., 1998; Yelon, 2001; Yelon et al., 2000). In wild type embryos, hand2, gata4, and gata5 are expressed in the ALPM at the 16S stage (Figure 5A,D,G). These genes are expressed at similar levels in ctr9 and rtf1 morphant embryos (Figure 5B,C,E,F,H,I), suggesting that the PAF1C is not required for the formation of the ALPM.
Figure 5. The PAF1C is required for the specification of cardiomyocytes from the anterior lateral plate mesoderm.
(A–L) Dorsal views of hand2 (A–C), gata4 (D–F), gata5 (G–I), and tbx20 (J–L) expression in 16S stage wild type and morphant embryos. In wild type embryos, expression of hand2 (A), gata4 (D), and gata5 (G) is detected in the anterior lateral plate mesoderm. In ctr9 and rtf1 morphants, hand2 (B,C), gata4 (E,F), and gata5 (H,I) are expressed in a domain similar to that of wild type embryos. (J–L) In wild type embryos, tbx20 is expressed by cardiomyocyte progenitors in the ALPM (J). Expression of tbx20 is reduced in ctr9 morphants (K) and strongly reduced in rtf1 morphants (L) at the 16S stage.
We also examined the expression pattern of the transcription factor tbx20 in ctr9 and rtf1 morphant embryos. Expression of tbx20 is detected in the ALPM at early stages, and becomes restricted to cardiac progenitors by the 15S stage (Griffin et al., 2000). At the 16S stage, we found that wild type embryos have strong expression of tbx20 in cardiac progenitors (Figure 5J), which occupy a domain of the ALPM. In contrast, ctr9 and rtf1 morphant embryos have reduced tbx20 expression (Figure 5K,L), indicating that despite the presence of ALPM, these embryos are defective in the process of cardiomyocyte specification. This is also consistent with the reduction in the expression level of the cardiomyocyte progenitor marker nkx2.5 we detected in PAF1C-deficient embryos, and suggests that the PAF1C is essential for the specification of cardiac tissue from the ALPM.
To test if the earliest steps in the differentiation of mature cardiomyocytes require PAF1C function, we examined expression of cmlc2 at the 16S stage in PAF1C component morphants. We found that cmlc2 expression has initiated in wild type embryos at 16S, but does not initiate in rtf1 morphants (Supplemental Figure 4K–N). In embryos injected with a combination of ctr9, cdc73, and paf1 morpholinos, we noted a significant delay in the initiation of cmlc2 expression (Supplemental Figure 4E,O). This indicates that in addition to the role of Rtf1 in generating cardiac progenitors, Rtf1 and other PAF1C components (excluding Leo1) are essential for the initial expression of the mature cardiomyocyte marker cmlc2. These data suggest that the PAF1C may be essential for two distinct phases of cardiac development: cardiac progenitor cell specification and the differentiation of mature cardiomyocytes.
The role of Rtf1 in cardiac specification is cell autonomous
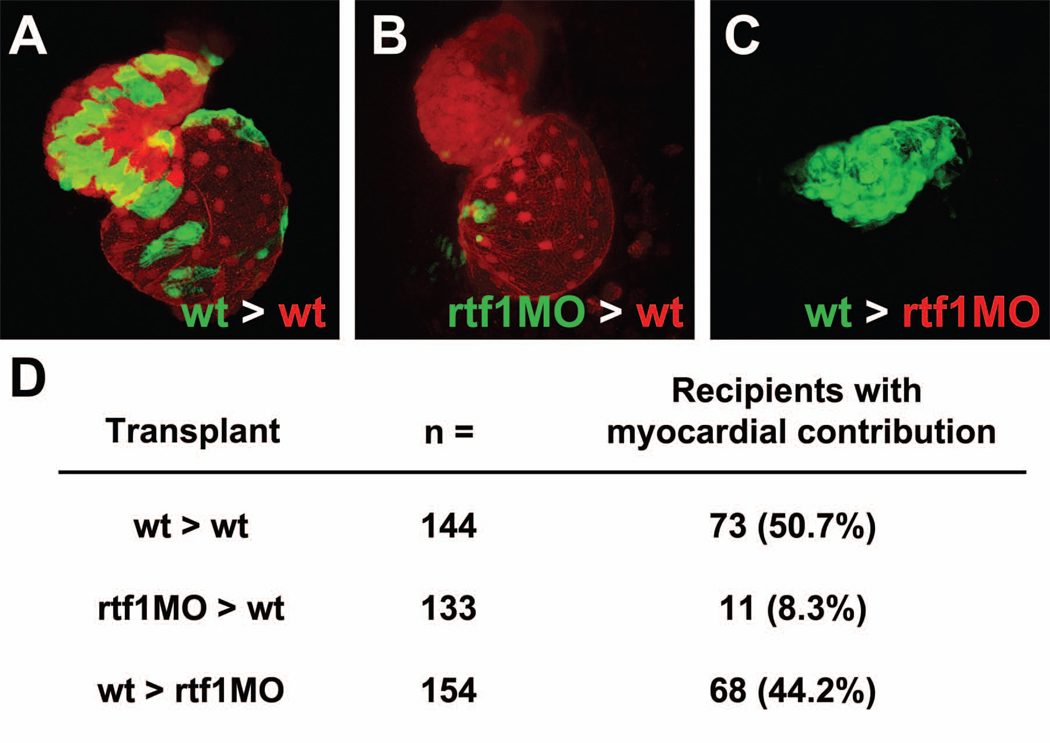
The heart field is patterned by signals originating both inside the LPM and in neighboring tissues (Chen and Fishman, 2000; Yelon, 2001; Yelon et al., 2002; Zaffran and Frasch, 2002), and therefore the activity of the PAF1C may be required in a cell intrinsic or extrinsic manner for the specification of cardiac cells. To determine in which tissue(s) the activity of the PAF1C is required we transplanted cells from rtf1 morphant embryos into wild type embryos and vice versa and analyzed the ability of the donor cells to contribute to the myocardium. In order to differentiate between cardiomyocytes descended from donor or host cells, we transplanted cells from Tg(myl7:EGFP) transgenic donors in which cardiomyocytes express GFP (green) into Tg(myl7:mCherry) transgenic host embryos in which cardiomyocytes express the red fluorescent protein mCherry. As a control, we first transplanted cells from wild type donor embryos to wild type recipients. When cells are transplanted from one wild type embryo to another, they robustly contribute to the heart in 51% (73/144) of recipient embryos (Figure 6A,D). Transplanted wild type cells were frequently found in clusters and were fully functional and integrated into the heart, displaying normal morphology and contractility (Figure 6A and data not shown).
Figure 6. Rtf1 regulates cardiomyocyte specification cell autonomously.
(A–C) Confocal projections showing a ventral view of the heart in two day-old transplanted embryos. Wild type donor cells contribute robustly to the heart when transplanted into wild type recipients (A), while rtf1 morphant cells exhibit limited contribution when transplanted into a wild type recipient (B). Wild type cells are also able to generate myocardial tissue when transplanted into an rtf1 morphant host. (D) Summary of the results from transplantation experiments showing the contribution of donor cells to the myocardium of recipient embryos.
On the contrary, we found that rtf1 morphant cells did not contribute to the heart at a high frequency when transplanted into wild type hosts. Donor rtf1 morphant cells were found in only 8% (11/133) of recipient hearts (Figure 6D), and unlike transplanted wild type cells, rtf1 morphant cardiomyocytes were small and dysmorphic and did not appear to contract (Figure 6B and data not shown). When visualized using confocal microscopy, it is apparent that these morphant cells are only loosely associated with the periphery of the myocardial layer and do not fully integrate into the heart (Supplemental Video 1). While this data shows that the activity of the PAF1C is required within potential myocardial precursors for their entry into the cardiogenic program, its activity may also be required in surrounding tissues. To test this we performed a reverse transplantation experiment, placing cells from a wild type embryo into an rtf1 morphant host. We found that although rtf1 morphants lack hearts at 2 dpf, wild type donor cells were able to form myocardium in 44% (68/154) of rtf1 morphant recipients (Figure 6D). Remarkably, wild type donor cells in 15 of these rtf1 morphant recipient embryos clustered to form a small beating heart-like structure. Taken together, our transplantation data suggest that PAF1C activity is required cell autonomously for the specification of myocardial progenitors, and that activity of the PAF1C in tissues surrounding the myocardium likely plays a lesser role in cardiac specification.
Discussion
The PAF1C is an RNA polymerase II-associated protein complex consisting of five proteins: Ctr9, Cdc73, Paf1, Rtf1, and Leo1. Genetic studies have revealed a requirement for the PAF1C in multiple biological processes including cell cycle regulation, somite and neural crest development, and maintenance of stem cell pluripotency (Akanuma et al., 2007; Ding et al., 2009; Koch et al., 1999; Nguyen et al., 2010). Here we show that the PAF1C is essential for multiple steps in the development of the heart. All PAF1C proteins are critical for specification of the atrioventricular boundary of the heart, and Ctr9, Cdc73, Paf1, and Rtf1 are required for establishing an appropriate number of cardiomyocyte precursors by promoting the specification of cardiac cells from the ALPM (Figure 7A). By transplanting cells between wild type and rtf1 morphant embryos, we found that Rtf1 functions in a cell autonomous manner. Transplanted wild type cells were capable of organizing into a beating structure in rtf1 morphants, whereas rtf1-deficient cells fail to incorporate into the myocardial layer when transplanted into wild type hosts. Our findings establish the PAF1C as a key regulator of cardiac cell fate.
Figure 7. Diagram of zebrafish PAF1C form and function.
(A) Cardiomyocyte progenitors (red) are derived from the anterior lateral plate mesoderm (ALPM, blue). The ALPM expresses markers including hand2, gata4, and gata5. The PAF1C is required for the specification of nkx2.5+/tbx20+ cardiomyocyte progenitors from the ALPM. (B) The PAF1C in zebrafish consists of at least 4 strongly interacting proteins: Ctr9, Paf1, Leo1, and Ctr9. Based on the phenotypic similarities between embryos lacking Rtf1 and other PAF1C components, we hypothesize that Rtf1 associates with the zebrafish PAF1C, albeit less strongly, as has been suggested in other studies on metazoans.
We found that PAF1C activity is dispensable for formation of the ALPM, but is required for determining the cardiogenic potential of the ALPM, likely via regulation of the expression of a set of cardiac determinants including nkx2.5 and tbx20. Previous studies in yeast, plants, flies and human cells have indicated critical roles of this complex in chromatin remodeling, transcription elongation and mRNA 3’-end formation (Kim et al., 2010; Krogan et al., 2003; Oh et al., 2004; Penheiter et al., 2005; Tenney et al., 2006; Zhang et al., 2009). Thus the PAF1C may regulate the transcription of target cardiac genes by influencing histone modifications or control the elongation and stability of cardiac gene transcripts. It is not yet known which cardiogenic determinants are directly regulated by PAF1C activity. This study paves the way for future examination of the direct binding and epigenetic effects of the PAF1C at cardiac gene promoters.
The reduction in cardiac cell number varied between ctr9, cdc73, paf1, and rtf1 morphants, with the most severe defect in cardiomyocyte specification being observed in rtf1 morphants, which usually produced no cardiomyocytes. Based on Western analysis, Rtf1 protein was almost completely absent in rtf1 morphants, whereas low levels of targeted proteins still existed in ctr9, cdc73 and paf1 morphants. This, along with the finding that cardiac progenitor cell specification was sensitive to the dosage of Rtf1 suggest that the variation in cardiac specification defects observed among PAF1C component morphants may reflect different efficacies of the morpholinos used in our study. Creating mutant embryos lacking both maternal and zygotic messages of PAF1C should provide further insights into the effect of complete loss of PAF1C function on cardiac development. Alternatively, the distinct cardiac phenotypes observed among PAF1C component morphants and mutants may be indicative of disparate roles of PAF1C proteins in heart development. Interestingly, simultaneous knockdown of ctr9, paf1, and cdc73 with the morpholino dosages used in our study induced a significantly weaker phenotype than that seen in rtf1 morphants. We have also previously shown that leo1 mutants generate a normal number of cardiomyocytes (Nguyen et al., 2010). These data argue in favor of the possibility that Rtf1 may play a unique role in cardiac development.
In yeast, Ctr9, Cdc73, Paf1, Rtf1 and Leo1 form a stable complex (Mueller and Jaehning, 2002). Our biochemical study shows that zebrafish Cdc73, Paf1, and Leo1, but not Rtf1, coimmunoprecipitate with Ctr9. While it is possible Rtf1 is not a component of the PAF1C in zebrafish, the abundant phenotypic similarities between rtf1 morphants and embryos deficient in other PAF1C proteins lead us to hypothesize that Rtf1 is an integral component of the PAF1C but its interaction with other members of PAF1C is too weak to detect by coimmunoprecipitation (Figure 7B). This hypothesis is consistent with earlier reports that Rtf1 fails to coimmunoprecipitate with the Drosophila and human PAF1Cs (Adelman et al., 2006; Kim et al., 2010; Rozenblatt-Rosen et al., 2005). Our findings suggest that a looser association between Rtf1 and other PAF1C members is a feature that may be conserved universally among metazoans.
Our analysis of the zebrafish PAF1C reveals that while all complex members are required in the specification of the atrioventricular boundary, individual PAF1C components have disparate roles in early cardiogenesis. Loss of each PAF1C member, excluding Leo1, causes a reduction in the number of cardiomyocytes specified from the LPM (Nguyen et al., 2010). The mechanisms responsible for differing roles of individual PAF1C components are not yet known. One potential explanation is that losing individual PAF1C components may have different effects on overall complex stability. Recent studies indicate that loss of human Paf1 or Ctr9, but not Leo1, destabilizes the interactions between other PAF1C members (Kim et al., 2010). This suggests that Paf1 and Ctr9 may serve as a scaffold for the formation of the PAF1C, and that loss of these proteins disrupts the stability of the complex as a whole. This idea is consistent with the severe defects in cardiogenesis noted in ctr9 and paf1 morphants compared to the later, mild cardiac defects noted in leo1 mutants. Another plausible explanation is that each PAF1C member mediates specific protein-protein interactions. Indeed, individual PAF1C members have been shown to interact with a number of different proteins involved in transcription regulation. For example, yeast Paf1 interacts directly with Bre1, linking the PAF1C to histone ubiquitination machinery (Kim and Roeder, 2009). Yeast Rtf1 directly interacts with the chromatin remodeling protein Chd1 (Simic et al., 2003). In mammalian cells, PAF1C interacts with the Wnt and Hh signaling pathways by the direct association of Cdc73 with beta-catenin and Gli proteins (Mosimann et al., 2006, 2009). Therefore, the specific effects on development we noted upon loss of individual PAF1C components could be a result of loss of interaction with particular genes and signaling pathways important for cardiogenesis. Further studies that map out all the interacting partners and target genes of the PAF1C will be needed to uncover the mechanisms by which this complex regulates specific signaling pathways and developmental processes in vertebrates.
Supplementary Material
Supplemental Video 1. Rtf1 morphant donor cells do not incorporate into the myocardial layer of a wild type recipient heart. Rotating three-dimensional confocal projection of a wild type heart (red) containing a cluster of rtf1MO donor cells (green).
Supplemental Figure 1. LA961 is a mutation in ctr9. (A–D) In contrast to wild type embryos (A), LA961 mutant embryos at 2 dpf (B) exhibit pericardial edema, a lack of pigmentation, and a curved body axis. Injection of 200 pg of ctr9WT mRNA rescues the cardiac, pigment, and body axis defects of LA961 mutants (C). Injection of 0.75 ng of ctr9MO phenocopies the LA961 mutant phenotype (D).
Supplemental Figure 2. Ctr9LA961 transcripts undergo nonsense-mediated decay. (A,B) Myc-Ctr9WT (A) and Myc-Ctr9LA961 (B) both localize to the nucleus. (C,D) Lateral views of ctr9 expression in 1 day post fertilization wild type and ctr9LA961 mutant embryos. Expression of ctr9 is very reduced in ctr9LA961 mutants (D) compared to wild type embryos (C).
Supplemental Figure 3. Morpholinos targeting PAF1C component genes reduce target protein levels. (A–D) Western blots detecting PAF1C component proteins and β-actin protein levels in uninjected control and morpholino injected embryo lysates. (A) Ctr9 protein is reduced to 38% of control levels in ctr9MO injected embryos. (B) Paf1 protein is reduced to 43% of control levels in paf1MO injected embryos. (C) Cdc73 protein is reduced to 39% of control levels in cdc73MO injected embryos. (D) Rtf1 protein is reduced to 7% of control levels in rtf1MO injected embryos.
Supplemental Figure 4. Specification of cardiac progenitor cells is sensitive to Rtf1 dosage. (A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas embryos injected with 0.1 ng, 0.25 ng, and 1.5 ng of rtf1MO have no detectable cardiac tissue (B–D). Embryos coinjected with ctr9MO, cdc73MO, and paf1MO (3KD) have small clump of cardiomyocytes situated at the midline (E). (F–J) Dorsal views of nkx2.5 expression in 16S stage wild type and morphant embryos. In wild type embryos, cardiac progenitors have been specified in the ALPM (F). Injection of rtf1MO produces a dosage-dependent effect on nkx2.5 expression. Embryos injected with 0.1 ng rtf1MO (G) have a reduction in nkx2.5 expression compared to wild type. Expression of nkx2.5 is further reduced in embryos injected with 0.25 ng rtf1MO (H), and absent in embryos injected with 1.5 ng rtf1MO (I). Simultaneous knockdown of ctr9, cdc73, and paf1 does not result in a phenotype as severe as that seen in rtf1 morphants. (K–O) Dorsal views of cmlc2 expression in 16S stage wild type and morphant embryos. Mature cardiomyocytes that express cmlc2 are beginning to be specified as early as the 16S stage in wild type embryos (K), but are not present in 0.1 ng, 0.25 ng, and 1.5 ng, rtf1MO and 3KD morphant embryos at this stage (L–O).
Acknowledgements
We are grateful to members of the Chen lab for experimental suggestions and critiques and to Yuan Dong for participating in the genetic screen. This work was supported by a grant from the NIH to JNC (HL081700), a National Science Foundation Graduate Research Fellowship to ADL, and predoctoral fellowships from the Training Program of Genetic Mechanisms to CTN and AMC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol. 2006;26:250–260. doi: 10.1128/MCB.26.1.250-260.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanuma T, Koshida S, Kawamura A, Kishimoto Y, Takada S. Paf1 complex homologues are required for Notch-regulated transcription during somite segmentation. EMBO Rep. 2007;8:858–863. doi: 10.1038/sj.embor.7401045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–237. doi: 10.1016/s0925-4773(00)00334-8. [DOI] [PubMed] [Google Scholar]
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, Zhou Y, Grunwald DJ, Lin S, Cantor AB, Orkin SH, Zon LI. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 1996;122:3809–3816. doi: 10.1242/dev.122.12.3809. [DOI] [PubMed] [Google Scholar]
- Chen JN, Fishman MC. Genetics of heart development. Trends Genet. 2000;16:383–388. doi: 10.1016/s0168-9525(00)02075-8. [DOI] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4:403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Fishman MC. Notochord regulates cardiac lineage in zebrafish embryos. Dev Biol. 1998;201:247–252. doi: 10.1006/dbio.1998.8976. [DOI] [PubMed] [Google Scholar]
- Griffin KJ, Stoller J, Gibson M, Chen S, Yelon D, Stainier DY, Kimelman D. A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev Biol. 2000;218:235–247. doi: 10.1006/dbio.1999.9571. [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev. 2004;18:2774–2784. doi: 10.1101/gad.1244504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzinger A, Evans T. Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol. 2007;312:613–622. doi: 10.1016/j.ydbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CJ, Tu CT, Hsiao CD, Hsieh FJ, Tsai HJ. Germ-line transmission of a myocardium-specific GFP transgene reveals critical regulatory elements in the cardiac myosin light chain 2 promoter of zebrafish. Dev Dyn. 2003;228:30–40. doi: 10.1002/dvdy.10356. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, Roeder RG. The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell. 2010;140:491–503. doi: 10.1016/j.cell.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Roeder RG. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem. 2009;284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Wollmann P, Dahl M, Lottspeich F. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 1999;27:2126–2134. doi: 10.1093/nar/27.10.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lin L, Czapiga M, Nini L, Zhang JH, Simonds WF. Nuclear localization of the parafibromin tumor suppressor protein implicated in the hyperparathyroidism-jaw tumor syndrome enhances its proapoptotic function. Mol Cancer Res. 2007a;5:183–193. doi: 10.1158/1541-7786.MCR-06-0129. [DOI] [PubMed] [Google Scholar]
- Lin X, Rinaldo L, Fazly AF, Xu X. Depletion of Med10 enhances Wnt and suppresses Nodal signaling during zebrafish embryogenesis. Dev Biol. 2007b;303:536–548. doi: 10.1016/j.ydbio.2006.11.034. [DOI] [PubMed] [Google Scholar]
- Marques SR, Lee Y, Poss KD, Yelon D. Reiterative roles for FGF signaling in the establishment of size and proportion of the zebrafish heart. Dev Biol. 2008;321:397–406. doi: 10.1016/j.ydbio.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell. 2006;125:327–341. doi: 10.1016/j.cell.2006.01.053. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mech Dev. 2009;126:394–405. doi: 10.1016/j.mod.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol. 2002;22:1971–1980. doi: 10.1128/MCB.22.7.1971-1980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen CT, Langenbacher A, Hsieh M, Chen JN. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Dev Biol. 2010;341:167–175. doi: 10.1016/j.ydbio.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Zhang H, Ludwig P, van Nocker S. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell. 2004;16:2940–2953. doi: 10.1105/tpc.104.026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol Cell. 2005;20:213–223. doi: 10.1016/j.molcel.2005.08.023. [DOI] [PubMed] [Google Scholar]
- Peterkin T, Gibson A, Patient R. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev Biol. 2007;311:623–635. doi: 10.1016/j.ydbio.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SE, Penheiter KL, Jaehning JA. Separation of the Saccharomyces cerevisiae Paf1 complex from RNA polymerase II results in changes in its subnuclear localization. Eukaryot Cell. 2005;4:209–220. doi: 10.1128/EC.4.1.209-220.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Kikuchi Y, Stainier DY. Multiple roles for Gata5 in zebrafish endoderm formation. Development. 2001a;128:125–135. doi: 10.1242/dev.128.1.125. [DOI] [PubMed] [Google Scholar]
- Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Dev Biol. 2001b;234:330–338. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Hughes CM, Nannepaga SJ, Shanmugam KS, Copeland TD, Guszczynski T, Resau JH, Meyerson M. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol Cell Biol. 2005;25:612–620. doi: 10.1128/MCB.25.2.612-620.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbedzija GN, Chen JN, Fishman MC. Regulation in the heart field of zebrafish. Development. 1998;125:1095–1101. doi: 10.1242/dev.125.6.1095. [DOI] [PubMed] [Google Scholar]
- Shi X, Chang M, Wolf AJ, Chang CH, Frazer-Abel AA, Wade PA, Burton ZF, Jaehning JA. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol Cell Biol. 1997;17:1160–1169. doi: 10.1128/mcb.17.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney K, Gerber M, Ilvarsonn A, Schneider J, Gause M, Dorsett D, Eissenberg JC, Shilatifard A. Drosophila Rtf1 functions in histone methylation, gene expression, and Notch signaling. Proc Natl Acad Sci U S A. 2006;103:11970–11974. doi: 10.1073/pnas.0603620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh LA, Yelon D, Stainier DY. Hand2 regulates epithelial formation during myocardial diferentiation. Curr Biol. 2005;15:441–446. doi: 10.1016/j.cub.2004.12.083. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. The University of Oregon Press; 2000. [Google Scholar]
- Yart A, Gstaiger M, Wirbelauer C, Pecnik M, Anastasiou D, Hess D, Krek W. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol Cell Biol. 2005;25:5052–5060. doi: 10.1128/MCB.25.12.5052-5060.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelon D. Cardiac patterning and morphogenesis in zebrafish. Dev Dyn. 2001;222:552–563. doi: 10.1002/dvdy.1243. [DOI] [PubMed] [Google Scholar]
- Yelon D, Feldman JL, Keegan BR. Genetic regulation of cardiac patterning in zebrafish. Cold Spring Harb Symp Quant Biol. 2002;67:19–25. doi: 10.1101/sqb.2002.67.19. [DOI] [PubMed] [Google Scholar]
- Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- Zaffran S, Frasch M. Early signals in cardiac development. Circ Res. 2002;91:457–469. doi: 10.1161/01.res.0000034152.74523.a8. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL, Schneider DA. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proc Natl Acad Sci U S A. 2009;106:2153–2158. doi: 10.1073/pnas.0812939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Video 1. Rtf1 morphant donor cells do not incorporate into the myocardial layer of a wild type recipient heart. Rotating three-dimensional confocal projection of a wild type heart (red) containing a cluster of rtf1MO donor cells (green).
Supplemental Figure 1. LA961 is a mutation in ctr9. (A–D) In contrast to wild type embryos (A), LA961 mutant embryos at 2 dpf (B) exhibit pericardial edema, a lack of pigmentation, and a curved body axis. Injection of 200 pg of ctr9WT mRNA rescues the cardiac, pigment, and body axis defects of LA961 mutants (C). Injection of 0.75 ng of ctr9MO phenocopies the LA961 mutant phenotype (D).
Supplemental Figure 2. Ctr9LA961 transcripts undergo nonsense-mediated decay. (A,B) Myc-Ctr9WT (A) and Myc-Ctr9LA961 (B) both localize to the nucleus. (C,D) Lateral views of ctr9 expression in 1 day post fertilization wild type and ctr9LA961 mutant embryos. Expression of ctr9 is very reduced in ctr9LA961 mutants (D) compared to wild type embryos (C).
Supplemental Figure 3. Morpholinos targeting PAF1C component genes reduce target protein levels. (A–D) Western blots detecting PAF1C component proteins and β-actin protein levels in uninjected control and morpholino injected embryo lysates. (A) Ctr9 protein is reduced to 38% of control levels in ctr9MO injected embryos. (B) Paf1 protein is reduced to 43% of control levels in paf1MO injected embryos. (C) Cdc73 protein is reduced to 39% of control levels in cdc73MO injected embryos. (D) Rtf1 protein is reduced to 7% of control levels in rtf1MO injected embryos.
Supplemental Figure 4. Specification of cardiac progenitor cells is sensitive to Rtf1 dosage. (A–E) Dorsal views of cmlc2 expression in 24 hpf wild type and morphant embryos. Wild type embryos have an elongated primitive heart tube at 24 hpf (A), whereas embryos injected with 0.1 ng, 0.25 ng, and 1.5 ng of rtf1MO have no detectable cardiac tissue (B–D). Embryos coinjected with ctr9MO, cdc73MO, and paf1MO (3KD) have small clump of cardiomyocytes situated at the midline (E). (F–J) Dorsal views of nkx2.5 expression in 16S stage wild type and morphant embryos. In wild type embryos, cardiac progenitors have been specified in the ALPM (F). Injection of rtf1MO produces a dosage-dependent effect on nkx2.5 expression. Embryos injected with 0.1 ng rtf1MO (G) have a reduction in nkx2.5 expression compared to wild type. Expression of nkx2.5 is further reduced in embryos injected with 0.25 ng rtf1MO (H), and absent in embryos injected with 1.5 ng rtf1MO (I). Simultaneous knockdown of ctr9, cdc73, and paf1 does not result in a phenotype as severe as that seen in rtf1 morphants. (K–O) Dorsal views of cmlc2 expression in 16S stage wild type and morphant embryos. Mature cardiomyocytes that express cmlc2 are beginning to be specified as early as the 16S stage in wild type embryos (K), but are not present in 0.1 ng, 0.25 ng, and 1.5 ng, rtf1MO and 3KD morphant embryos at this stage (L–O).