Abstract
Worldwide structural genomics projects are increasing structure coverage of sequence space but have not significantly expanded the protein structure space itself (i.e. number of unique structural folds) since 2007. Discovering new structural folds experimentally by directed evolution and random recombination of secondary-structure blocks is also proved rarely successful. Meanwhile, previous computational efforts for large-scale mapping of protein structure space are limited to simple model proteins and led to an inconclusive answer on the completeness of the existing, observed protein structure space. Here, we build novel protein structures by extending naturally occurring circular (single-loop) permutation to multiple-loop permutations (MLP). These structures are clustered by structural similarity measure called TM-Score. The computational technique allows us to produce different structural clusters on the same naturally occurring, packed, stable core but with alternatively connected secondary-structure segments. A large-scale MLP of 2936 SCOP domains reproduces those existing structural clusters (63%) mostly as hubs for many non-redundant sequences and illustrates newly discovered novel clusters as islands adopted by a few sequences only. Results further show that there exist a significant number of novel, potentially stable clusters for medium or large-size single-domain proteins, in particular (>100 amino-acid residues) that are either not yet adopted by nature or adopted only by a few sequences. This study suggests that MLP provides a simple yet highly effective tool for engineering and design of novel protein structures (including naturally knotted proteins). The implication of recovering CASP new-fold targets by MLP on template-based structure prediction is also discussed. Our MLP structures are available for download at the publication page of the website http://sparks.informatics.iupui.edu.
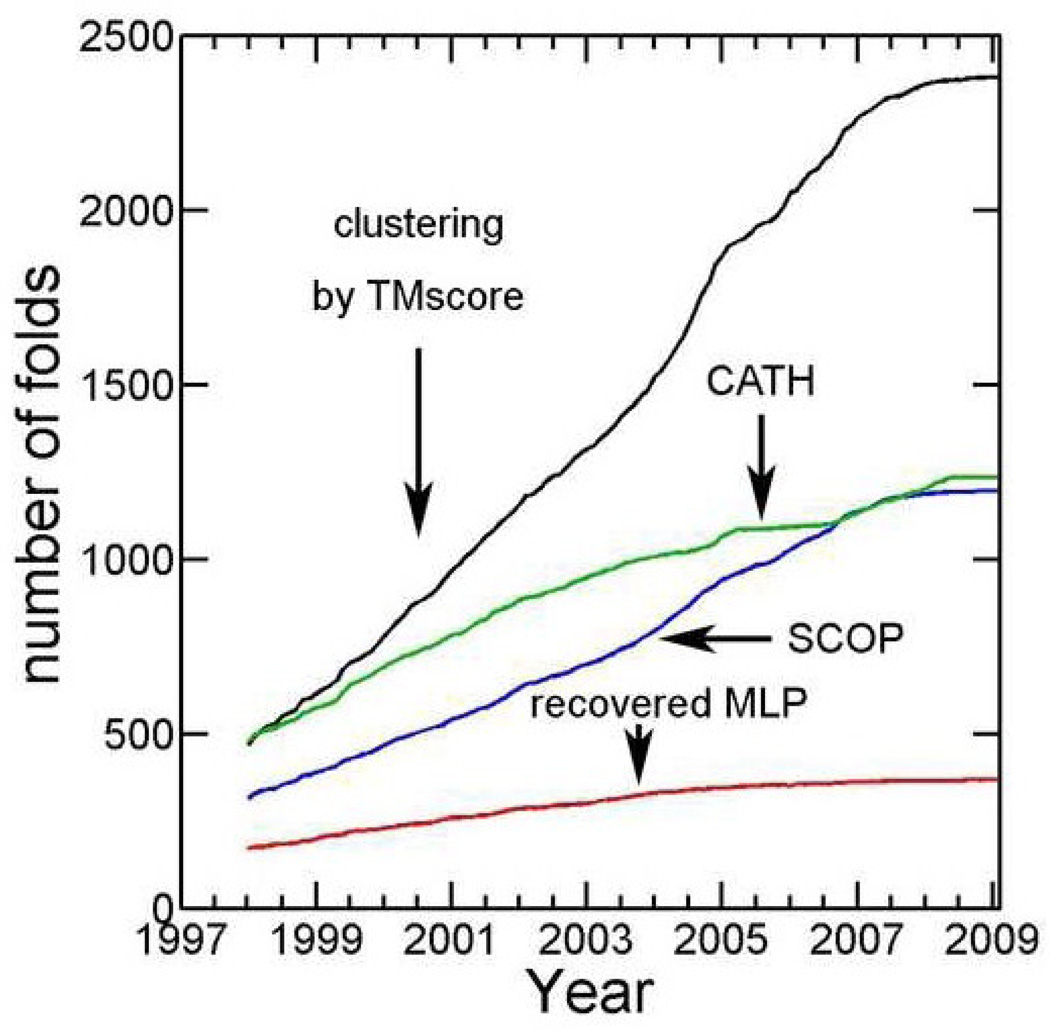
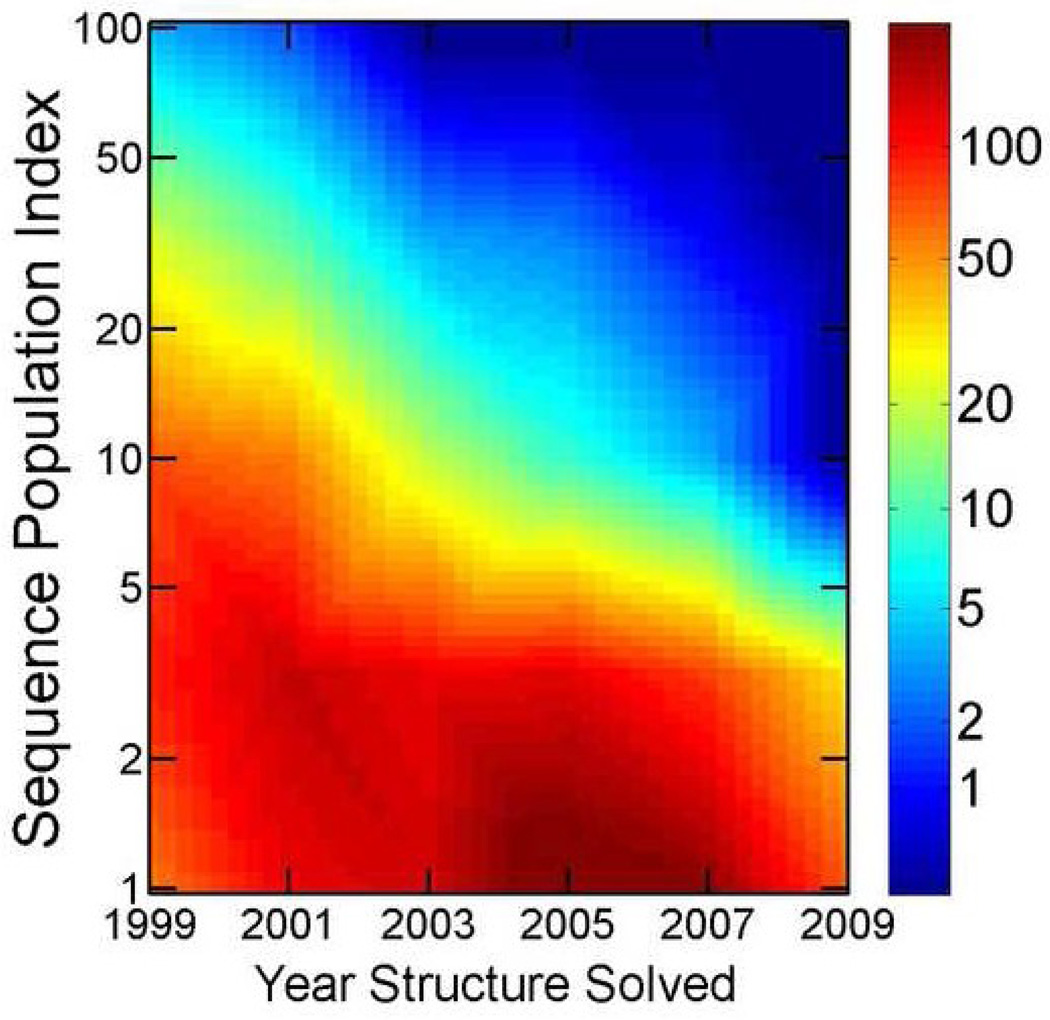
Despite of exponential increase in the number of protein sequences due to genome projects, the structure coverage of single domain sequence families has continued to expand from 21% in 2004 to 26% in 2009. This expansion is largely because of systematic, worldwide effort of large-scale, protein-structure determination (structural genomics projects) 1. Growth in structure coverage of sequence space, however, did not translate into a meaningful expansion of protein structure space. In fact, the total number of structural folds appears to have converged since 2007. As shown in Fig. 1, this trend, is clear from either manual classification of protein structures (SCOP 2 or CATH 3) or from automatic clustering based on pairwise structural similarity (TM-Score from TMalign at a cutoff of 0.5 -- an approximate measure to determine whether or not protein pairs belong to the same structural fold or cluster).
Fig. 1.
Number of structural folds from SCOP annotations (2/23/09 release, in Blue), structural topologies from CATH annotations (7/7/09 release, in green), and structure clusters derived from 16712 ASTRAL domains (95% cutoff) according to TM-Score cutoff of 0.5 (in black) as a function of time (evaluated monthly). Multiple loop permutations (MLP) on 2936 SCOP domains generated 820 new structure clusters that are significantly different from original native templates (TM-Score<0.5). More and more these clusters (MLP in Red) recover known native structure clusters discovered over time.
What does this convergence in the number of structural folds or clusters mean? Zhang and Skolnick 6 found that existing protein structures are self-contained such that one can always find a non-homologous (<35% sequence identity) native structure within 2.5Å rms deviation and with about 80% alignment coverage from any given structure of a medium size protein (less than 200 amino acid residues). They 7 further proposed that the existing structure space is complete because all structures in the protein databank (PDB) can be mapped on to randomly generated homo-peptide compact structures and, conversely, all randomly generated compact structures have their corresponding PDB structures. On the other hand, a more recent study of medium-size, lattice-model protein with idealized secondary structure found only one in ten predicted structures can be matched to existing structures and proposed the existence of excess amount of "dark matter" in the protein structure universe. The conflicting conclusions drawn on the completeness of structure space are likely due to different coarse-grained models employed to generate artificial structures that may or may not represent the actual structure space of proteins.
To deepen our understanding of what we have seen and yet to see in the protein structure universe, we need a method that generates protein structures that are likely foldable. Generating novel folds has been attempted experimentally by employing combinatorial sequence 10 or secondary-structure block libraries. However, the occurrence of stably folded proteins is rare. For example, initial 4×1012 random sequences followed by many iterations of in vitro selections and directed evolution 10 yielded only several functional proteins 13 while in vitro random recombination of secondary structure elements (blocks) does not lead to uniquely folded proteins.
Computationally, Kuhlman et al. 14 designed a novel fold by ab initio backbone design coupled with Rosetta de novo structure prediction. This technique is too time consuming for large-scale mapping of possible structure space. Pseudo-native structures (artificial decoys) have been generated by connecting spatially contacting residues 15 and by combining reverse-sequence order, cyclic permutation, and three-point switching (loop crossover) 16. They were introduced for the purpose of improving threading and score normalization for structural comparison, respectively. Yang and Sharp 17 employed the elastic network models to generate alternative protein structures by perturbation along low-frequency normal modes. The method was used to improve homology modeling. These computational methods, however, have not been employed to characterize the structural space of proteins.
It is known that changing protein structural topology can occur naturally through circular permutation that involves closing the N and C termini with a short loop and opening another surface location (often in a loop region) for new termini (i.e., opening and closing of a single loop). Because circular permutation usually yields a structure that is not that different from its original structure , we propose a computational method that generates novel structures by multiple loop permutations (MLP, i.e. opening and closing of multiple rather than single loops) while leaving the core packing of secondary-structure segments unperturbed. The proposed MLP technique for generating novel structures with naturally occurring stable core is supported by the following observations. Most circular permutated structures retain their stability and function , suggesting that core packing is dominant determinant for protein stability22. Likewise, fixing core packing will maintain the stability and function of their original native templates at least for some MLP structures and potentially for all MLP structures with appropriately designed sequences. Moreover, it has been observed that nearly identical core packing naturally occurs between secondary structure elements of different structural folds through different loop connections and relates newly discovered, novel structural folds to known folds.
In this paper, MLP is applied to 2936 SCOP domains and yields 2843 structures significantly different from their respective original templates. Analysis and clustering of these 2843 structures by TM-Score allows us to characterize the existing as well as potential structural clusters of proteins. Results highlight the potential utility of MLP for generating novel structure clusters for engineering and design and for template-based structure prediction.
RESULTS
We made multiple loop permutations to 2936 SCOP domains (one domain per family chosen from the domains in ASTRAL 1.75 release26 with no missing residues or backbone atoms). To reduce the number of new structures generated and have an initial assessment of structure space, we limited to a maximum of 100 permutated structures per domain and a permutation of up to five loops whose end-to-end distances are less than 15Å (See methods). This leads to a total of 96131 loop permutated structures. After removing overlaps and incorrectly built loops and the structures with nearly completely buried termini and having less than 60 residues, we obtained 2843 structures that differ significantly from their respective original structures (with TM-Score<0.5 from structural alignment program TM-align 4; TM-Score=0.5 indicates, for example, 85% residues aligned with 2.6Å RMSD for a protein of 200 residues long). These significantly altered structures (MLP-D2843) can be clustered with a threshold of TM-Score=0.5 into 820 unique MLP structure clusters (MLP-C820).
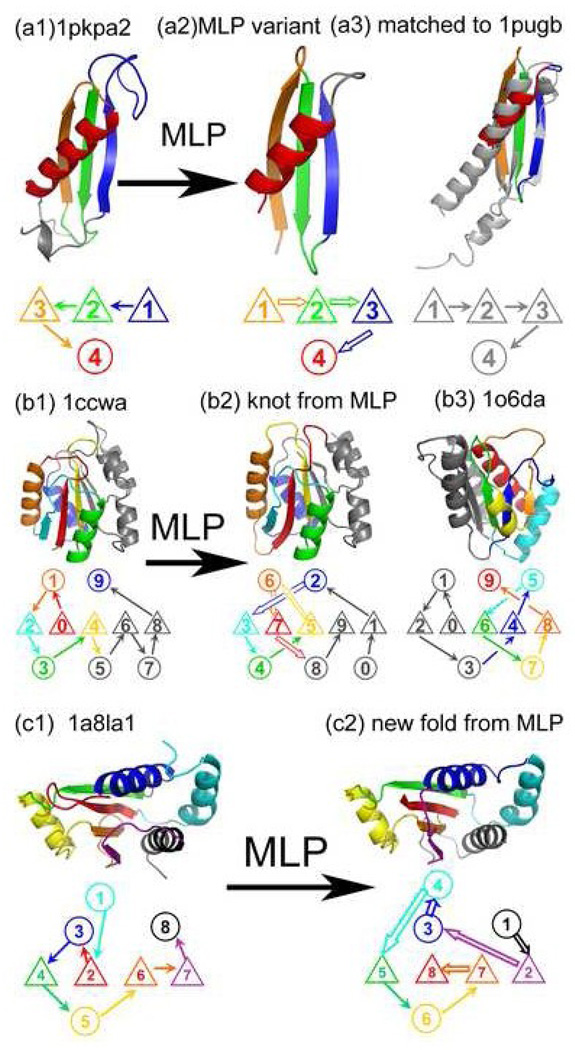
Because MLP-D2843 structures differ significantly from their respective native templates (based on structural alignment), high structural similarity between a MLP generated structure and any existing structure in a different cluster indicates the ability of MLP to change from one known structural cluster to another. Fig. 2a shows an example of a native structure (SCOP ID#1pkpa2, left) whose three-loop permutated structure (center) matches to another native structure (SCOP ID#1pugb, right, in gray) with a TM-Score of 0.66 (aligning 42 of 46 residues with RMSD of 1.6Å). By comparison, the TM-Score between 1pugb and 1pkpa2 (prior to permutations) is only 0.38. [The TM-Score between random structure pairs is size-independent and distributed between 0.08 and 0.3 5.] Proteins 1pugb (hypothetical UPF0133 protein ybaB) and 1pkpa2 (ribosomal S5 protein, N-terminal domain) belong to separate SCOP folds (YbaB and dsRBD-like, respectively) and have biological functions of DNA and RNA binding, respectively. The ability to link two different folds by MLP indicates that some native structural folds (clusters) can be reproduced (or recovered) by MLP of a structure in a different fold cluster.
Fig. 2.
A multiple-loop permutated structure has an identical core packing as its original native structure but with different loop connections between secondary structures. Three examples are shown. (a) A three-loop permutation (open-arrow) of the native structure of wild-type (wt) sequence (SCOP ID#1pkpa2, left) leads to a new structure (center, with terminal coil regions removed) that is substantially different from its original native structure with a TM-Score of 0.49 but matches a native structure 1pugb of a different SCOP fold (in gray, right) with a TM-Score of 0.66 (aligning 42 of 46 residues with RMSD of 1.6Å). (b) The similarity in knotted regions between the knotted structure generated from four-loop permutation (center) of 1ccwa (left) to the naturally occurring knotted structure (SCOP#1o6da) (right). (c) An example of new structure: Native domain structure (1a8La1, left) and its five-loop permutated structure (right).
Fig. 1 compares MLP-C820 structure clusters to a total of 2379 existing native clusters up to early 2009 from the latest ASTRAL 1.75 release (in black). 173 clusters out of the MLP-C820 set (21%) have one or more structures that have a TM-Score of 0.5 or above with one or more native structures in the structure clusters experimentally solved before 1998. 195 additional MLP clusters (another 24%) can be mapped to the clusters discovered from 1998 to 2009. In other words, a total of 368 clusters of the MLP-C820 set (45%) can be mapped on to 1495 existing structure clusters, 63% (1495/2379) of all existing clusters of native structures. This large-scale mapping confirms that MLP can produce (or reproduce) existing stable, structural clusters.
The similarity between MLP structures and native structures can be extended to knotted regions. Fig. 2b shows an example of knotted MLP structure generated from four-loop permutation of non-knotted domain 1ccwa that has a knot in the region made of three strands enclosed by two helices each side, resembling the knot region in a naturally occurring knotted protein 1o6da in terms of how knots are composed of although two knots occur in different parts of the two proteins. Both knots can be described as the simplest trefoil type 3127. There are a total of 7 natively knotted structures out of 2936 domains before loop permutation (0.2%). MLP of these 2936 domains leads to 38 knotted structures (1.3%) in MLP-D2843. None of the 38 knotted MLP structures is from 7 knotted native proteins prior to permutation. According to SCOP classification, 0, 7, 23, 8 of these knotted structures are all-α, all-β, α/β, and α+β proteins, respectively. Interestingly, the protein knot server27 listed 40 naturally occurring knotted proteins. Among them, there are 31 SCOP-annotated, knotted proteins consisting of 1 all-α, 9 all-β, 15 α/β and 7 α+β proteins, respectively. There are 8 additional naturally occurring knotted proteins not yet annotated by SCOP (1x7p, 1y7w, 1yr1, 1z93, 1zjr, 2ha8, 2i6d, 3kzc). Manual examination of these 8 structures suggests 6 α/β and 2 α+β proteins. Thus, the overall distribution of knotted MLP structures in different structure classes is similar to those naturally occurring knotted structures. The latter have 36 of 31, 2 of 41 and 2 of 52 knots27. Due to the limitation of up to five loop permutations permitted in this study, all knotted, MLP structures are the simplest trefoil type 31. The result above further highlights the ability of MLP to recover naturally occurring structural clusters, even for the most complex knotted regions.
The existence of both knots and slip knots in original 2936 native domains and MLP-D2843 can be compared. Among native domains, there are 7 knotted proteins (0.2%) and 237 deep slip-knotted proteins (8.1%) with depth of 10 residues or more. By comparison, there are 38 knotted proteins (1.3%) and 420 deep slip-knotted proteins (14.7%) in MLP-D2843. There is an increase in number of knotted and slip-knotted structures in multiple loop permutated structures. This is a result of unconstrained loop building during multiple loop permutation. Nevertheless, the majority of MLP structures do not have knot or deep slip knot. Shallow slip knotted proteins are also common in native proteins (23.9% for depths of 1 to 9 residues) and thus, not considered as unusual here. Fig. 2c shows one example where five loop permutations change the overall linkage of the secondary structure of protein disulfide oxidoreductase (SCOP ID#1a8l1). There is nothing obviously unusual after five loop permutations while maintaining an identically packed core.
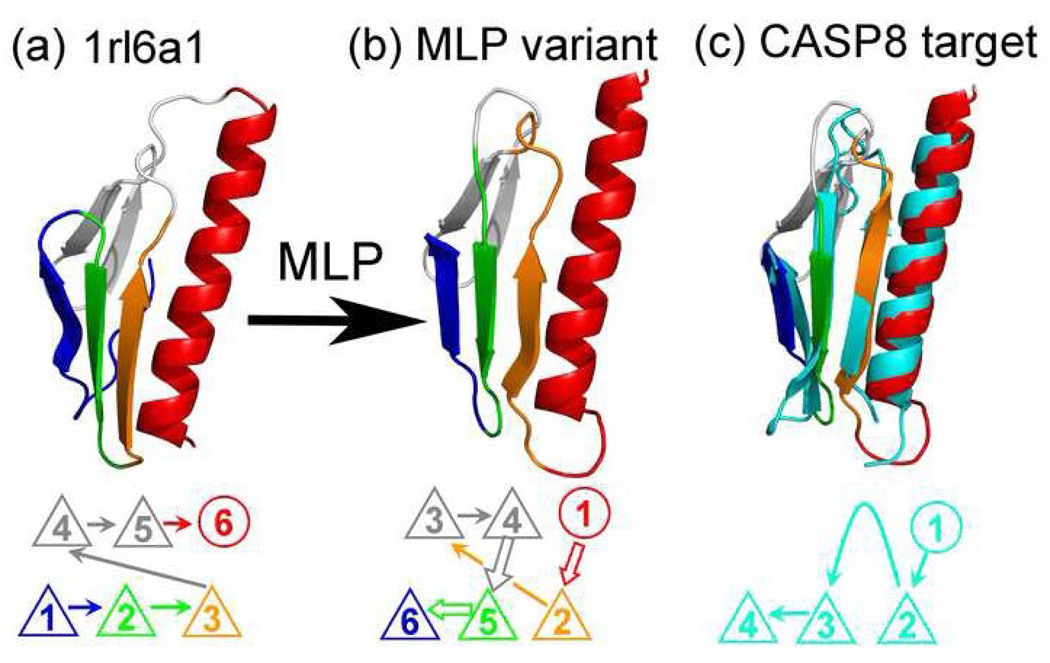
We further compared MLP structures in MLP-D2843 with CASP 8 free-modeling (new-fold) targets released in 200828. New-fold targets in CASP (Critical Assessment of Structure Prediction techniques) are those targets without high-quality, recognizable templates at the time of release. They are employed for testing template-free structure prediction techniques [20]. We compared MLP structures with CASP targets after removing original templates solved on or after year 2008 and having TM-Score≥0.5 with CASP 8 targets. We found that there are 178 MLP structures having TM-Score≥0.5 to seven (out of 12) CASP 8 new-fold targets. An example is shown in Fig. 3. The three-loop permutated structure of ribosomal protein l6 (SCOP ID#1rl6a1, solved by X-ray structure determination in 1993) has a TM-Score of 0.73 with CASP 8 target T0443-D2 (56 of 60 residues in T0443-D2 are aligned with RMSD of 1.75Å). By comparison, the ribosomal protein l6 itself has TM-Score of only 0.29 with T0443-D2 and the best predicted model for this target in CASP 8 has TM-Score of 0.4 only.
Fig. 3.
The three-loop permutated structure of ribosomal protein l6 (SCOP ID#1rl6a1, center) has a TM-Score of 0.73 with the CASP 8 target T0443-D2 (56 of 60 residues in T0443-D2 are aligned with RMSD 1.75Å, Right in Cyan). By comparison, the ribosomal protein l6 itself (Left) has TM-Score of only 0.29 with T0443-D2.
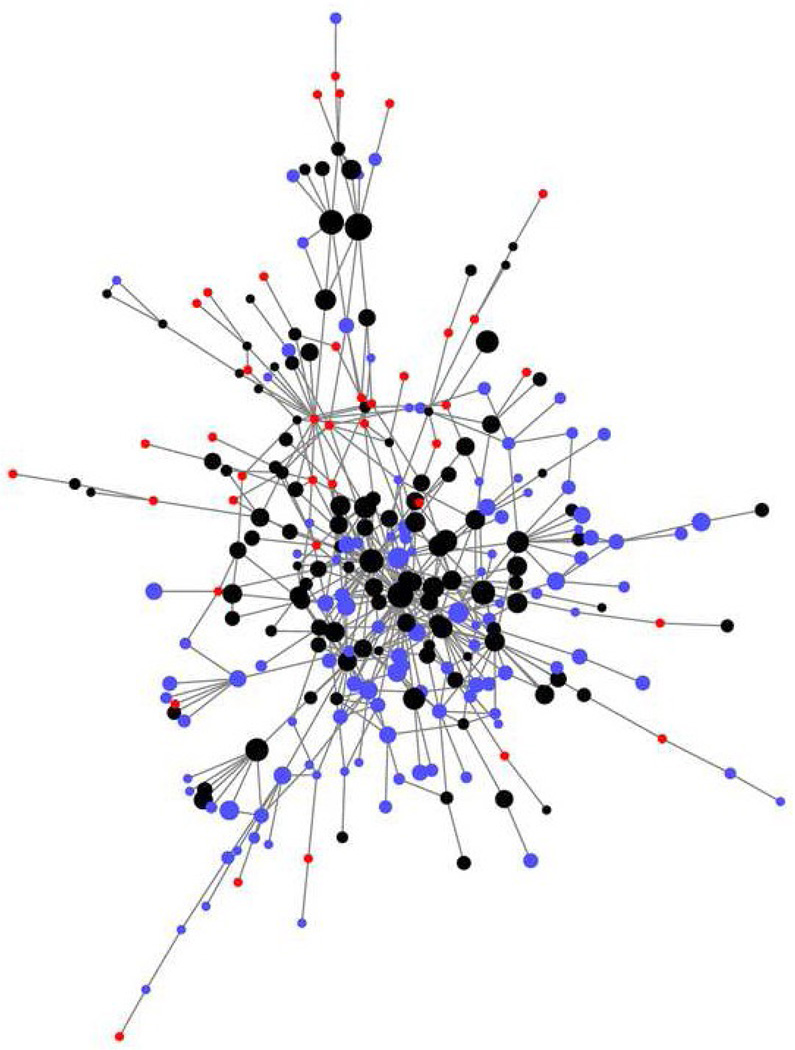
To reveal the underlying difference between MLP new-fold clusters and existing clusters of native structures, we define that a native-structure cluster is recovered by MLP structures if one or more members of the cluster have a TM-Score≥0.5 with one or more MLP structures. Because two structures can be considered as structurally related with a TM-Score of 0.4 or above [17], we can further define a "structural popularity" index (PIstruc) (or degree in graph theory) of a structure cluster based on the number of other neighboring structure clusters within TM-Score of 0.4 from the structure cluster (i.e. within a super-structure cluster). For simplicity, only the structural similarity between the representative structures of each cluster is evaluated here. The average PIstruc is 9.4 for native structure clusters recovered by MLP, 6.1 for unrecovered native structure clusters and 2.0 for MLP new-fold clusters. The PIstruc difference between recovered and unrecovered native-structure clusters and that between recovered and MLP new-fold clusters are statistically significant (p-value<0.00001 for both cases). As an example, the largest structural similarity network for mixed α/β proteins is shown in Fig. 4. Obviously, this TM-Score=0.4 threshold is somewhat arbitrary for defining structural popularity. Any other threshold (less than the threshold TM-Score=0.5 for defining structural clusters) would yield the same qualitative result. In fact, for clarity, TM-Score=0.45 or above is employed for drawing a link between two clusters in Fig. 4. It is clear that most recovered native-structure clusters (in black) are located at the center of the network while most MLP new-fold clusters (in Red) at the outskirt of the network with few links.
Fig. 4.
The largest network for alpha/beta structure clusters containing 231 nodes. The sizes of nodes are scaled according to the number of non-redundant sequences within each cluster. Black nodes denote the native structure clusters that have one or more structures similar to one or more MLP structures (TM-Score≥0.5, denoted as recovered by MLP) while the blue nodes are those native-structure clusters not yet recovered by MLP structures generated in this study. Red nodes are MLP new fold clusters that are structurally different from all existing structures (TM-Score<0.5). We used the smallest size for red nodes because the number of non-redundant sequences for them is unknown. For clarity of the graph, a link between two nodes is made only if TM-Score between the representative structures of the two cluster centers is greater than or equal to 0.45. A threshold of 0.4 is employed to calculate PIstruc in the paper.
To confirm the above finding, we also make a sequence-based definition of popularity index (PIseq): the number of unique (non-redundant) sequences within a structure cluster (sequence identity < 30%). A structure cluster is popular if it is adopted by many non-redundant sequences. Indeed, there is a statistically significant difference (p-value <0.00001) in PIseq values between the clusters recovered by MLP (12.0) and other native-structure clusters (4.5). Moreover, there is a continuous reduction of PIseq for newly discovered structure clusters over the time (Fig. 5). The average PIseq decreases from 11.4 before and in 1998 to 1.5 in 2007–2009. Similarly, the average PIstruc (number of neighboring clusters) decreases from 12.5 before and in 1998 to 3.9 between 2007–2009 (the difference is statistically significant with a p-value <0.0001). Thus, the number of experimentally solved structural folds (or clusters) converges in Fig. 1 because highly "popular" structures (adopted by many sequences) are mostly solved and less popular structures are more difficult to find by random sampling of protein sequence space. This result is somewhat expected because highly popular structures in sequence space are more probable to be located for experimental structure determination by random search in sequence space. Nevertheless, the result provides the first quantitative evidence that converging in number of new folds (or structure clusters) solved experimentally does not necessary mean the completeness of the structure space.
Fig. 5.
The number of native-structure clusters as a function of the age and sequence popularity index (PIseq) of each structure cluster. The age of a cluster is defined as the oldest (deposit date) structure within the cluster. PIseq of a structure cluster is defined as the number of non-redundant sequences (30% sequence identity or less) within each cluster. (Statistics is based on 5 bins each for time and PIseq (in a logarithmic scale). Newly discovered structural clusters are less and less popular (adopted by fewer and fewer sequences) over the time.
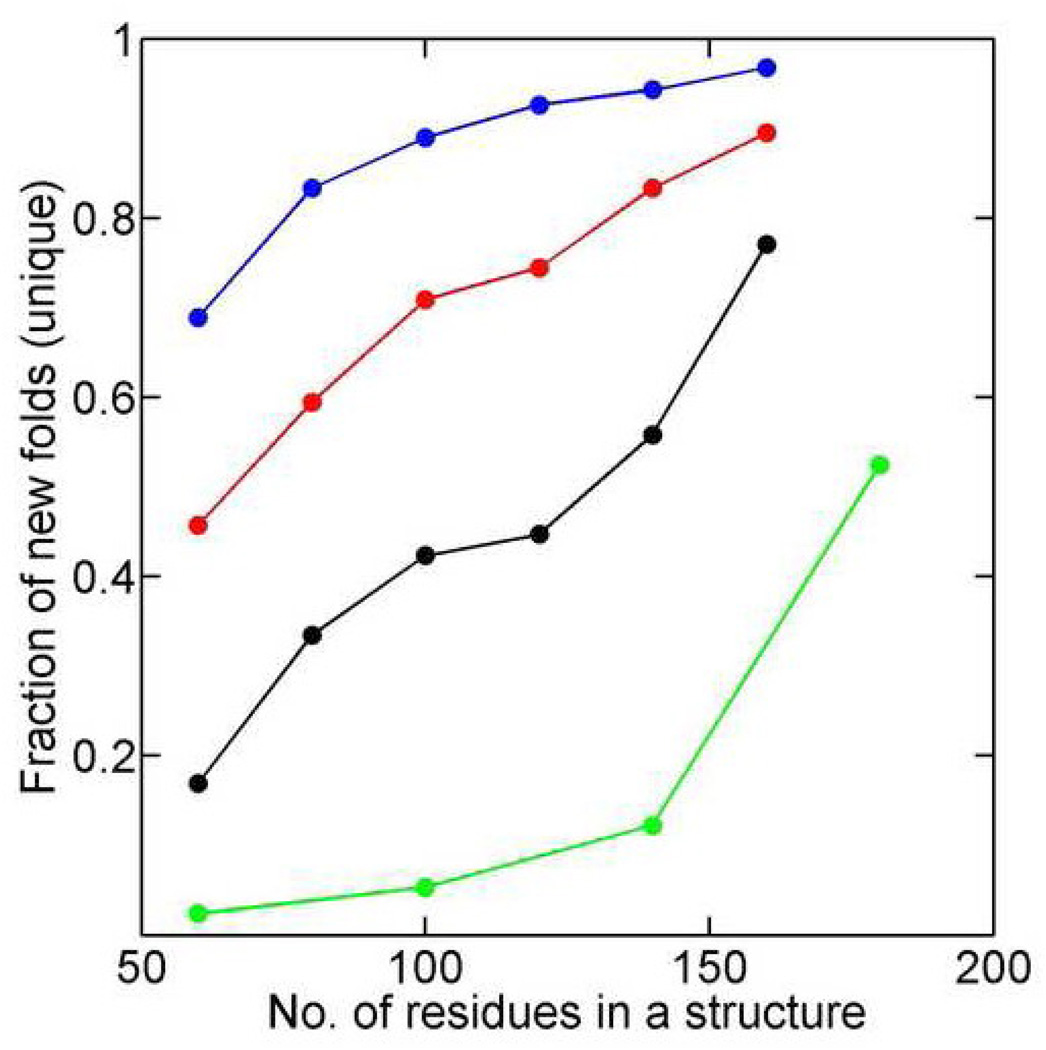
While popular structure "hubs" of proteins have been mostly solved as shown in Fig. 5, what about those less popular island-like structural clusters? One can gain a feel about the completeness of existing structure space by calculating the fraction of new-fold structure clusters in all MLP-C820 structure clusters generated. We found that only 23% of MLP structure clusters of small proteins (between 60 and 80 residues long) but 82% of MLP structure clusters of medium-size proteins (between 180–200 residues long) belong to new-fold clusters (See Fig. 6 in Black). This suggests that the observed structure space for small-domain proteins is about 80% complete. However, considering that this study has limited to a maximum permutation of five loops and this limitation has significant impact on large-size proteins that often have more than five loops, the potential structural space for medium and large-size domains is far from complete. This result is based on a TM-Score cutoff of 0.5 as a definition of clusters. Fig. 6 also shows that different cutoff values will move the curve up or down but more than 50% MLP structures for medium-size proteins (180–200) are new for all cutoff values from 0.45 to 0.6. That is, a significant number of new structure clusters exists for large domain proteins, in particular, regardless the threshold employed for defining clusters.
Fig. 6.
Fraction of new-fold structure clusters in all MLP clusters (based on the sizes of representative proteins of each cluster) as a function of protein size (the center of each bin) at different TM Score cutoffs [0.6 (Blue), 0.55 (Red), 0.5 (Black), and 0.45 (Green) from top to bottom]. A smaller cutoff value leads to a reduction of new structure clusters as expected. Even at TM-Score cutoff of 0.45, the fraction of new clusters for medium-size proteins (180–200) is >50%.
DISCUSSION
In this paper, we have developed a multiple loop permutation technique to explore the potential structural space of proteins. This method is developed so that the explored structure space is as realistic as possible while permitting a large-scale study. This is accomplished by fixing the core region of a given native template and generating alternatively connected secondary structure segments. A large-scale but limited MLP study of 2936 SCOP domains (limited to 5 loop permutations) confirmed its ability to generate realistic structural clusters by recovering 63% of existing clusters and 7/12 new-fold CASP 8 targets. Analysis of recovered and not-recovered MLP clusters indicates that the observed number of structural clusters converges because most popular structural clusters have already been solved.
We would like to note that rearranging secondary structure elements for generating new potential folds is not a new idea. Taylor et al. has performed combinatorial search of secondary structure arrangement on the 2-D lattice model and discovered the existence of significant number of new folds based on inability to map native structures to most discovered idealized forms. Here, we proposed a different approach that achieves the rearrangement of secondary structure by making loop permutation on existing native structures while fixing naturally occurring core packing. Despite significantly more restrictive in structure generation, our results confirm the existence of a significant number of new structural clusters with a naturally occurring core.
This study also touches upon an unsolved problem: how to define a fold computationally. While the definition of folds made by manually curated SCOP 2 and semi-manually curated CATH 3 are widely accepted, there are no widely accepted automatic techniques for defining folds that are required for large-scale classification of newly generated structures. We have employed TM-Score from TMalign4 because it has been employed to demonstrate that the existing structural space is complete7 and it provides a uniform standard for classifying both native structures and MLP structures. A threshold of TM-Score=0.5 for clustering structures was suggested by Xu and Zhang5. This threshold leads significantly more (about twice more) structural clusters than the number of folds defined by either SCOP or CATH. A more relaxed TM-Score threshold (e.g. TM-Score=0.45) would lead to a smaller number of structural clusters (1700) that is closer in line with the number of folds defined by SCOP or CATH (about 1200). However, different thresholds will not change the overall conclusion drawn in this paper as demonstrated in Fig. 6. This figure shows that a significant number of new structure clusters exists for large domain proteins regardless the threshold employed. In this paper, to avoid confusion, “fold” is reserved for SCOP or CATH classification while “cluster” is reserved for classification made by TM-Score.
There is an on-going discussion regarding whether or not the protein structure space is discrete or continuous . The result reported here supports the dual characteristics of the structure space : all structures can be continuously linked with a low structural similarity threshold 34, but, the distribution of structures is far from even as found in SCOP/CATH classifications and revealed in Fig. 4. Outlying unpopular island-like structural clusters (with few links) display the discrete side of the structure space. They constitute either yet-to-be-seen or yet-to-be-adopted (potential) structures in the protein structure space.
What causes these potential clusters to be unpopular "islands" that are rarely (or not) adopted by nature? The majority of uncovered new structural clusters are free of knots or deep slipknots. We examine if these MLP new-fold clusters are as optimized as their original in term of their contact orders , which are defined as a summation of absolute difference in sequence positions for all pairs of residues in contact, normalized by the number of contacts and the number of residues. It is well known that folding rates depend on the amount of nonlocal contacts (contacts with large sequence separation) in a protein structure. The average contact order for MLP new-fold clusters (based on cluster centers) is 0.238±0.06, statistically insignificantly different from 0.235±0.06, the average for their original native structures prior to loop permutations (p-value=0.17). In other words, the amount of nonlocal contacts of native and that of MLP new-fold structures are similar in average.
Other probable causes are evolution (initially larger families grow faster due to the higher probability to be duplicated) 38 or functional requirement (rapid expansion of certain functionalities 39 or suitability of some structures for multiple functionality). It may also be caused by that some structures can be formed by many more sequences than others (more designable) . More likely, it is a combination of these factors 43 that are responsible for the uneven distribution of the number of non-redundant sequences in different structure clusters and for the rare occurrence or absence of potentially stable structure clusters uncovered by MLP.
These potential structural clusters, however, have significant implication on protein engineering and design because MLP provides a simple yet highly effective tool for generating new structural topologies that are likely stable due to their naturally occurring packed cores. This is significant because producing new structures experimentally by directed evolution and random recombination of secondary-structure blocks is proved rarely successful while ab initio backbone design coupled with Rosetta de novo structure prediction for de novo design is complicated and time consuming 14. The proposed method will expand our capability to design novel proteins and biomaterials that have useful nano-scale and biological properties. For example, MLP of some non-knotted proteins can generate naturally knotted structures that possess unique functional properties.
While the proposed MLP can serve as a new technique for generating novel structural clusters with naturally occurring stable core, a slight modification of the method employed here is needed in order to generate stable structures for experimental verification because the main purpose of this paper here is to demonstrate that new structure clusters exist even with a restriction of fixing the naturally occurring core in a large-scale study. Structural classification depends mainly on how secondary structural elements are linked and not so much on the types of amino-acid residues linking them. Thus, polyalanine linkers are employed in this paper for computational efficiency. If the purpose is to produce experimentally stable proteins, more hydrophilic linkers optimized with necessary protein design tools (e.g.) are necessary in order to produce a more soluble protein. Different types of links will not change the assessment of structural similarity by TM-Score.
The results reported here also have significant implication in protein structure and function prediction. Hamprecht et al.15 showed the potential utility of using pseudo-native structures for threading-based structure prediction. Here, significant match between MLP structures and CASP targets indicates that these "new-fold" targets may be predicted by template-based structure prediction with MLP structures as templates. This is important because template-based structure prediction is significantly more reliable and accurate than template-free structure prediction, for proteins with 100 residues or longer, in particular 45. Furthermore, the matches between native structural folds and MLP structures (conservation of core packing) could suggest possible functional link between two previously unconnected folds.
Methods
Protein structure dataset
The domain structures with sequence identity cutoff at 95% from the ASTRAL 1.75 release26 were downloaded. This database is based on SCOP 1.75 for all protein structures in PDB released by Feb 20092. It contains 16712 domains. After removing the structures with missing atoms, we obtained 11291 domains belonging to 2936 SCOP families. One random structure was chosen from each SCOP family. This leads to 2936 domain structures (SCOP2936) as the starting structures for subsequent multiple loop permutations.
Multiple loop permutations (MLP)
Multiple loop permutations were performed in the following steps:
Secondary structure for each domain was assigned by the program STRIDE46. Short helical and strand segments (<4 residues) were treated as coils to decrease the number of loops for a given protein by reducing the number of secondary structure segments (SSSs).
The distances between the N-terminus of one SSS and the C-terminus of another SSS were calculated for all SSS pairs. The N and C termini of two SSSs were allowed to connect by building a new loop between them if their distance is less than a cutoff distance (15Å initially). The connection between two N (or C) termini of two SSSs was not allowed in order to maintain the original N to C direction. The original loops longer than 15Å were unchanged.
A combinatorial search was made for all possible loop permutations allowed. If two SSSs change from sequence neighbor to non-neighbor after rearrangement, their connection loop will be removed. Meanwhile, new loops will be built to connect two SSSs that become sequence neighbors after rearrangement. For example, a protein with 6 SSSs is arranged in a native structure as 1-2-3-4-5-6. One possible rearrangement of this sequence is 6-5-2-3-4-1. This rearrangement requires retaining two native loops for unchanged neighboring SSSs between 2-3 and 3-4, removing three native loops (1-2, 4-5 and 5-6) because they are no longer sequence neighbors (5-6 is not same as 6-5 because of the N to C direction), and building three new loops between 6-5, 5-2, and 4-1. In this study, we limited ourselves to generate 100 MLP structures and a maximum of five permutated loops per proteins. If the number of permutations is greater than 100, we decreased the cutoff distance with a step size of 0.5Å to reduce the number of loops allowed to permute until the number of permutations is less than or equal to 100.
All new loops were built by the program Modloop47. We estimated the number of residues for a new loop by dividing the end-to-end distance with 2.5Å. This approximate formula was obtained from a statistical analysis of the end-to-end distances of short loops. Because the maximum end-to-end distance for a loop to be permutated is 15Å, the maximum number of residues for a rebuilt loop is 6. That is, we have avoided building potentially unrealistic long loops (>6) 47. All loops were built with alanine residues for computational efficiency.
The above procedure generated 96131 MLP structures. We first removed any MLP structures with severe steric clashes and abnormal bond lengths as identified by the program PROCHECK48 (30714 structures remaining). We then removed any MLP structures whose terminal residues are less than 20% exposed based on their solvent accessible surface areas from STRIDE [21] (22296 structures remaining). We further removed the structures that are similar (TM-Score>0.5) to the original native structures prior to loop permutations (3374 structures remaining). Finally, we removed MLP structures with less than 60 residues and obtained 2843 structures. These 2843 MLP structures can be separated into 1048 novel new-fold structures and 1795 "old-fold" structures that have TM-Score≥ 0.5 with at least one of the 16712 structure domains from the ASTRAL 1.75 release.
Clustering protein structures
Because MLP structures are not annotated by either SCOP or CATH, a fully automatic technique is needed to compare MLP structures with existing structures and cluster them to remove redundancy. We use TM-Score because it has been found to be a reasonable measure to define structural folds5. As in Ref. 5, we considered that two structures are similar when TM-Score ≥ 0.5. Structures are clustered as follows: 1) the number of similar structures (TM-Score ≥ 0.5) for each structure in the dataset is calculated; 2) the structure with the largest number of similar structures is chosen as the representative structure for the first cluster, 3) the structures in the first cluster are removed and the remaining structures are subjected to steps 1) and 2) until all structures are clustered. The above steps produce the representative structures, i.e., cluster center for all structure clusters. The TM-Scores between these cluster centers and each structure are compared. All structures are re-assigned according to the highest TM-Score between a structure and a cluster center. The same clustering technique is applied to existing native and MLP structures. When we compare two structure clusters, we calculate the TM-Score between the representative structures in each structure cluster.
Knot detection
We detect whether or not a protein has a knot by using the algorithm developed by Khatib et al49. A protein chain is treated as a series of segments connecting two adjacent Cα atoms. A protein containing N residues has N nodes, N-1 segments and N-2 triangles, while each triangle consists of two adjacent segments. A triangle, consisting of three nodes ith, (i+1)th and (i+2)th Cα atoms, can be simplified to a segment connecting ith and (i+2)th nodes by removing the (i+1)th node, if this triangle satisfies the condition that none of segments connecting any pair of nodes in the protein chain crosses the triangle. In this way, a non-knotted protein chain can be simplified into four nodes by iteratively removing nodes, while the knotted protein chain is stuck with more than 4 nodes during the simplification process.
Slip Knot detection
The algorithm for slip-knot detection follows the method developed by Khatib et al 50. It has the following three steps. First, a “closed” loop is defined when the distance between Cα atoms of two ending residues of the loop is less than 7Å and their sequence separation along the chain is between 3 and 33 residues. Second, the closed loop is broken down into a number of small triangles and each triangle is formed by three Cα atoms in the closed loop. Third, for all lines that connect two adjacent Cα atoms, we examine whether the line intersects any triangle of the closed loop. The residue index of each intersection is recorded. A slip-knot corresponds to two intersections between the chain and a closed loop, while the first intersection indicates the chain enters the closed loop and the second intersection indicates the chain leaves the closed loop. The depth of a slip-knot is the difference between residue indices of two intersections.
Acknowledgement
This work was supported by the National Institutes of Health grants GM R01 085003.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levitt M. Nature of the protein universe. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Research. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH--a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Research. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu JR, Zhang Y. How significant is a protein structure similarity with TM-score=0.5? Bioinformatics. 2010;26:889–895. doi: 10.1093/bioinformatics/btq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor WR, Bartlett GJ, Chefliah V, Klose D, Lin K, Sheldon T, Jonassen I. Prediction of protein structure from ideal forms. Proteins-Structure Function and Bioinformatics. 2008;70:1610–1619. doi: 10.1002/prot.21913. [DOI] [PubMed] [Google Scholar]
- 9.Taylor WR, Chelliah V, Hollup SM, MacDonald JT, Jonassen I. Probing the "Dark Matter" of Protein Fold Space. Structure. 2009;17:1244–1252. doi: 10.1016/j.str.2009.07.012. [DOI] [PubMed] [Google Scholar]
- 10.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graziano JJ, Liu WS, Perera R, Geierstanger BH, Lesley SA, Schultz PG. Selecting folded proteins from a library of secondary structural elements. Journal of the American Chemical Society. 2008;130:176–185. doi: 10.1021/ja074405w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuji T, Onimaru M, Doi N, Miyamoto-Sato E, Takashima H, Yanagawa H. In vitro selection of GTP-binding proteins by block shuffling of estrogen-receptor fragments. Biochemical and Biophysical Research Communications. 2009;390:689–693. doi: 10.1016/j.bbrc.2009.10.029. [DOI] [PubMed] [Google Scholar]
- 13.Lo Surdo P, Walsh MA, Sollazzo M. A novel ADP- and zinc-binding fold from function-directed in vitro evolution. Nature Structural & Molecular Biology. 2004;11:382–383. doi: 10.1038/nsmb745. [DOI] [PubMed] [Google Scholar]
- 14.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 15.Hamprecht FA, Scott W, vanGunsteren WF. Generation of pseudonative protein structures for threading. Proteins-Structure Function and Genetics. 1997;28:522–529. [PubMed] [Google Scholar]
- 16.Taylor WR. Decoy models for protein structure comparison score normalisation. Journal of Molecular Biology. 2006;357:676–699. doi: 10.1016/j.jmb.2005.12.084. [DOI] [PubMed] [Google Scholar]
- 17.Yang QY, Sharp KA. Building alternate protein structures using the elastic network model. Proteins-Structure Function and Bioinformatics. 2009;74:682–700. doi: 10.1002/prot.22184. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham BA, Hemperly JJ, Hopp TP, Edelman GM. Favin versus concanavalin A: Circularly permuted amino acid sequences. Proc Natl Acad Sci U S A. 1979;76:3218–3222. doi: 10.1073/pnas.76.7.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindqvist Y, Schneider G. Circular permutations of natural protein sequences: Structural evidence. Current Opinion in Structural Biology. 1997;7:422–427. doi: 10.1016/s0959-440x(97)80061-9. [DOI] [PubMed] [Google Scholar]
- 20.Iwakura M, Nakamura T, Yamane C, Maki K. Systematic circular permutation of an entire protein reveals essential folding elements. Nature Structural Biology. 2000;7:580–585. doi: 10.1038/76811. [DOI] [PubMed] [Google Scholar]
- 21.Hennecke J, Sebbel P, Glockshuber R. Random circular permutation of DsbA reveals segments that are essential for protein folding and stability. Journal of Molecular Biology. 1999;286:1197–1215. doi: 10.1006/jmbi.1998.2531. [DOI] [PubMed] [Google Scholar]
- 22.Dill KA, Stigter D. Modeling Protein Stability as Heteropolymer Collapse. Advances in Protein Chemistry, Vol 46. 1995;46:59–104. doi: 10.1016/s0065-3233(08)60332-0. [DOI] [PubMed] [Google Scholar]
- 23.Yuan X, Bystroff C. Non-sequential structure-based alignments reveal topology-independent core packing arrangements in proteins. Bioinformatics. 2005;21:1010–1019. doi: 10.1093/bioinformatics/bti128. [DOI] [PubMed] [Google Scholar]
- 24.Guerler A, Knapp EW. Novel protein folds and their nonsequential structural analogs. Protein Science. 2008;17:1374–1382. doi: 10.1110/ps.035469.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abagyan RA, Maiorov VN. An Automatic Search for Similar Spatial Arrangements of Alpha-Helices and Beta-Strands in Globular-Proteins. Journal of Biomolecular Structure & Dynamics. 1989;6:1045–1060. doi: 10.1080/07391102.1989.10506535. [DOI] [PubMed] [Google Scholar]
- 26.Brenner SE, Koehl P, Levitt R. The ASTRAL compendium for protein structure and sequence analysis. Nucleic Acids Research. 2000;28:254–256. doi: 10.1093/nar/28.1.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolesov G, Virnau P, Kardar M, Mirny LA. Protein knot server: detection of knots in protein structures. Nucleic Acids Research. 2007;35:W425–W428. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moult J, Fidelis K, Kryshtafovych A, Rost B, Tramontano A. Critical assessment of methods of protein structure prediction - Round VIII. Proteins-Structure Function and Bioinformatics. 2009;77(Suppl 9):1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- 29.Xie L, Bourne PE. Detecting evolutionary relationships across existing fold space, using sequence order-independent profile-profile alignments. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:5441–5446. doi: 10.1073/pnas.0704422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolodny R, Petrey D, Honig B. Protein structure comparison: implications for the nature of 'fold space', and structure and function prediction. Current Opinion in Structural Biology. 2006;16:393–398. doi: 10.1016/j.sbi.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 31.Sadreyev RI, Kim BH, Grishin NV. Discrete-continuous duality of protein structure space. Current Opinion in Structural Biology. 2009;19:321–328. doi: 10.1016/j.sbi.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pascual-Garcia A, Abia D, Ortiz AR, Bastolla U. Cross-Over between Discrete and Continuous Protein Structure Space: Insights into Automatic Classification and Networks of Protein Structures. Plos Computational Biology. 2009;5:E1000331. doi: 10.1371/journal.pcbi.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrison A, Pearl F, Mott R, Thornton J, Orengo C. Quantifying the similarities within fold space. Journal of Molecular Biology. 2002;323:909–926. doi: 10.1016/s0022-2836(02)00992-0. [DOI] [PubMed] [Google Scholar]
- 34.Skolnick J, Arakaki AK, Lee SY, Brylinski M. The continuity of protein structure space is an intrinsic property of proteins. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15690–15695. doi: 10.1073/pnas.0907683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaxco KW, Simons KT, Baker D. Contact order, transition state placement and the refolding rates of single domain proteins. Journal of Molecular Biology. 1998;277:985–994. doi: 10.1006/jmbi.1998.1645. [DOI] [PubMed] [Google Scholar]
- 36.Zhou HY, Zhou YQ. Folding rate prediction using total contact distance. Biophysical Journal. 2002;82:458–463. doi: 10.1016/S0006-3495(02)75410-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai YW, Zhou HY, Zhou Y. Critical nucleation size in the folding of small apparently two-state proteins. Protein Science. 2004;13:1173–1181. doi: 10.1110/ps.03587604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. doi: 10.1038/nature01256. [DOI] [PubMed] [Google Scholar]
- 39.van Nimwegen E. Scaling laws in the functional content of genomes. Trends in Genetics. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- 40.Govindarajan S, Goldstein RA. Why are some protein structures so common? Proceedings of the National Academy of Sciences of the United States of America. 1996;93:3341–3345. doi: 10.1073/pnas.93.8.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiana G, Shakhnovich BE, Dokholyan NV, Shakhnovich EI. Imprint of evolution on protein structures. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2846–2851. doi: 10.1073/pnas.0306638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Helling R, Tang C, Wingreen N. Emergence of preferred structures in a simple model of protein folding. Science. 1996;273:666–669. doi: 10.1126/science.273.5275.666. [DOI] [PubMed] [Google Scholar]
- 43.Goldstein RA. The structure of protein evolution and the evolution of protein structure. Current Opinion in Structural Biology. 2008;18:170–177. doi: 10.1016/j.sbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Dai L, Yang Y, Kim HR, Zhou Y. Improving computational protein design by using structure-derived sequence profile. Proteins-Structure Function and Bioinformatics. 2010;78:2338–2348. doi: 10.1002/prot.22746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y. Protein structure prediction: when is it useful? Current Opinion in Structural Biology. 2009;19:145–155. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins-Structure Function and Genetics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 47.Fiser A, Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics. 2003;19:2500–2501. doi: 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 49.Khatib F, Weirauch MT, Rohl CA. Rapid knot detection and application to protein structure prediction. Bioinformatics. 2006;22:e252–e259. doi: 10.1093/bioinformatics/btl236. [DOI] [PubMed] [Google Scholar]
- 50.Khatib F, Rohl CA, Karplus K. Pokefind: a novel topological filter for use with protein structure prediction. Bioinformatics. 2009 25;:I281–I288. doi: 10.1093/bioinformatics/btp198. [DOI] [PMC free article] [PubMed] [Google Scholar]