SUMMARY
The encapsidated genome in all double strand DNA bacteriophages is packaged to liquid crystalline density through a unique vertex in the procapsid assembly intermediate, which has a portal protein dodecamer in place of five coat protein subunits. The portal orchestrates DNA packaging and exit, through a series of varying interactions with the scaffolding, terminase, and closure proteins. Here we report an asymmetric cryo-EM reconstruction of the entire P22 virion at 7.8-Å resolution. X-ray crystal structure models of the full-length portal and of the portal lacking 123 residues at the C-terminus in complex with gene product 4 (Δ123portal-gp4) obtained by Olia et al. were fitted into this reconstruction. The interpreted density map revealed that the 150-Å, coiled-coil, barrel portion of the portal entraps the last DNA to be packaged and suggests a mechanism for head-full DNA signaling and transient stabilization of the genome during addition of closure proteins.
INTRODUCTION
Bacteriophages provide exceptionally tractable model systems to understand the fundamental mechanistic principles by which large, macromolecular complexes assemble and function. Genetic studies of P22 (family Podoviridae), one of many dsDNA bacteriophages, provided detailed cause and effect relationships among the eleven viral gene products that participate in particle assembly and the subset of nine gene products resident in the mature virion (Botstein et al., 1973). Mutational analyses firmly established the order in which P22 gene products interact during assembly, the folding and assembly pathway of the capsid protein, and the regulation of dsDNA packaging (King et al., 1973). These resulted in flow charts of gene product relationships, but transmission electron micrographs of negatively stained P22 samples yielded insufficient resolution to correlate the detailed genetics with structure (King et al., 1976). The development and increasingly routine use of cryo-electron microscopy (cryoEM) and icosahedral image reconstruction methods in the mid 1990’s led to more detailed pictures of the capsid (Prasad et al., 1993; Thuman-Commike et al., 1996), and more recently, structures of whole (asymmetric) virions at comparable resolution (17-20 Å) (Chang et al., 2006; Lander et al., 2006). Detailed segmentation of the isolated tail machine at sub nanometer resolution was additionally achieved, and pseudo-atomic structures were derived by homology modeling for three viral proteins: portal (gp1:82.6kDa), gp4 (18.0 kDa), and gp10 (52.5 kDa) (Lander et al., 2009). Models of the tail needle (plug) (gp26:24.6 kDa) (Olia et al., 2007b, 2009) and tailspike (gp9:71.9 kDa) (Steinbacher et al., 1996; Steinbacher et al., 1997) proteins based on their crystal structures closely matched features in that cryoEM density map. One region of density extending from the portal to near the particle center was misinterpreted in the earlier asymmetric reconstruction of P22(Lander et al., 2006) as corresponding to the three ejection (“pilot”) proteins known to be in the particle (Casjens and King, 1974).
P22 is a meta-stable assembly, poised to inject its genome into salmonella host bacteria. A total of 415 copies of gp5, the major capsid protein (46.7 kDa), assemble into a nearly icosahedral, T=7 laevo, quasi-symmetric head. A 12-fold symmetric dodecamer of gp1 (“portal”) occupies one of the vertices of the icosahedron, forming an axial channel through which the dsDNA genome enters and exits the capsid. The portal also serves as the attachment site for the tail machine, formed by four proteins (gp4, gp9, gp10, and gp26), each of which assumes a different oligomeric state with respective subunit stoichiometries in a 12:18:6:3 ratio.
Here we report the asymmetric cryoEM-reconstruction of the infectious P22 virion at 7.8-Å resolution and the icosahedrally symmetrized map at 5.0 Å. Models derived from the crystal structures of full length portal and the Δ123portal-gp4 complex (“core-gp4 complex”) described in Olia et al. now provide an accurate density assignment for the entire reconstruction and a compelling description of the particle dynamics that accompany dsDNA packaging and tail-machine morphogenesis.
RESULTS AND DISCUSSION
Asymmetric Cryo-Reconstruction of Virion
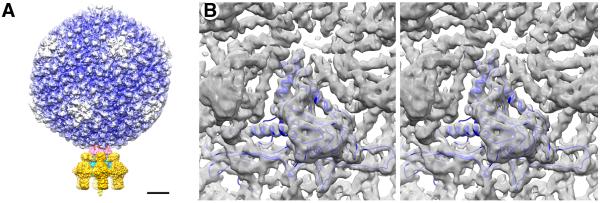
Three-dimensional (3D) reconstructions of the P22 virion (Figure 1A) and capsid (Figure 1B) were computed, respectively, from 21,645 and 18,602 images of unstained, vitrified virions (see Experimental Procedures). The density map of the icosahedrally-symmetrized, T=7 laevo capsid showed numerous long, tubular structures with a right-handed helical twist, characteristic of α-helical polypeptide segments rendered at ~5-Å resolution (Figure 1B). These correspond to the homologous “spine” helices prevalent in the core of the coat protein of bacteriophage HK97 (Wikoff et al., 2000). The capsid reconstruction provided a means to closely monitor steps taken to produce the final asymmetric virion reconstruction, which was computed without imposing any global or local symmetry (Tang et al., 2010).
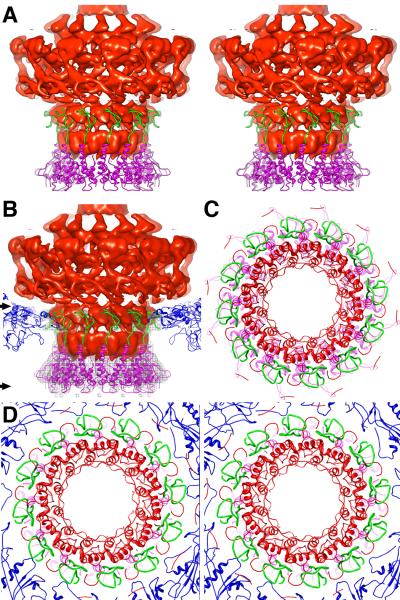
Figure 1.
Cryo-reconstructions of P22. (A) Asymmetric reconstruction of P22 virion at 7.8-Å resolution, with density map thresholded and segmented to highlight individual components: gp4 (pink), gp5 (color-cued radially from inner (blue) to outer (white) radii), gp9 (dark yellow), gp10 (cyan), and gp26 (yellow). Scale bar is 100Å. (B) A stereo view of the icosahedrally-symmetrized P22 capsid at 5.0-Å resolution. A modified capsid protein model (blue) nicely fits the reconstructed density.
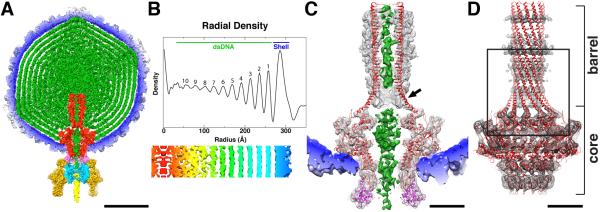
The atomic model for the full-length portal (Olia et al., 2011) nicely fit the asymmetric virion density map, with the 150-Å coiled-coil barrel structure redefining the scabbard-like tube of density (previously interpreted as ejection proteins(Lander et al., 2006)) that extends from the portal core to near the center of the head (Figure 2A; the entire portal, including the core and barrel, are colored red). The organization of the bulk dsDNA in the head (Figure 2A; DNA is green) closely matched what was previously reported (Chang et al., 2006; Lander et al., 2006), and includes eight distinct, concentric layers, all of which are ~22 Å apart (Figure 2B). The bulk DNA in our P22 structure appears to be ~15Å resolution or worse because that is the intrinsic resolution of the DNA within the capsid due to the dynamic variation that occurs between particles, regardless of the resolution of the capsid.
Figure 2.
P22 virion structure. (A) Cross-section (~60-Å thick) through the center of the P22 virion cryo-reconstruction, segmented to highlight individual components: gp1 (red), gp4 (pink), gp5 (color-cued radially from inner (blue) to outer (white) radii), gp9 (dark yellow), gp10 (cyan), gp26 (yellow), and DNA plus pilot proteins (green). Scale bar is 200 Å. (B) Radial density plot of the spherically averaged phage reconstruction highlights at least ten distinct layers of dsDNA, separated by about 22 Å, within the capsid shell. A thin, central section slab from a map averaged with 12-fold symmetry about the portal axis, aligned with and at the same magnification as the radial density plot above it and depicted with a radial color-ramp (red-to-blue, low-to-high radii), shows a close-up view of the DNA layers. (C) Magnified view of a slab of the virion density map, segmented to highlight the portal/gp4 complex (grey mesh) wedged inside the capsid (blue, solid density), the DNA (green, solid density) contained within the portal channel, and the ribbon model of the portal/gp4 crystal structure (gp1 and gp4 in red and magenta, respectively) fitted into the portal/gp4 density. The gp1 crystal structure clearly does not fit the density near the base (proximal end) of the barrel (black arrow) and extends ~10 Å above the reconstructed density at the distal end. Scale bar is 50Å. (D) Ribbon model of entire gp1 crystal structure (red) fit into the segmented portal density (grey mesh). Close-up view of boxed region appears in stereo in Figure 4. Scale bar is 50Å.
The DNA adopts dramatically different structures in the region immediately surrounding the scabbard and inside it (Figure 2A and C). Near the center of the head where the distal end of the barrel terminates, the bulk DNA is unconstrained and disordered whereas, inside the barrel, the DNA is confined and follows a linear path. The density ascribed to DNA again changes at the transition between the proximal end of the barrel and the portal core where it becomes considerably wider than expected for a single duplex. Density assigned to DNA extends ~20Å below the portal core and the gp4 ring, ending just inside the gp10 ring. This final ~ 20 Å of DNA may correspond to the portion of the genome that is not fully inserted into the capsid when the gp2/3 terminase complex disengages from the portal. As described below, we suggest that conformational changes in the barrel of gp1 may function as a “valve” to retain the DNA in the capsid until the gp26 tail needle (plug), which is the primary gene product known to keep the DNA within the virion (Lander et al., 2009; Olia et al., 2007b), is inserted into the tail machine. This occurs after gp4 and gp10 have associated with the portal, implying that intermediate stabilization of the packaged DNA is required.
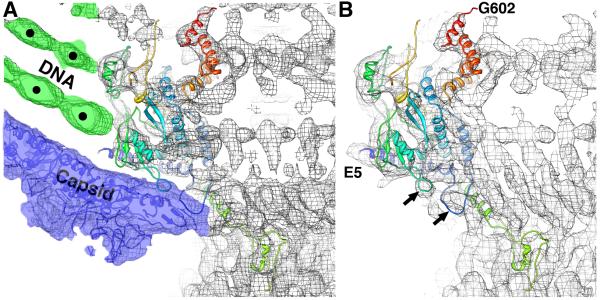
Near the portal-capsid interface, the individual dsDNA strands are hexagonally close-packed (marked by dots in Figure 3A) and an obvious, dense and circular ring of dsDNA abuts tightly against the portal subunits just inside the capsid (Figure 3A, green density marked by dot closest to the center of the panel). Comparison of rigid body fits of the portal X-ray model into cryo-reconstructions of the virion (Figure 3A) and isolated tail (Figure 3B) show that the model best fits the isolated tail. Portions of the portal whose conformations change in apparent response to the presence of gp5 subunits, include two loops (Figure 3B, arrows) and residues near the N-terminus, which clearly fit density present in the isolated tail (Figure 3B; E5 is first residue in the X-ray model) but not in the virion map. All residues on the portal subunit that interact with dsDNA in the virion are identified in Table 1.
Figure 3.
Conformational changes in portal occur after DNA packaging. (A) Fit of unmodified gp1 core ribbon model into P22 virion cryo-reconstruction (grey mesh) highlights interactions that occur with the packaged DNA (green colored mesh) and capsid subunits (blue-colored mesh and blue model). The most significant changes in portal conformation occur at the N-terminus and in the loops highlighted by arrows in (B). (B) Ribbon model of gp1 core crystal structure (E5 to G602, ramped in color from blue to red) nicely fits the reconstructed density (grey mesh) of the isolated tail machine (Lander et al., 2009). Black arrows point to two loops that change conformation upon interaction with capsid. Density threshold (~1σ) was used for the surface rendering.
Table 1.
A list of amino acid residues in portal that contact the first ring of the dsDNA genome. Each tail spike interacts with three adjacent gp4 subunits (lists of contact residues in the three gp4 subunits). Gp4 subunits A, B, and C correspond, respectively, to the green, red and magenta subunits in Figure 6.
| Portal residues in contact with dsDNA |
Gp4 residues in contact with gp9 tail spike |
||
|---|---|---|---|
| Subunit A | Subunit B | Subunit C | |
| LEU 8 | PRO 30 | THR 5 | THR 5 |
| LYS 228 | GLN 31 | LYS 6 | LYS 6 |
| ARG 249 | GLN 34 | GLY 7 | GLY 7 |
| ASP 250 | ALA 68 | ASP 8 | ASP 8 |
| ILE 251 | GLU 69 | VAL 10 | |
| LYS 252 | ARG 11 | ||
| ASP 253 | LEU 14 | ||
| VAL 254 | ASP 27 | ||
| ASP 256 | GLU 29 | ||
| ASP 257 | PRO 30 | ||
| GLN 270 | GLN 31 | ||
| ARG 275 | MET 33 | ||
| TYR 277 | GLN 34 | ||
| LYS 290 | ASN 65 | ||
| PRO 66 | |||
| PRO 67 | |||
| ALA 68 | |||
| GLU 69 | |||
| ASP 71 | |||
Portal Barrel Exhibits Large Conformational Changes
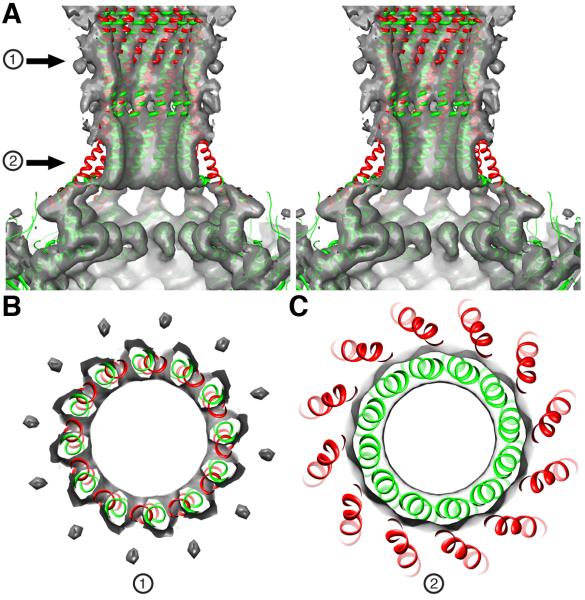
The structure of the portal barrel clearly differs between virions and crystals. A rigid body fit of the full-length portal X-ray model into the virion cryo-reconstruction positions the distal end of the barrel ~10 Å beyond the scabbard density (Figure 2C). Also, at the base of the scabbard where it meets the portal core, the X-ray model splays radially outwards beyond the EM density (Figure 2 C (arrow) and D). An additional local 12-fold symmetry averaging step was carried out to improve the signal-to-noise ratio of the reconstructed barrel density. When the EM map is rendered at a higher density threshold, the twelve helices in the X-ray model are seen to fall out of phase (i.e. rotated ~10° clockwise as viewed down the barrel axis) with the corresponding tube-like features in the virion map (Figure 4). All three regions of inconsistency between the X-ray model and cryoEM density are accommodated if the model is twisted and the bundle of helices is compressed slightly inwards towards the axis of the barrel near its base (Movie M1, Supporting Material). This concerted rearrangement shifts the distal end of the barrel towards the core of the portal by ~10 Å, positioning the helices in register with the tube-like features near the proximal end of the scabbard (Figure 4). As the head fills during packaging, the mounting pressure exerted by the DNA may induce these rearrangements in the barrel structure, which we posit functions as a valve to retain the DNA until the tail machine assembles.
Figure 4.
Remodeling of the gp1 crystal structure to fit the P22 virion density map. (A) Stereo, close-up view of the back half of the proximal part of the portal barrel (grey surface) into which the gp1 crystal structure (red, ribbon model) is fitted. The model fits the portal core density quite well (“core” consists of residues 1-602 and only the topmost portion appears in this view at the bottom of the panel). Above the core, the model deviates significantly from the density map. For example, the helices traverse empty (low density) space and then line up out of register with tubular density features that follow a slewed path and clearly define the wall of the barrel. The adjusted model (green) fits the density map better with regions pointed by two arrows (1) and (2) highlighted in the cross-sections shown in (B) and (C). See also supplemental Movie M1.
Interaction of Closure Protein (gp4) with Capsid
The atomic model of the core-gp4 complex (Olia et al., 2011) fit the reconstructed 8-Å density map of the isolated tail machine (Lander et al., 2009) with virtually no need for adjustment, indicating that release of the tail machine from virions has no deleterious effect on the portal core structure and associated proteins. Although SDS page analysis of the proteins in the isolated tail machine indicates that the barrel residues (602-725) of the portal are present, there was no density that corresponds to a fully-formed barrel, indicating that, in the process of tail machine release from the virions, the coiled-coil structure is disrupted and does not reform in the tail machine storage conditions in vitro.
Fitting the X-ray model of the core-gp4 complex into the 7.8-Å virion density map revealed the presence of an unexpected interaction between the extended C-terminus of gp4 with both the portal and capsid (Figure 5). A stretch of about 21 gp4 C-terminal residues (a.a. 130-150) lies wedged between the capsid and portal. A potential multi-step mechanism that explains this observation includes: 1) The packaging complex of the small terminase (gp3: 18.6 kDa), and gp2 (57.6 kDa), the ATPase that provides the driving force, associates with the portal and packages the DNA. 2) A head full of DNA generates the signal that allows gp2 to cut the DNA and terminate packaging (Casjens et al., 1992). The pressure created by the highly condensed DNA in the head is likely responsible for changing the barrel conformation and this in turn closes the valve that retains the DNA, transducing the signal to the terminase proteins. 3) Next, gp4 monomers compete with the terminase complex for its attachment site on the portal and displace it. This induces or permits a final change in portal conformation that allows the C-terminus of gp4 to invade the portal-capsid interface and thereby anchor it securely to the particle. The recently reported P22 procapsid structure clearly shows scaffold protein wedged between the capsid protein and the scaffold (Chen et al., 2011). Comparing the portal position in the procapsid and the virion shows that the portal increases its contact with the capsid shell during maturation (see figure S1). We propose that this portion of the scaffold remains in place during dsDNA packaging, allowing access of the gp4 C-terminus to the bottom of the portal. When gp4 binds, the scaffold protein is displaced allowing the final conformational change implied by the position gp4-C terminal polypeptide that is wedged between the capsid and portal.
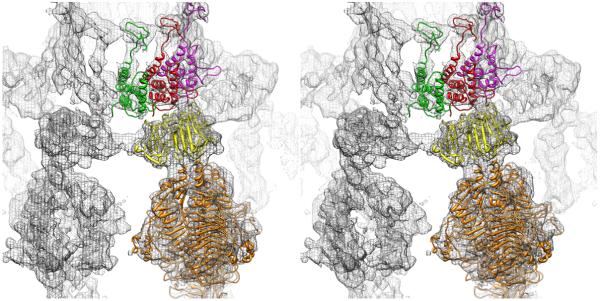
Figure 5.
Interactions among P22 proteins gp1, gp4, and gp5. (A) Stereo, side view of the portal density (red surface) and the fitted gp4 crystal structure (magenta). Density threshold (~2σ) was selected for the surface rendering. The C-terminal loop portion of gp4 (amino acids 122-150 highlighted in green) extends up the portal core. (B) Same as (A) with ribbon model (blue) for the capsid and density mesh for gp4 added to show that the C-terminal loop of gp4 wedges between the portal and capsid subunit interface. Arrows on the left of panel B delineate the slab shown in panels C and D. (C) Top-down view of the modeled portal/gp4 complex for the region defined by the two arrows shown in B. (D) Same as (C), but in stereo and including the capsid model (blue ribbon). Overlap between the gp4 (green) and gp5 (blue) models clearly suggests that portions of gp4 must undergo significant conformational changes when the tail structure is added during phage assembly. See also Figure S1.
After twelve gp4 subunits bind to the bottom of the portal, a preformed hexamer of gp10 attaches to the gp4 ring (Olia et al., 2007a). The exposed gp4-gp10 interfaces form the sites to which six trimers of the tail spike protein (gp9) attach. Spike trimers are bi-functional, having both receptor-binding activity for O-antigenic repeats of polysaccharides on the salmonella cell surface as well as endoglycosidase activity for cleaving glycosidic bonds after attachment (Baxa et al., 1996). A pseudo-atomic model of the gp9-gp4 interaction (Figure 6) indicates that this symmetry-mismatched interface is rigid (Table 1, columns 2,3,4). However, a flexible hinge on gp9 confers significant adaptability of the gp9 trimers in their various roles. The gp26 tail needle (plug), the final component added to the tail assembly, interacts only with gp10 and prevents escape of DNA from the capsid (Strauss and King, 1984).
Figure 6.
Stereo view of interactions between P22 proteins gp4 and gp9. Portion of the P22 virion density map (grey mesh) with rigid body-fitted models of three gp4 subunits from the dodecamer crystal structure (colored green, red, and magenta) and a gp9 trimer (yellow for the spike head domain and orange for the spike tail domain) for one of the six tail spikes.
Biological Role of the Portal Barrel
The realization that the barrel is such a prominent component in the phage structure prompted us to use bioinformatics to search for sequences likely to form such structures. A strong coiled-coil propensity (Lupas et al., 1991) occurs in the C-terminal ~130 residues of all sequenced P22-like podoviruses, and we find that 20 of 25 podovirus portal proteins (chosen to span the diversity within the Podoviridae) have similar C-terminal, coiled-coil predictions (Figure 7A shows a sampling of these). In contrast, the barrel domain is not found in φ29-like phage (Simpson et al., 2000) and is not predicted to be present in the T7-like phages. A random sample of 30 portal proteins from long-tailed sipho- and myovirus bacteriophages revealed that only two had such a prediction (Figure 7A shows one of these, for phage T4). Thus, portal proteins with a C-terminal barrel appear to be preferred by podoviruses and are found less frequently in other tailed phages.
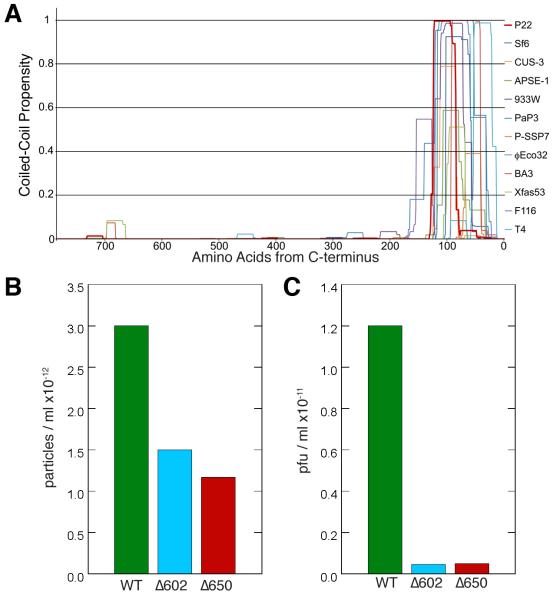
Figure 7.
(A) Plot of the conserved, predicted coiled-coil motif in the C-termini of portal proteins for several members of the Podoviridae family, as determined by the software COILS (Lupas et al., 1991) with a 21 amino acid scan window. The C-termini of the proteins are aligned at the right side of the plot. The phages encoding these portal proteins include: P22, Sf6, CUS-3 and APSE-1, divergent members of the P22-like phages; 933W, F116, BA3, Xfas53, P-SSP7, PaP3 and φEco32, podoviruses that are all distantly related to one another; and T4, a long-tailed myovirus. Each of these portal proteins shows a strong peak of coiled-coil probability within its C-terminal 150-100 a.a. For some, including P22 (in dark red), the probability drops in the terminal 70 a. a., even though the crystal structure clearly shows an uninterrupted coiled-coil extending up to within 6 a.a. of the C-terminus (Olia et al., 2011). The reason for this discrepancy between predicted and observed coiled-coil in P22 is not known. (B and C) C-terminal deletions of portal protein reduce phage assembly and infectivity. Phage particles/ml (B) and plaque forming units/ml (C) produced by trans-complementation of a P22 nonsense portal mutant with full length and portal proteins truncated from residues 603-725 (Δ602) and 651-725 (Δ650) as previously described (Moore and Prevelige, 2002). Fractions from a CsCl gradient that displayed peak infectivity were collected. The number of particles/ml was determined by DNA absorbance and the titer was determined on a permissive host. EM and gel analysis indicated that the particles produced with truncated portal were properly tailed and contained full length DNA as occurs in wild-type (WT) phage.
The barrel structure also suggested a series of C-terminal deletion experiments to explore its role(s) in the P22 infection cycle. A mutational study of the P22 barrel was monitored by SDS-PAGE and Western blot analysis and showed that these truncated portals were incorporated into phage that stably packaged full-length DNA and the ejection proteins. A naturally occurring deletion of the 48 C-terminal residues (a.a. 677-725) has no apparent deleterious effect on virus infectivity (Bazinet et al., 1988). Larger deletions, however, starting between residues 650 and 602, all showed ~10-fold decrease in the percentage of phage that were infectious as determined from the ratio of plaque forming units (pfu) to particles (Figure 7 B,C). Thus, truncated barrels appear defective in delivering DNA to the host. Collectively, the above observations suggest two, potentially related, functions for the portal barrel. First, as suggested by Olia et al., it may help direct ordered packaging of the genome. Although particles with truncated forms of the barrel still package DNA, this may lead to disordered DNA arrangements that interfere with efficient release. Alternatively, a complete or nearly complete barrel may be required to deliver the DNA and pilot proteins to the bacterium efficiently. We suggest that the barrel supports both functions.
Conclusions
Crystallography of the full length P22 portal and the barrel-truncated portal in complex with the closure protein gp4 combined with a sub nanometer, asymmetric cryoEM reconstruction of P22, dramatically changed the static description of the P22 virion and the dynamic requirements for its assembly. Density previously interpreted as the internal ejection proteins (gp7, gp16, gp20) is now seen to be the coiled-coil, portal barrel (residues 602-725). Remarkably such a dramatic and functionally important extension of the portal was not anticipated and was fully recognized only after the crystal structure of the isolated, full-length portal was determined. The adjustments required to fit the crystal structure of the barrel to the cryoEM density provide a satisfying mechanism for portal-sensing of head-full packaging pressure and transient portal restriction of DNA release during the addition of the closure proteins. Still not known is the role of the barrel during DNA packaging and release. Does it function as a nozzle that gyrates to guide the layering of the DNA into the shells that are strikingly visible in the cryoEM reconstruction (Figure 2B)? Does it maintain its extended structure during DNA release? These questions can be addressed through further cryoEM studies. Particles with portals containing truncated barrels display 10-fold reduction in infectivity, yet normal DNA content. Moderate resolution cryoEM studies of these particles will show if the ordering of the DNA is disturbed in these particles. Likewise, particles lacking the gp26 tail-needle (protein plug), spontaneously lose their DNA after packaging (Strauss and King, 1984) and cryoEM reconstructions of these particles will determine if the coiled-coil conformation is retained after DNA release. Finally, it is intriguing that a substantial change is likely to occur in the capsid-portal interaction following addition of the gp4 closure protein to the DNA-containing particle. The C-terminal region of gp4 is likely to bind to the portal followed by a change in interaction with the capsid, that traps the C-terminal residues in the final conformation. As discussed above, scaffolding protein may play a role in this final conformational adjustment (Chen et al., 2011). The strong association of 12 gp4 subunits with the portal via their C-terminal regions provides a robust platform for the addition of the other gene products in the tail machine. After more than 50 years, P22 continues to reluctantly reveal it’s structural strategies and reward those investigating them.
EXPERIMENTAL PROCEDURES
P22 virion isolation, purification, and cryoEM imaging were described previously (Lander et al., 2006). The original set of particle images used by Lander (Lander et al., 2006) were automatically boxed from the raw micrograph data. In this study, we visually screened each micrograph to identify and manually box out only those particles most likely to be intact virions. This provided a complete data set of 21,645 images that were used to compute the 3D reconstructions. The icosahedral reconstruction of P22 was computed using the AUTO3DEM program package (Yan et al., 2007) and a sub-set of these images (18,602 particles). This also included automatic reboxing of all particle images to assure that each particle was centered in the binned 5332 pixel dimension image. Unbinned versions (10232 pixels) of these particle image data were then used to carry out an asymmetric P22 reconstruction using a strategy similar to that employed to compute 3D reconstructions of bacteriophage φ29 virions and ghosts (Tang et al., 2008; Tang et al., 2010). The earlier, 17-Å resolution, P22 asymmetric reconstruction (Lander et al., 2006) was used as the starting model to expedite the data processing. Consistent with the icosahedral processing strategy, particle origins were set to the center of the capsid and initial particle orientations were estimated with the program FREALIGN (Grigorieff, 2007). Particle origins and orientations were then subjected to extensive refinement in AUTO3DEM. The estimated resolutions of the final icosahedral and asymmetric reconstructions, as determined by Fourier-Shell Correlation criteria (0.5 threshold (van Heel and Schatz 2005)), were 5.0 and 7.8 Å, respectively. The capsid region has the nominal resolution, but the flexible regions such as the bulk DNA and portal barrel domain have a lower resolution (~15 Å) due to the variations in different particles. It is clear that the barrel domain is flexible because its structure has escaped us until recently.
Crystal structure fitting and modeling were carried out with program O (Jones et al., 1991) and figures were prepared with the graphics program Chimera (Goddard et al., 2007). The published P22 capsid model (Chen et al., 2011) was modified to fit the density of the icosahedral reconstruction. The portal boundary estimation was guided by both the crystal structure and the position of the scabbard-like tube density previously identified and mis-interpreted as ejection proteins. The initial reconstruction of the scabbard-like tube density showed the constriction at the barrel base and the shrinking at the top of the barrel. Only after an additional local averaging step employing the 12-fold symmetry as found in the barrel crystal structure, did the barrel reconstruction reveal the helical rod separation, thus showing the twist of the helices from the crystal structure.
Supplementary Material
Acknowledgements
This work was supported by NIH Grants R01 GM054076 to JEJ; R37 GM-033050 and 1S10 RR-020016 to TSB; AI074825 to SC and R56 AI076509 to GC. Image data used in this study were recorded at the National Resource for Automated Molecular Microscopy at TSRI, which is supported by the National Institutes of Health (NIH) through the National Center for Research Resources’ P41 program (RR17573).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers The 3D density maps of the asymmetric and icosahedral reconstruction of P22 virion have been deposited in the EMDB at EBI with accession codes EMD–5231 (asymmetric) and EMD–5232 (icosahedral).
References
- Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys J. 1996;71:2040–2048. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet C, Benbasat J, King J, Carazo JM, Carrascosa JL. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- Botstein D, Waddell CH, King J. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 1973;80:669–695. doi: 10.1016/0022-2836(73)90204-0. [DOI] [PubMed] [Google Scholar]
- Casjens S, King J. P22 morphogenesis. I: Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- Casjens S, Wyckoff E, Hayden M, Sampson L, Eppler K, Randall S, Moreno ET, Serwer P. Bacteriophage P22 portal protein is part of the gauge that regulates packing density of intravirion DNA. J Mol Biol. 1992;224:1055–1074. doi: 10.1016/0022-2836(92)90469-z. [DOI] [PubMed] [Google Scholar]
- Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14:1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chen DH, Baker ML, Hryc CF, Dimaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc Natl Acad Sci U S A. 2011;108:1355–1360. doi: 10.1073/pnas.1015739108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. FREALIGN: high-resolution refinement of single particle structures. J Struct Biol. 2007;157:117–125. doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallog. sect. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- King J, Botstein D, Casjens S, Earnshaw W, Harrison S, Lenk E. Structure and assembly of the capsid of bacteriophage P22. Philos Trans R Soc Lond B Biol Sci. 1976;276:37–49. doi: 10.1098/rstb.1976.0096. [DOI] [PubMed] [Google Scholar]
- King J, Lenk EV, Botstein D. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J. Mol. Biol. 1973;80:697–731. doi: 10.1016/0022-2836(73)90205-2. [DOI] [PubMed] [Google Scholar]
- Lander GC, Khayat R, Li R, Prevelige PE, Potter CS, Carragher B, Johnson JE. The P22 tail machine at subnanometer resolution reveals the architecture of an infection conduit. Structure. 2009;17:789–799. doi: 10.1016/j.str.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Moore SD, Prevelige PE., Jr. Bacteriophage p22 portal vertex formation in vivo. J Mol Biol. 2002;315:975–994. doi: 10.1006/jmbi.2001.5275. [DOI] [PubMed] [Google Scholar]
- Olia AS, Bhardwaj A, Joss L, Casjens S, Cingolani G. Role of gene 10 protein in the hierarchical assembly of the bacteriophage P22 portal vertex structure. Biochemistry. 2007a;46:8776–8784. doi: 10.1021/bi700186e. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structure of phage P22 cell envelope-penetrating needle. Nat Struct Mol Biol. 2007b;14:1221–1226. doi: 10.1038/nsmb1317. [DOI] [PubMed] [Google Scholar]
- Olia AS, Casjens S, Cingolani G. Structural plasticity of the phage P22 tail needle gp26 probed with xenon gas. Protein Sci. 2009;18:537–548. doi: 10.1002/pro.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, et al. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci U S A. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss H, King J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J Mol Biol. 1984;172:523–543. doi: 10.1016/s0022-2836(84)80021-2. [DOI] [PubMed] [Google Scholar]
- Tang J, Olson N, Jardine PJ, Grimes S, Anderson DL, Baker TS. DNA poised for release in bacteriophage phi29. Structure. 2008;16:935–943. doi: 10.1016/j.str.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Sinkovits RS, Baker TS. Three-dimensional asymmetric reconstruction of tailed bacteriophage. Methods Enzymol. 2010;482:209–234. doi: 10.1016/S0076-6879(10)82008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuman-Commike PA, Greene B, Jakana J, Prasad BV, King J, Prevelige PE, Jr., Chiu W. Three-dimensional structure of scaffolding-containing phage p22 procapsids by electron cryo-microscopy. J Mol Biol. 1996;260:85–98. doi: 10.1006/jmbi.1996.0383. [DOI] [PubMed] [Google Scholar]
- van Heel M, Schatz M. Fourier shell correlation threshold criteria. J. Struct. Biol. 2005;151:250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- Yan X, Dryden KA, Tang J, Baker TS. Ab initio random model method facilitates 3D reconstruction of icosahedral particles. J Struct Biol. 2007;157:211–225. doi: 10.1016/j.jsb.2006.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.