SUMMARY
Hepatitis E virus (HEV) infection is an important cause of acute viral hepatitis in several developing countries, but has recently been shown to cause chronic hepatitis in immunosuppressed persons. Other hepatotropic viruses that cause chronic infection have been shown to infect peripheral blood mononuclear cells (PBMCs) and to persist in those cells. We therefore decided to look for evidence of replication of HEV in PBMCs obtained from patients with acute hepatitis E, using strand-specific assays for positive and negative HEV RNA. Of the 44 patients with acute hepatitis E during an outbreak in India, including 27 with detectable IgM anti-HEV and 19 with detectable serum HEV RNA, 11 had detectable HEV RNA in their PBMCs. However, of the six PBMC specimens with strong HEV RNA signal, none had detectable negative-strand-HEV RNA, a marker of viral replication. These findings indicate the presence of HEV RNA but the absence of its replication in PBMCs from patients with acute hepatitis E.
Keywords: Hepatitis E, peripheral blood mononuclear cells, replication, disease pathogenesis
INTRODUCTION
Hepatitis E virus (HEV) is a major cause of epidemic and sporadic acute hepatitis in several developing countries [1]. The virus is transmitted by the fecal-oral route, often through contaminated water. The infection is usually self-limited, but fulminant hepatic failure and death occur in about 0.1% of cases. The illness is particularly severe among pregnant women, with mortality rates of 15% to 25% [1]. Based on genomic sequence analysis, HEV isolates have been classified into four genotypes [2]; genotypes 1 and 2 are found in HEV endemic areas and affect humans, whereas genotypes 3 and 4 have been found in non-endemic and endemic areas and also infect several animal species.
It has been the belief that HEV infection is cleared rapidly and that chronic infection with HEV does not occur [1]. We have shown that HEV RNA disappears from the serum of patients with acute hepatitis E within 4 weeks of the onset of acute illness [3]. However, recently, Kamar et al. found evidence of chronic infection with genotype 3 HEV in French transplant recipients receiving immunosuppressive drugs [4]. Chronic HEV infection has since also been reported in persons with HIV infection. These chronic HEV infections could arise from newly acquired infection. However, the frequency of transmission of HEV in the areas where chronic HEV infection has been reported is quite low. Another possibility is that previously dormant HEV in an as yet unidentified compartment, such as peripheral blood mononuclear cells (PBMCs), might start replicating during immunosuppression. PBMCs have been shown to serve as a reservoir for other hepatotropic viruses that cause chronic infection, such as hepatitis B virus (HBV) and hepatitis C virus (HCV) [5, 6]. No data are available on the presence and replication of HEV in PBMCs. We therefore decided to investigate this.
MATERIAL AND METHODS
Specimens
Blood specimens were collected from 44 patients (age = 13–57 [mean ± SD 26 ± 10] years; 32 male) with acute hepatitis during an outbreak of hepatitis E in Hyderabad, Andhra Pradesh, India. The duration of illness varied from 2 to 10 (7.2 ± 2.3) days. The patients included five pregnant women. PBMCs were isolated from EDTA blood (10 ml) by density gradient centrifugation using Histopaque-1077 (Sigma, St. Louis, USA) as per standard protocol, washed four times with phosphate buffer saline (pH 7.4) and stored in Trizol (Invitrogen, Carlsbad, CA, USA) at −80°C. In addition, serum was stored at −80°C.
All study subjects provided an informed consent. An institutional ethics committee approved the study protocol.
IgM anti-HEV testing
Sera were tested for IgM anti-HEV antibodies using a commercial enzyme immunoassay (Genelabs, Singapore), which detects antibodies against recombinant proteins corresponding to open reading frames (ORF) 2 and ORF 3 of HEV. A cut-off value was calculated as recommended by the manufacturer.
Detection of HEV RNA in serum
RNA was extracted from serum (100 µl) using the QIAamp viral RNA kit (Qiagen, Valencia, CA, USA) in 50 µl of water, as described previously [7]. In brief, a reverse transcription was done using superscript II reverse transcriptase (SS II RT; Invitrogen, Carlsbad, CA, USA) and a reverse primer (PR3, antisense: 5’-TAA TAA CGG CCA TAT TCC AGA CAG TAT TCC-3’), followed by addition of the forward primer (PF2, sense: 5’-TGT TTG AGA ATG ACT TTT CTG AGT TTG ACT-3’) and 35 cycles of PCR amplification [7]. A 1:1000 dilution of 10% suspension (in 0.01M PBS, pH 6.8) of feces from a patient with acute epidemic hepatitis E and RNase free water were used as positive and negative controls, respectively. The PCR products were subjected to 1% agarose gel electrophoresis and Southern hybridization using a digoxigenin-labelled oligonucleotide probe (5’-CTA GAG TGT GCT ATT ATG GAG GAG TGT GGG ATG CCG CAG TGG C-3’) to detect a 237-bp band [7].
Detection of HEV RNA in PBMCs
RNA was extracted from PBMCs [~ 3 million] from each patient using Trizol reagent (Invitrogen). The extracted RNA was initially tested for HEV RNA, as described above. For specimens testing positive, another aliquot was tested using strand-specific RT-PCR separately for positive and negative strand HEV RNA, as described below.
Development of strand-specific assays for positive and negative strand HEV RNA
A recombinant plasmid generated by insertion of a 744 bp (nt 5131 to 4388) ORF1 fragment of human HEV genotype I into pGBKT7 vector was digested with EcoRI (New England Biolabs, Beverly, MA, USA) to produce DNA templates for run-off transcription. Transcription reactions (20 µl) were set up to generate RNA corresponding to HEV negative strand using a MEGAscript kit (Ambion, Austin, TX, USA). Transcribed RNA was digested with 5 U/µg DNA of DNase I (Ambion) for 60 min at 37°C and followed by two rounds of purification using Trizol. After verifying the absence of residual plasmid DNA by PCR without reverse transcription, this RNA was used as a control in assays for detection of negative strand HEV RNA.
Initially, we tried a modification of the RT-PCR assay described above to detect negative-strand HEV RNA, by replacing the primer PR3 with PF2 during reverse transcription. However, since this assay using SS II RT showed only limited discrimination between the amplification of positive and negative HEV RNA strands, we resorted to a RT-PCR assay that used the rTth polymerase. This enzyme being thermostable, carries out reverse transcription at 70°C and minimizes false priming. Using synthetic RNAs, this method has previously been shown to provide about 10,000-fold discrimination in sensitivity for detection of opposing strands of HCV RNA [8].
In brief, for rTth-based assays, cDNA synthesis was carried out in 20 µl reactions containing 5.4 pmol of the appropriate primer (PR3 for positive strand RNA, and PF2 for negative strand RNA) from the ORF1 region of the HEV genome, 1X RT buffer (Applied Biosystems, Foster City, CA, USA), 1 mM MnCl2, 200 µM of each deoxynucleotide triphosphate, and 5 U of rTth DNA polymerase (Applied Biosystems). After 20 min at 70°C, Mn+2 was chelated out with 8 µl of the supplied 10X chelating buffer (Applied Biosystems). The opposite primer (5.4 pmol) was then added, followed by adjustment of the reaction volume to 100 µl and of the MgCl2 concentration to 2.2 mM. Amplification conditions were similar to those for the non-strand-specific PCR. The products of RT-PCR (10 µl) were subjected to a semi-nested second round PCR using a forward primer (5'- CTT TTC TTT GGG TCT AGA GTG TGC- 3'), and PR3 for 40 cycles. PCR products (192 bp) were visualized using 1% agarose gel electrophoresis and Southern hybridization with a digoxigenin-labelled HP1 probe, as described above [7]. For standardization, the strand-specific rTth assays were performed on 10-fold serial dilutions (1:10 to 1:109) of HEV RNA extracted from a stool specimen containing positive-strand RNA and on in vitro generated negative-strand RNA.
PBMC specimens that showed strong amplification signals by non-strand-specific RT-PCR were tested using the strand-specific rTth RT-PCR assay for positive and negative strand HEV RNA in separate tubes, simultaneously. The assays were done on undiluted RNA as well as on serial 10-fold dilutions of each RNA specimen.
RESULTS
Of the 44 sera, 27 (61%) tested positive for IgM anti-HEV and 19 (43%) had detectable HEV RNA. Overall, 35 (80%) of the 44 sera were positive for one or both of these markers.
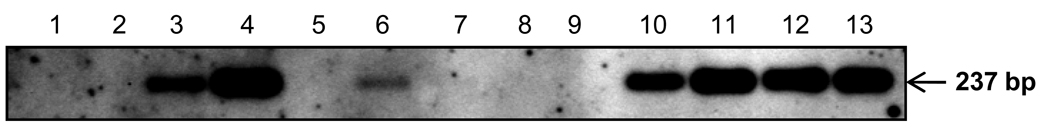
Of the 44 PMBC specimens, 11 (25%) tested positive for HEV RNA in the SS II RT based assay (Fig. 1). Of these, 10 were positive for serum IgM anti-HEV and/or HEV RNA, but one was negative for both these markers in the serum.
Fig. 1.
HEV-RNA positive-strand detection in PBMCs by RT-PCR using SS II RT. Lane 1: negative control; lanes 2–12: PBMC lane 13: positive control.
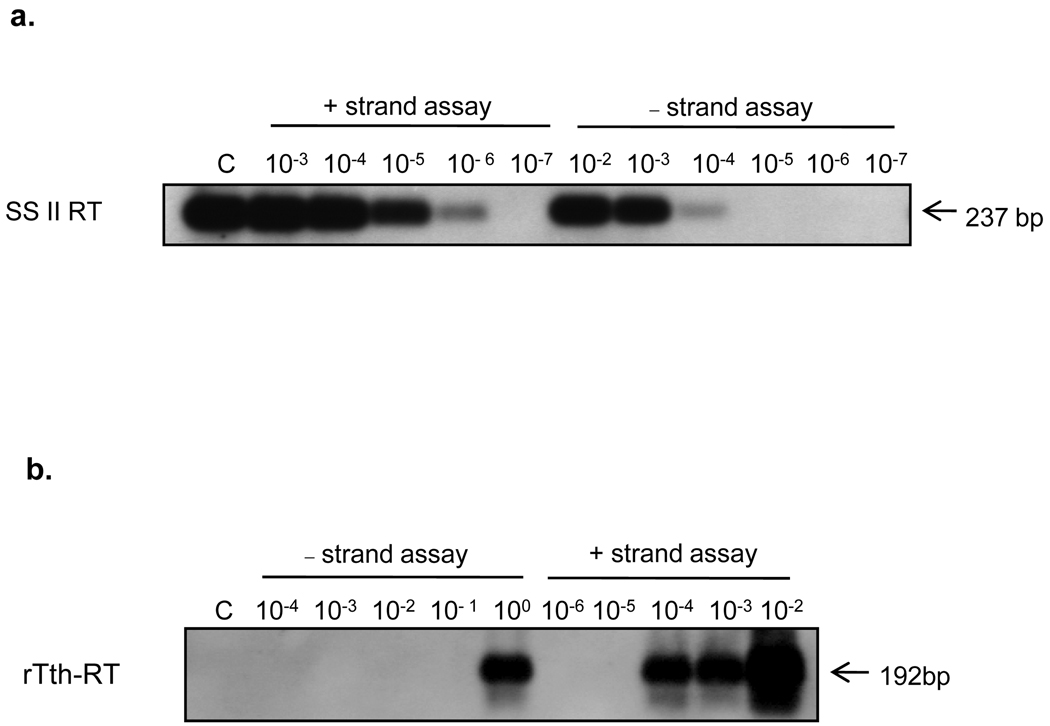
Using HEV RNA derived from serial dilutions of a stool suspension as template, a signal could be detected up to dilutions of 1:106 and 1:104 using the SS II RT assays for positive-strand and negative-strand HEV RNA, respectively. This 100-fold strand-specific discrimination was considered inadequate. In addition, in SS II RT based assays, an amplification signal was seen even when no primer was added during the reverse transcription step (Fig. 2a).
Fig. 2.
Sensitivity and specificity of RT-PCR using SS II RT and rTth (a). Strand-specific RT-PCR assays on 10-fold serial dilutions of positive-strand HEV RNA using SS II RT. Lane C, control-reaction where primer for cDNA synthesis is omitted (b). Strand-specific RT-PCR assays on 10-fold serial dilutions of positive-strand HEV RNA using rTth. Lane C, control-reaction where primer for cDNA synthesis is omitted
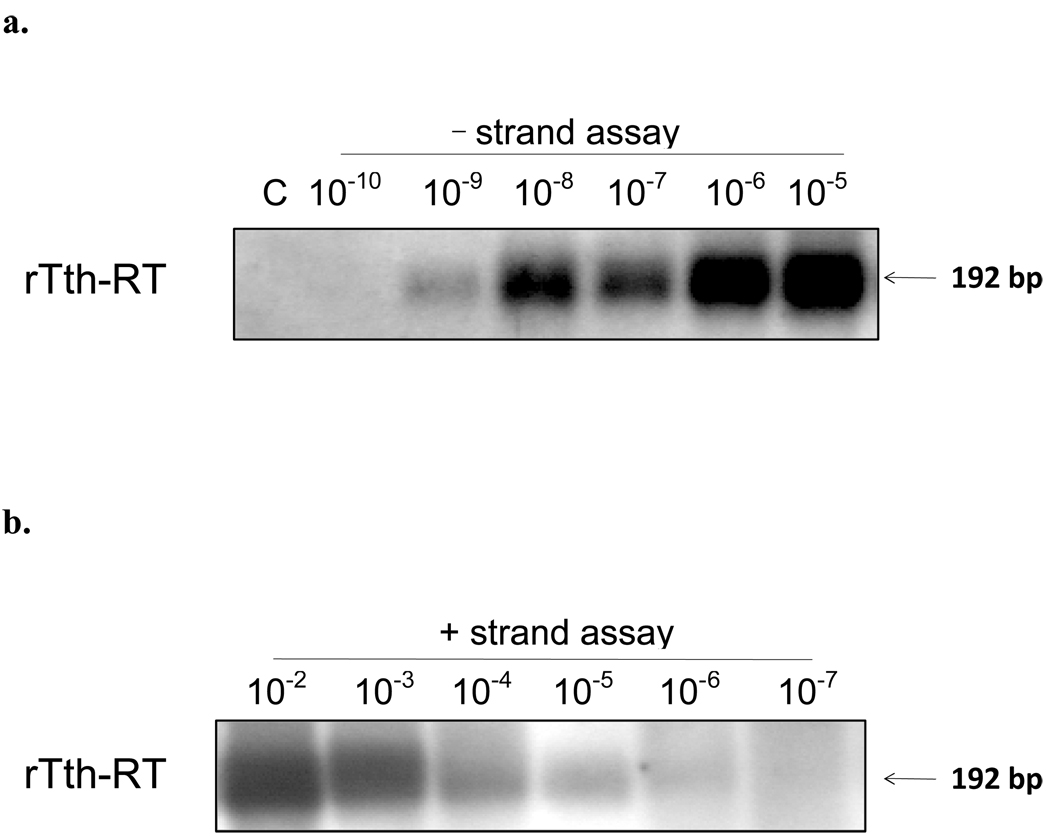
In comparison, the rTth-based assays showed a better strand specificity. Using the stool suspension as HEV RNA template, a positive signal was detected up to a dilution of 1:104 in the rTth-based assay for positive-strand HEV RNA and up to 100 using the assay for negative-strand HEV RNA (Fig. 2b). Similarly, using the in vitro generated negative-strand HEV RNA as a template, a signal could be detected up to a dilution of 1:109 using the assay specific for negative strand HEV RNA (Fig. 3a), and up to 1:105 using the positive-strand detection assay (Fig. 3b). Thus, the rTth assays for the positive and the negative strand HEV RNA showed a strand-specific discrimination of 10,000-fold. In addition, the rTth-based assay did not show amplification in the control reaction in which the primer had been omitted during the reverse transcription step.
Fig. 3.
Sensitivity and specificity of RT-PCR using rTth (a) Negative-strand-specific assay on 10-fold serial dilutions of negative-strand HEV RNA using rTth. Lane C, control-reaction where primer for cDNA synthesis is omitted (b). Positive-strand-specific assay on 10-fold serial dilutions of negative-strand HEV RNA using rTth.
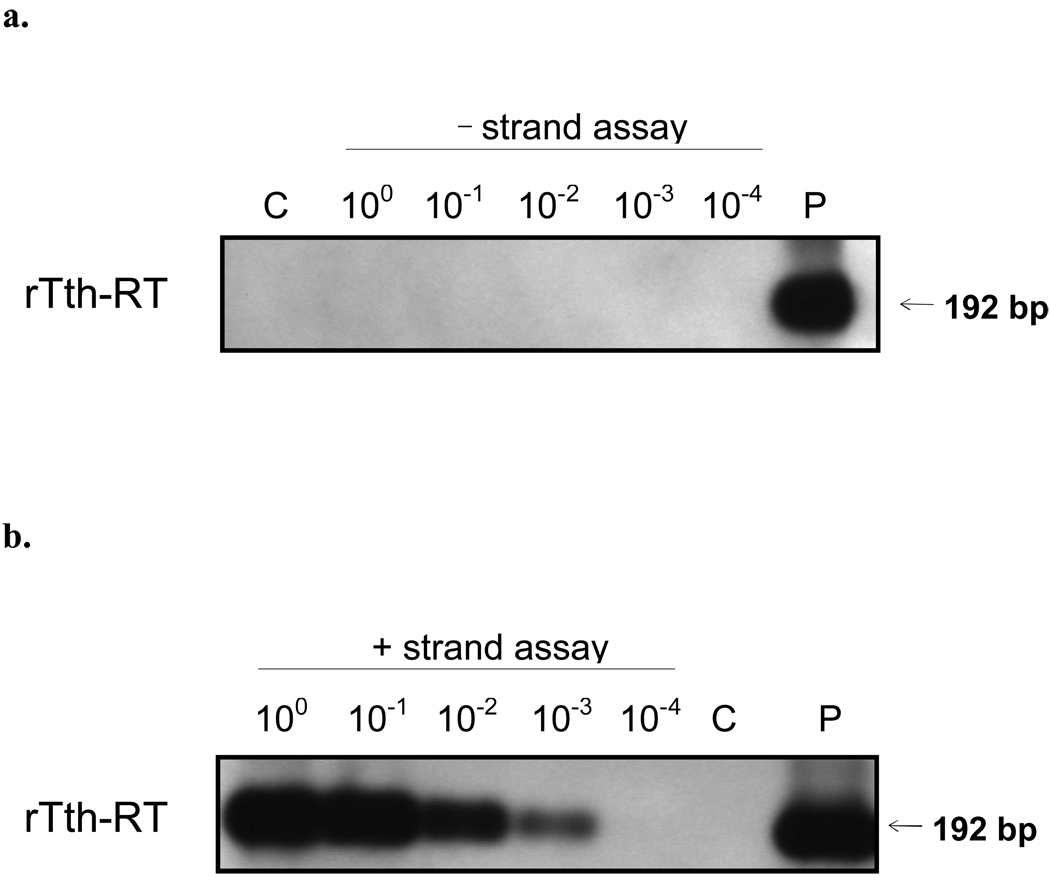
RNA extracted from PBMCs from 6 patients were tested for HEV RNA using strand-specific rTth RT-PCR assays for positive and negative strand HEV RNA. Of these, one tested positive in rTth PCR assay for positive-strand HEV RNA up to a dilution of 103 (Fig. 4a) but was negative in the rTth PCR assay for negative-strand HEV RNA (Fig. 4b). The other five specimens tested positive in assay for positive-strand HEV RNA up to a dilution of 102, but were negative in the assay for negative-strand HEV RNA, even when tested undiluted.
Fig. 4.
HEV-RNA detection in PBMC by strand-specific rTth RT- PCR. (a) Negative-strand-specific assay on 10 fold serial dilutions of total RNA. Lane C, control-reaction where primer for cDNA synthesis is omitted; lane P, positive control. (b) Positive-strand-specific assay on 10 fold serial dilutions of total RNA. Lane C, control-reaction where primer for cDNA synthesis is omitted; lane P, positive control.
DISCUSSION
In the present study, HEV RNA was detected in repeatedly-washed PBMCs from nearly one-fourth of patients with acute hepatitis E. However, these PBMC specimens tested negative in a strand-specific assay for negative-strand HEV RNA, indicating that viral replication in PBMCs was unlikely.
Other hepatotropic viruses that cause chronic infection, namely HBV and HCV, have been shown to replicate in PBMCs, in addition to liver, the main site of infection and viral replication. Laskus et al. reported integration of HBV DNA in PBMCs obtained from patients with chronic hepatitis B [5]. Similarly, presence of HCV RNA has been shown in PBMCs from patients with chronic HCV infection [9–12]. This has prompted several studies to look for evidence of HCV replication, i.e. presence of negative-strand HCV RNA, in these cells. Using sensitive and highly strand-specific assays, several workers have shown the presence of negative-strand HCV RNA in PBMCs [13–19] though, admittedly, some studies have failed to detect such RNA [8, 20]. Thus, though the issue remains somewhat controversial, it appears that replication of HBV or HCV in extrahepatic sites, in particular PBMCs, may play a role in the pathogenesis of these infections and in reactivation of these infections in persons receiving immunosuppressive drugs, as also for infection of naïve allograft organs following liver transplantation [21, 22].
Replication of HEV in host cells is poorly understood due to the lack of an efficient cell-culture system. However, since HEV has a positive-stranded RNA genome that codes for a putative RNA-dependent RNA polymerase, its replication is likely to involve a negative-stranded RNA intermediate [23]. Tam et al. showed the presence of both positive and negative strand HEV RNA in hepatocytes infected in vitro with HEV [24]. In a swine HEV model, negative-strand HEV RNA was detected in the small intestine, lymph node, colon and liver [25]. In a recent study, Billam et al. identified replicative viral RNA in gastrointestinal tissues, including the colorectal, cecal, jejunal, ileal, duodenal, and cecal tonsil tissues, in addition to the liver in chickens experimentally infected with avian HEV [26]. Negative-strand HEV RNA has also been shown in HepG2 cells transfected in vitro with an infectious full-length HEV cDNA clone [27]. Thus, the presence of negative strand RNA appears to indicate ongoing viral replication in a tissue.
Following acute HEV infection, viral RNA can be detected in serum and stool for only a limited period [3]. Chronic HEV infection has not been shown to occur in regions where hepatitis E is endemic. In recent years, however, chronic HEV has been detected in developed countries among persons receiving immunosuppressive drugs [4]. Further, HEV RNA has been detected in a small proportion of healthy blood donor sera. These findings raise the possibility of ongoing low-level viral replication in a compartment such as PBMCs, which serves as a source for infection of the liver once immunosuppression sets in. Thus, whether HEV persists and multiplies in PBMCs appears to be an important issue.
To address this question, we developed a strand-specific rTth assay, with 10,000-fold discrimination between detection of HEV RNA of the desired strand and the alternative strand. In our initial assay, we could detect the presence of HEV RNA in repeatedly-washed PBMCs from a proportion of subjects with acute hepatitis E, including one who tested negative for HEV RNA in the serum. However, using the strand-specific assays, the HEV RNA detected in PBMCs was shown to be exclusively positive-stranded, indicating absence of HEV replication in PBMCs. This finding suggests that HEV RNA does not replicate in PBMCs in patients with uncomplicated acute hepatitis. However, our findings may not exclude persistence of HEV RNA in PBMCs and its replication in the presence of immune suppression.
Our data may be limited by the relatively lower sensitivity of the strand-specific rTth RT-PCR assay. Our rTth assay for positive-strand HEV RNA could detect positive-strand RNA in a stool suspension at a dilution of 1:104 compared to detection at a dilution of 1:106 by a SS II RT based assay; however, the latter assay was not strand-specific and hence could not be used to look for negative-strand HEV RNA. Development of more sensitive assays specific for negative-strand HEV RNA will be useful for confirming our findings.
In conclusion, our data indicate that PBMCs from patients with acute hepatitis E do not contain negative-strand HEV RNA. Hence, these cells are unlikely to be a site for replication of HEV in immunocompetent persons with HEV infection. Further studies are needed to determine whether PBMCs can serve as a reservoir site for reactivation of HEV infection in immunosuppressed persons.
ACKNOWLEDGEMENTS
This study was supported by an intramural research grant from the Sanjay Gandhi Postgraduate Institute of Medical Sciences, Lucknow to RA, and in part by a grant from the National Institutes of Health, USA (1R011076192) to SJ. SKI was supported by a Senior Research Fellowship from Indian Council of Medical Research, New Delhi, India.
Contributor Information
Sirish Kumar Ippagunta, Email: ippaguntask@gmail.com.
Sita Naik, Email: sitanaik@gmail.com.
Shahid Jameel, Email: jameelshahid@gmail.com.
Rakesh Aggarwal, Email: aggarwal.ra@gmail.com.
REFERENCES
- 1.Aggarwal R, Naik S. Epidemiology of hepatitis E: current status. J Gastroenterol Hepatol. 2009;24:1484–1493. doi: 10.1111/j.1440-1746.2009.05933.x. [DOI] [PubMed] [Google Scholar]
- 2.Schlauder GG, Mushahwar IK. Genetic heterogeneity of hepatitis E virus. J Med Virol. 2001;65:282–292. doi: 10.1002/jmv.2031. [DOI] [PubMed] [Google Scholar]
- 3.Aggarwal R, Kini D, Sofat S, Naik SR, Krawczynski K. Duration of viraemia and faecal viral excretion in acute hepatitis E. Lancet. 2000;356:1081–1082. doi: 10.1016/S0140-6736(00)02737-9. [DOI] [PubMed] [Google Scholar]
- 4.Kamar N, Selves J, Mansuy JM, et al. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358:811–817. doi: 10.1056/NEJMoa0706992. [DOI] [PubMed] [Google Scholar]
- 5.Laskus T, Radkowski M, Wang LF, Nowicki M, Rakela J. Detection and sequence analysis of hepatitis B virus integation in peripheral blood mononuclear cells. J Virol. 1999;73:1235–1238. doi: 10.1128/jvi.73.2.1235-1238.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muller HM, Pfaff E, Goeser T, Kallinowski B, Solbach C, Theilmann L. Peripheral blood leukocytes serve as a possible extrahepatic site for hepatitis C virus replication. J Gen Virol. 1993;74:669–676. doi: 10.1099/0022-1317-74-4-669. [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal R, McCaustland KA. Hepatitis E virus RNA detection in serum and stool specimens with the use of microspin columns. J Virol Methods. 1998;74:209–213. doi: 10.1016/s0166-0934(98)00049-4. [DOI] [PubMed] [Google Scholar]
- 8.Lanford RE, Chavez D, Chisari FV, Sureau C. Lack of detection of negative-strand hepatitis C virus RNA in peripheral blood mononuclear cells and other extrahepatic tissues by the highly strand-specific rTth reverse transcriptase PCR. J Virol. 1995;69:8079–8083. doi: 10.1128/jvi.69.12.8079-8083.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kao J, Chen P, Lai M, Wang T, Chen D. Positive and negative strand of hepatitis C virus RNA sequences in peripheral blood mononuclear cells in patients with chronic hepatitis C: no correlation with viral genotypes 1b, 2a, and 2b. J Med Virol. 1997;52:270–274. [PubMed] [Google Scholar]
- 10.Taliani G, Badolato M, Lecce R, et al. Hepatitis C virus RNA in peripheral blood mononuclear cells: relation with response to interferon treatment. J Med Virol. 1995;47:16–22. doi: 10.1002/jmv.1890470105. [DOI] [PubMed] [Google Scholar]
- 11.Boisvert J, He X, Cheung R, Keeffe E, Wright T, Greenberg H. Quantitative analysis of hepatitis C virus in peripheral blood and liver: replication detected only in liver. J Infect Dis. 2001;184:827–835. doi: 10.1086/323391. [DOI] [PubMed] [Google Scholar]
- 12.Lerat H, Berby F, Trabaud MA, et al. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–851. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackard J, Smeaton L, Hiasa Y, et al. Detection of hepatitis C virus (HCV) in serum and peripheral blood mononuclear cells of HCV-monoinfected and HIV/HCV-coinfected persons. J Infect Dis. 2005;192:258–265. doi: 10.1086/430949. [DOI] [PubMed] [Google Scholar]
- 14.Castillo I, Rodríguez-Iñigo E, Bartolomé J, et al. Hepatitis C virus replicates in peripheral blood mononuclear cells of patients with occult hepatitis C virus infection. Gut. 2005;54:682–685. doi: 10.1136/gut.2004.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mazur W, Mazurek U, Jurzak M, Wilczok T, Bulanowski Z, Gonciarz Z. Positive and negative strands of HCV-RNA in sera and peripheral blood mononuclear cells of chronically hemodialyzed patients. Med Sci Monit. 2001;7:108–115. [PubMed] [Google Scholar]
- 16.Pham T, MacParland S, Mulrooney P, Cooksley H, Naoumov N, Michalak T. Hepatitis C virus persistence after spontaneous or treatment-induced resolution of hepatitis C. J Virol. 2004;78:5867–5874. doi: 10.1128/JVI.78.11.5867-5874.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Radkowski M, Gallegos-Orozco J, Jablonska J, et al. Persistence of hepatitis C virus in patients successfully treated for chronic hepatitis C. Hepatology. 2005;41:106–114. doi: 10.1002/hep.20518. [DOI] [PubMed] [Google Scholar]
- 18.Laskus T, Radkowski M, Wang L, Vargas H, Rakela J. The presence of active hepatitis C virus replication in lymphoid tissue in patients coinfected with human immunodeficiency virus type 1. J Infect Dis. 1998;178:1189–1192. doi: 10.1086/515682. [DOI] [PubMed] [Google Scholar]
- 19.Laskus T, Operskalski EA, Radkowski M, et al. Negative-strand hepatitis C virus (HCV) RNA in peripheral blood mononuclear cells from anti-HCV-positive/HIV-infected women. J Infect Dis. 2007;195:124–133. doi: 10.1086/509897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskus T, Radkowski M, Wang L, Cianciara J, Vargas H, Rakela J. Hepatitis C virus negative strand RNA is not detected in peripheral blood mononuclear cells and viral sequences are identical to those in serum: a case against extrahepatic replication. J Gen Virol. 1997;78:2747–2750. doi: 10.1099/0022-1317-78-11-2747. [DOI] [PubMed] [Google Scholar]
- 21.Feray C, Samuel D, Thiers V, et al. Reinfection of liver graft by hepatitis C virus after liver transplantation. J Clin Invest. 1992;89:1361–1365. doi: 10.1172/JCI115723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskus T, Radkowski M, Wilkinson J, et al. The origin of hepatitis C virus reinfecting transplanted livers: serum- derived versus peripheral blood mononuclear cell- derived virus. J Infect Dis. 2002;185:417–421. doi: 10.1086/338635. [DOI] [PubMed] [Google Scholar]
- 23.Tam AW, Smith MM, Guerra ME, et al. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam AW, White R, Reed E, et al. In vitro propagation of hepatitis E virus from in vivo infected primary macacque hepatocytes. Virology. 1996;215:1–9. doi: 10.1006/viro.1996.0001. [DOI] [PubMed] [Google Scholar]
- 25.Williams TPE, Kasorndorkbua C, Halbur PG, et al. Evidence of extrahepatic sites of replication of the hepatitis E virus in a swine model. J Clin Microbiol. 2001;39:3040–3046. doi: 10.1128/JCM.39.9.3040-3046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Billam P, Pierson FW, Li W, LeRoith T, Duncan RB, Meng XJ. Development and validation of a negative-strand-specific reverse transcription-PCR assay for detection of a chicken strain of hepatitis E virus: identification of nonliver replication sites. J Clin Microbiol. 2008;46:2630–2634. doi: 10.1128/JCM.00536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panda SK, Ansari IH, Durgapal H, Agrawal S, Jameel S. The in vitro synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol. 2000;74:2430–2437. doi: 10.1128/jvi.74.5.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]