Abstract
Coenzyme Q (ubiquinone or Q) is a lipid electron and proton carrier in the electron transport chain. In yeast Saccharomyces cerevisiae eleven genes, designated COQ1 through COQ9, YAH1 and ARH1, have been identified as being required for Q biosynthesis. One of these genes, COQ8 (ABC1) encodes an atypical protein kinase, containing six (I, II, III, VIB, VII, and VIII) of the twelve motifs characteristically present in canonical protein kinases. Here we characterize seven distinct Q-less coq8 yeast mutants, and show that unlike the coq8 null mutant, each maintained normal steady state levels of the Coq8 polypeptide. The phosphorylation states of Coq polypeptides were determined with two-dimensional gel analyses. Coq3p, Coq5p, and Coq7p were phosphorylated in a Coq8p dependent manner. Expression of a human homolog of Coq8p, ADCK3(CABC1) bearing an amino-terminal yeast mitochondrial leader sequence, rescued growth of yeast coq8 mutants on medium containing a nonfermentable carbon source and partially restored biosynthesis of Q6. The phosphorylation state of several of the yeast Coq polypeptides was also rescued, indicating a profound conservation of yeast Coq8p and human ADCK3 protein kinase function in Q biosynthesis.
Keywords: coenzyme Q, ubiquinone, Saccharomyces cerevisiae, mitochondria, lipid metabolism, protein kinase
1. Introduction
Coenzyme Q (ubiquinone or Q)3 is an essential lipid component and plays a well-known role in respiratory electron and proton transport in energy metabolism. Q, the oxidized quinone accepts electrons from NADH via complex I or succinate via complex II, and the reduced hydroquinone (QH2) is oxidized by complex III where the electrons are transferred to cytochrome c. Q also functions as the electron acceptor in a number of metabolic oxidation reactions including amino acid catabolism and fatty acid beta oxidation via acyl-CoA dehydrogenases [1], glycerol-3-phosphate, dihydroorotate [2], and the oxidation of sulfide [3]. QH2 is an important lipid-soluble chain-terminating antioxidant [4], and may also function as a co-antioxidant to maintain levels of vitamin E in membranes of cells and lipoproteins [5]. Q and Q analogs have been shown to be capable of inhibiting, activating, or occupying sites of the mitochondrial permeability transition pore [6-8]. The mitochondrial permeability transition pore is involved in apoptosis through the opening of a non-selective channel that disrupts membrane potential and causes hypotonic swelling of the inner membrane, a process that appears to be conserved from yeast to plants and animals [9, 10].
Cells utilize isoprenoid biosynthetic pathways (mevalonate or methylerythritol) to produce the polyisoprenyl tail of Qn, where n designates the number of isoprene units; Q10, Q9, Q8 and Q6 in human, Caenorhabditis elegans, Escherichia coli, and Saccharomyces cerevisiae, respectively [11]. Coq1p in S. cerevisiae, IspB in E. coli, or a complex of PDSS1 and PDSS2 in human cells synthesize the polyisoprene diphosphate tail precursor of Qn. The aromatic ring precursors supplying the quinone ring of Q include 4-hydroxybenzoic acid (4-HB) derived from either chorismic acid or tyrosine [12]. Recently, para-aminobenzoic acid (pABA) was discovered to function as a ring precursor of Q in S. cerevisiae [13, 14]. Coq2p is required for the prenylation of either 4-HB or pABA to form 3-hexaprenyl-4-hydroxy benzoic acid or 3-hexaprenyl-4-amino-benzoic acid [13]. S. cerevisiae requires at least nine additional polypeptides (Coq3-Coq9, Yah1, and Arh1) for biosynthesis of Q6 [11, 14, 15].
Although much progress has been made in determining the enzymatic functions of the Coq proteins, the functions of Coq4p and Coq9p in Q biosynthesis are not known, and new information on the putative kinase function of Coq8p is described in this work. In S. cerevisiae, Coq4p and Coq9p are members of a high molecular mass multi-subunit mitochondrial complex together with the O-methyltransferase Coq3p, the C-methyltransferase Coq5p, and the hydroxylases Coq6p and Coq7p, required for ring modifications forming Q [16-18]. A zinc-binding motif identified in Coq4p was postulated to be important for Coq4p function as a scaffolding protein [17]. Recently, Rea et al., [19] derived a model of yeast Coq4p by taking advantage of the structure determined by the Northeast Structural Genomics consortium of a Coq4p homolog from Nostoc sp. PCC7120 (Alr8543) that crystallized with a bound geranylgeranyl monophosphate and a magnesium ion. The predicted Coq4p structure is consistent with the idea that Coq4p may bind the polyisoprene tail of a Q-intermediate, possibly serving as an important anchor for the Q-multisubunit biosynthetic complex.
The yeast COQ8 gene was originally identified as ABC1 (activator of the bc1 complex) in a multicopy suppression screen of a specific cbs2 mutant allele (cbs2-223) and proposed to function as a cytochrome b chaperone [20], necessary for bc1 complex function [21]. However, the decrease in bc1 complex in coq8 mutants can be explained fully by the requirement of COQ8 for Q biosynthesis [22]. Yeast coq8 mutants lack Q6 and the growth defect in media containing a nonfermentable carbon source can be rescued by the addition of exogenous Q6 to the growth medium. The suppression of the cbs2 mutation was shown to be due to a neighboring tRNA and not to COQ8 [23]. E. coli UbiB and Provendencia stuartii AarF are prokaryotic Coq8 homologs required for Q biosynthesis; mutants accumulate octaprenyl-phenol, the Q-intermediate expected from a block at the first hydroxylase step [24, 25]. Patients with mutations in ADCK3/CABC1 (a homolog of yeast COQ8) exhibit progressive neurological disorders with childhood onset cerebellar ataxia and atrophy and have decreased Q10 content in muscle, fibroblast, and lymphoblast cells [26] [27]. Thus, in prokaryotes, yeast, and human cells, the principal and conserved function of the Coq8 polypeptide is its requirement for Q biosynthesis.
Yeast Coq8p, E. coli UbiB, and human ADCK3 are members of an atypical kinase family, first identified by Leonard et al. [28], as potential protein kinases harboring motifs I, II, VIB and VII. Subsequent analyses identified ADCK1-5 as novel human kinases [29], containing six (I, II, III, VIB, VII, and VIII) of the twelve motifs characteristically present in canonical protein kinases [27]. Patients with Q10 deficiencies harboring mutations in ADCK3 indicated it is required for Q10 biosynthesis in humans. In these studies, expression of human ADCK3 did not rescue yeast coq8 mutants. However, introduction of the human mutations into the corresponding yeast COQ8 gene impaired growth of yeast on nonfermentable carbon sources and resulted in decreased Q6 content [26, 27]. These findings suggested that yeast Coq8p and human ADCK3 may function as kinase required for Q biosynthesis. There is also evidence that Coq8p may function to regulate Q biosynthesis, as overexpression of yeast Coq8p has been shown to rescue a yeast coq9 nonsense mutant [18, 30], a yeast coq10 null mutant [31, 32], and to restore synthesis of DMQ6 in a coq7 null mutant [33]. Tauche et al. [34] utilized two dimensional IEF-SDS PAGE and tagged forms of yeast Coq polypeptides and determined that phosphorylation of yeast Coq3p is dependent on Coq8. They also demonstrated that the phosphorylation state of Coq3 affects its association with the Coq high molecular mass polypeptide complex. Thus, Coq8p appears to be essential for the formation of a multi-subunit Q-biosynthetic complex. It is intriguing that the first O-methylation step catalyzed by yeast Coq3 was identified as a potential regulated site of Q biosynthesis in response to high glucose, and it was speculated that a cAMP-dependent protein kinase might be involved in mediating this regulation [35].
In this study we have analyzed mutations in Coq8 that result in Q biosynthetic defects. Initially, we employed site directed mutagenesis to target the catalytic lysine in kinase motif II of Coq8p. Although yeast coq8 null mutants expressing the Coq8-K216A polypeptide lacked Q6, the Coq8-K216A polypeptide was not stable, and the phenotype of this strain mirrored the coq8 null mutant. Several Coq polypeptides are unstable in the coq8 null mutant (including Coq4p, Coq6p, Coq7p and Coq9p) [18]. To assess the role of Coq8p as a potential kinase, our goal was to analyze coq8 mutants that retained normal steady state levels of Coq8p. Therefore we examined the collection of yeast coq8 mutants in order to identify those that retained normal steady state levels of Coq8p. Seven distinct yeast coq8 amino acid substitution mutants have been characterized and a subset of these mutants used to investigate the phosphorylation state of Coq3 and other Coq polypeptides. We show that expression of human ADCK3 bearing an amino-terminal mitochondrial leader sequence in yeast coq8 mutants rescues both synthesis of Q6 and the phosphorylation state of several of the yeast Coq polypeptides, indicating a profound conservation of protein kinase function in Q biosynthesis.
2. Materials and Methods
2.1 Strains and growth media
The yeast strains used in this study are listed in Table 1. Growth media were prepared as described [36]. Media included YPD (1% yeast extract, 2% peptone, 2% dextrose), YPG (1% yeast extract, 2% peptone, 3% glycerol) and YPGal + 0.1% Dextrose (1% yeast extract, 2% peptone, 2% galactose, 0.1% dextrose). SDC consisted of 0.18% yeast nitrogen base without amino acids, 2% dextrose, 0.14% NaH2PO4, 0.5% (NH4)2SO4 and amino acids were added at final concentrations as described in [37]. SD–ade, SD–his, SD–leu, SD–met, SD–trp and SD–ura consisted of SDC media minus adenine, histidine, leucine, methionine, tryptophan and uracil respectively. Solid media contained 2% agar. All materials were obtained from Difco, Fisher or Sigma.
Table 1.
Genotypes and Sources of S. cerevisiae Strains
| Strain | Genotype | Source |
|---|---|---|
| W303-1A | MAT a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 | R. Rothsteina |
| W303ΔABC1 | MAT a ade2-1 his3-1,15 leu2-3,112 trp1-1 ura3-1 coq8::HIS3 |
[22] |
| FY251 | MAT a ura3-52 his3Δ200 leu2Δ1 trp1Δ63 | F. Winstonb |
| FYΔABC1 | MAT a ura3-52 hisΔA200 leu2Δ1 trp1Δ63 coq8::TRP1 | [22] |
| CH130-A1 | MAT a his3-1,15 leu2-3,112 trp1-1 ura3-1 coq8-1 | [22] |
| CF130-2B | MAT α his3Δ200 ura3-52 trp1Δ63 coq8-1 | C130c × FY251 |
| C177 | MAT α met6 coq8-2 | A. Tzagoloff c |
| W177-3B | MAT a his3-1,15 leu2-3,112 met6 ura3-1 coq8-2 | C177 × W303-1A |
| C183 | MAT α met6 coq8-3 | A. Tzagoloff c |
| W183-2A | MAT a his3-1,15 trp1-1 ura3-1 coq8-3 | C183 × W303-1A |
| C194 | MAT α met6 coq8-4 | A. Tzagoloff c |
| W194-3A | MAT α ade2-1 his3-1,15 met6 ura3-1 coq8-4 | C194 × W303-1A |
| C222 | MAT α met6 coq8-5 | A. Tzagoloff c |
| W222-7D | MAT a ade2-1 his3-1,15 leu2-3,112 ura3-1 coq8-5 | C222 × W303-1A |
| C240 | MAT α met6 coq8-6 | A. Tzagoloff c |
| W240-1A | MAT a leu2-3,112 ura3-1 coq8-6 | C240 × W303-1A |
| C275 | MAT α met6 coq8-9 | A. Tzagoloff c |
| C308 | MAT α met6 coq8-7 | A. Tzagoloff c |
| W308-4C | MAT α ade2-1 his3-1,15 met6 trp1-1 ura3-1 coq8-7 | C308 × W303-1A |
| C315 | MAT α met6 coq8-8 | A. Tzagoloff c |
| W315-4A | MAT α trp1-1 ura3-1 coq8-8 | C315 × W303-1A |
| JM6 | MAT a, his4 ρ0 | [78] |
| JM8 | MAT α ade1 ρ0 | [78] |
Dr. Rodney Rothstein, Department of Human Genetics, Columbia University.
Dr. Fred Winston, C. Dollard, and S. Ricapero-Hovasse, Department of Genetics, Harvard University.
Dr. Alex Tzagoloff, Department of Biological Sciences, Columbia University.
2.2 Sporulation and Tetrad Analysis
Diploid cells were grown to a cell concentration of OD600 = 2.5 in YPD. Yeast cells from 1 ml of culture were pelleted by centrifugation 12,000×g, 30 seconds. The cell pellet was washed with sterile water and pelleted by centrifugation again. Cells were then resuspended in 1 ml sporulation medium (1% potassium acetate, 0.1% yeast extract, 0.05% dextrose) and incubated at 30°C for 2 to 3 days. Cells were pelleted by centrifugation, and then resuspended in zymolyase buffer (1 M sorbitol, 50 mM Tris-Cl pH 7.5, 6 mg/ml zymolyase-20T (MP Biomedicals)). Cells were incubated for 15 or 30 min at 30°C. The zymolyase digestion was stopped by diluting the sample with four volumes of water and placing it on ice. The procedure for analysis and dissection of haploid spores was as described [36]. Yeast cells were transformed with plasmid DNA as described [38].
2.3 Subcloning of COQ8 and ADCK3
The plasmids used in this study are listed in Table 2. To construct COQ8 with a biotin accepting sequence at its C-terminus, a DNA segment was amplified with the primers pBC8-1 (5′ CGCGGAATTCACCAGCATTTTGGATAATTC 3′ −600 to −589 of COQ8) and pBC8-2 (5′ CGCGGAATTCAACTTTATAGGCAAAAATCT 3′ +1503 to +1484 of COQ8). The DNA fragment was digested with EcoRI (sites underlined) and then ligated into the EcoRI site of the yeast/E. coli shuttle vector containing the biotinylated Propionibacterium shermanii transcarboxylase sequence, Yep352-Bio6 [39] [37] [40]. The resulting plasmid, pBT8-1, contains COQ8 in frame with a carboxyl-terminal biotinylation site.
Table 2.
Plasmid constructs used in this study
| Plasmid | Genes | Copy number | Source |
|---|---|---|---|
| pRS316 | Yeast shuttle vector only | Low copy | [79] |
| pRS426 | Yeast shuttle vector only | Multi copy | [80] |
| pAH01 | Yeast vector with CYC1 promoter | Low copy | [81] |
| pCH1 | Yeast vector with CYC1 promoter | Multi copy | [81] |
| pQM | pAH01 with COQ3 mito leader | Low copy | [41] |
| pRCM | pCH1 with COQ3 mito leader | Multi copy | Saiki, R. and Morvaridi, S. |
| p3HN4 | Yeast ABC1/COQ8 | Low copy | [82] |
| pK216A | Yeast abc1/coq8-K216A | Low copy | This work |
| p4HN4 | Yeast ABC1/COQ8 | Multi copy | [23] |
| p4K216A | Yeast abc1/coq8-K216A | Multi copy | This work |
| pBT8-1 | Yeast ABC1/COQ8-biotin | Multi copy | This work |
| pBT8KA | Yeast abc1/coq8-K216A-biotin | Multi copy | This work |
| psCABC1 | pAH01/Human CABC1/ADCK3 with CYC1 promoter |
Low copy | This work |
| pmCABC1 | pCH1/Human CABC1/ADCK3 with CYC1 promoter |
Multi copy | This work |
| plcADCK3 | pQM/Human CABC1/ADCK3 with CYC1 promoter and COQ3 mito leader |
Low copy | This work |
| pmcADCK3 | pRCM/ Human CABC1/ADCK3 with CYC1 promoter and COQ3 mito leader |
Multi copy | This work |
The plasmid MHS1011 containing human ADCK3 cDNA was obtained from Open Biosystems. To generate a low copy plasmid containing the open reading frame of ADCK3 subcloned behind the S. cerevisiae CYC1 promoter, MHS1011 was digested with XhoI and EcoRI. The DNA fragment containing ADCK3 was purified with a Qiaquick Gel Extraction Kit (Qiagen) and the purified DNA treated with Klenow. A low copy plasmid containing the CYC1 promoter, pAH01 [41], was digested with HindIII, treated with Klenow and then treated with bacterial alkaline phosphatase. The purified DNA fragment containing ADCK3 was then ligated into the treated pAH01 plasmid. The plasmid with ADCK3 in the correct orientation was designated psCABC1. To generate a high copy plasmid containing ADCK3 subcloned behind the CYC1 promoter, psCABC1 was digested with KpnI and SpeI to excise a DNA fragment containing the CYC1 promoter and ADCK3. This DNA fragment was then ligated into the high copy plasmid pCH1 [41], also digested with KpnI and SpeI. This plasmid was designated pmCABC1.
Two other yeast expression plasmids were constructed so that the ORF of human ADCK3 would contain an amino-terminal mitochondrial-targeting leader sequence (from yeast COQ3), and expressed from the yeast CYC1 promoter (see Table 2). A forward primer, ADCK3For (5′-clamp-Cla1-(+1 to +31)-3′ of ADCK3) and a reverse complement primer, ADCK3Rev (5′-clamp-Kpn1-(+1944 to +1927)-3′ of ADCK3) were used with MHS1011 as template DNA to generate a PCR-amplifed segment of DNA containing the ORF of ADCK3. The resulting DNA was digested with ClaI and KpnI and inserted into the corresponding restriction sites of pQM [41] or pRCM (Table 2), to generate plcADCK3 (low copy) and pmcADCK3 (multi copy), respectively.
2.4 Site Directed Mutagenesis
Primer mediated site directed mutagenesis was performed as described [42]. Primer pairs used to generate coq8-K216A were KAbc1AF4 (5′ TCTTAAGAACTATACAGAACGGGAGAATTTC 3′ +55 to +89 bp) with KAbc1AR3 (5′ CAGGATATTGAATTGCGACAACCACTCTTT 3′ +661 to +632 bp) and KAbc1AF3 (5′ AAAGAGTGGTTGTCGCAATTCAATATCCTG 3′ +632 to 661 bp) with KAbc1AR4 (5′ CATTTCGTACGCCCCTTTCCTATCTCTCAATGTGG 3′ +1194 to 1160 bp). Nucleotides that are underlined are the mutated nucleotides. The PCR fragments were digested with AflII and BsiWI. The plasmids p3HN4 [22], p4HN4 [23] and pBT8-1 were treated with AflII and BsiWI to remove a fragment of COQ8. The AflII/BsiWI digested PCR fragment containing the K216A mutation was then ligated into the digested plasmids to generate pK216A, p4K216A and pBT8KA, respectively. pK216A and p4K216A are single copy and multicopy plasmids that contain the coq8 gene with the K216A mutation. The pBT8KA plasmid contains the K216A mutation of coq8 and a carboxyl-terminal biotinylation site.
2.5 Genomic DNA Purification and Sequencing
Yeast strains were grown overnight in 5 ml YPD at 30°C. Yeast cells from 0.5 ml of culture were pelleted by centrifugation at 12,000×g for 1 min. Genomic DNA was purified using the Wizard Genomic DNA Kit (Promega). For sequencing, the COQ8 locus was amplified in two overlapping segments. The first segment was amplified with the primers Abc1Seq-F (5′ ACTCGAGAAAAGCAATCTGGTAGATTTATGGG 3′ −300 to −276 (underlined) relative to the COQ8 ATG) and Abc1Seq-R (5′ CCTCGAGTTAAACTTTATAGGCAAAAATCTCTT 3′ +1506 to +1481 (underlined) relative to the COQ8 ATG). The second segment was amplified with the primers KAbc1A-F1 (5′ CGTCTCTTAAGAACTATACAGAACG 3′ +55 to +74 relative to the COQ8 ATG) and Abc1Seq-R2 (5′ TCTGTTTTATCTTTTTTTTTTGTTCTTCGAGATT 3′ +1640 to +1607 from the COQ8 ATG). The UCLA Sequencing and Genotyping Core determined the nucleotide sequence of each of the PCR products.
2.6 Purification of Mitochondria
Yeast cells were grown in 1 liter of YPGal + 0.1% Dextrose and harvested at an OD600 of approximately 1. Crude mitochondria were isolated and purified on a Histodenz (Sigma) gradient as described in [43]. Protein concentration was measured by bicinchoninic acid protein assay (Pierce).
2.7 Submitochondrial localization
Mitochondria were hypotonically shocked for 20 min on ice, with 5 volumes of 20 mM HEPES-KOH pH 7.4, to generate mitoplasts. Mitoplasts were pelleted by centrifugation (14,000×g, 10 min) and the supernatant saved as the intermembrane space fraction (IMS). Mitoplasts were sonicated and the matrix (supernatant) and membrane (pellet) components separated by centrifugation (100,000×g, 30 min). Mitoplasts were also alkaline extracted with 0.1 M Na2CO3 pH 11.5 for 20 min at 4°C, and subjected to centrifugation to separate matrix (supernatant), peripheral membrane (supernatant) and integral membrane (pellet) proteins. Proteinase K protection assays were as described [44]. Mitoplasts were pelleted and resuspended in isotonic buffer (0.6 M sorbitol, 20 mM HEPES-KOH pH 7.4) with or without the addition of 100 μg/ml proteinase K (Fisher Scientific) and 1% Triton X-100, for 1 h at 4°C. Proteins were precipitated with 10% trichloroacetic acid and resuspended in SDS-PAGE sample buffer. SDS-polyacrylamide denaturing gels were prepared containing 10% acrylamide as described [45].
2.8 2D-Isoelectric focusing (IEF)/SDS-PAGE
IEF-PAGE was performed as described [34]. Approximately 200 μg of purified mitochondria from each strain were sedimented by centrifugation (14000×g, 10 min) at 4°C, and resuspended in standard Rehydration Solution (7 M urea, 2 M thiourea, 4 % CHAPS, 0.5 % Ampholytes 3-10, 0.0002% bromophenol blue) to a final concentration of 1.0 μg/μl. The sample was sedimented again (14000×g, 10 min) and de-streak reagent (GE Healthcare; final concentration of 1.25%) was added to the sample as a reducing agent. For in-gel hydration, 200 μl of diluted samples were applied to Immobiline DryStrips (GE Healthcare, 13cm, pH 3-11 NL) and incubated overnight at room temperature. IEF was performed in the Bio-Rad Protean Isoelectric Focusing Cell (Bio-Rad) with four steps of voltage, (4 h at 100 V, 2 h at 250 V, 5 h gradient up to 4000 V, and a further 80,000 Volt-h at constant 4000 V) at 20°C with a maximum current of 50 μA. The strips were equilibrated first for 20 min in Equilibration Buffer (75 mM Tris-HCl, pH 8.8, 6 M urea, 29.3% glycerol, 2% SDS, 0.002% bromophenol blue, 20 mg/ml dithiothreiotol), and then another 20 min in Equilibration Buffer with 25 mg/ml iodoacetamide. For the second dimension, the strips were applied to Criterion 8-16% PAGE gels in a Bio-Rad Criterion Dodeca Electrophoresis Cell. For phosphatase treatment, isolated mitochondria (200 μg protein) were resuspended in 19 μL λ-phosphatase buffer (New England Bio Labs; 50 mM HEPES, pH 7.4, 100 mM NaCl, 2 mM dithiothreitol, 0.01% Brij 35) containing complete EDTA-free Protease Inhibitor Cocktail (Roche). Samples were incubated for 45 min at 30°C after addition of 1 uL λ-phosphatase (100 U/ uL) (New England Bio Labs).
2.9 Immunoblot analyses
Proteins were transferred from SDS-polyacrylamide gels to polyvinylidene difuloride (PVDF) membrane, and then incubated in 2% non-fat milk and probed with primary antibodies (generated in rabbits) to yeast mitochondrial proteins at the following dilutions: Coq1p, 1:10,000; Coq2p, 1:1000; Coq3p, 1:1000; Coq4p, 1:1000; Coq5p, 1:10,000; Coq6p: 1:250; Coq7p: 1:1000; Coq8p: 1:100; Coq9p, 1:1000; Secondary antibody goat anti-rabbit IgG, heavy and light chain specific peroxidase conjugate (Calbiochem) were used at 1:10,000. A mouse monoclonal antibody for Rip1p was used at 1:10,000 and the secondary antibody was goat anti-mouse peroxidase conjugate. Supersignal West Pico Chemiluminescent kit (Pierce) was used for detection of polypeptides on Western blots.
2.10 Lipid Extraction and Analysis
Yeast cells were grown in YPGal + 0.1% dextrose. Q was extracted from yeast whole cells as described [13, 46]. Lipids were separated and analyzed essentially as described [13]. In brief, reverse-phase HPLC with tandem mass spectrometry was used to detect and quantify Q6 and Q4 was used as an internal standard. The mass spectrometer was an Applied Biosystems-MDS Sciex 4000 Q Trap (Hybrid triple-quad linear ion trap analyzer with Autosampler, and a Turbo-V source equipped with ESI and APCI sources).
3. Results
3.1 Coq8p is a peripheral membrane protein located on the matrix side of the inner membrane
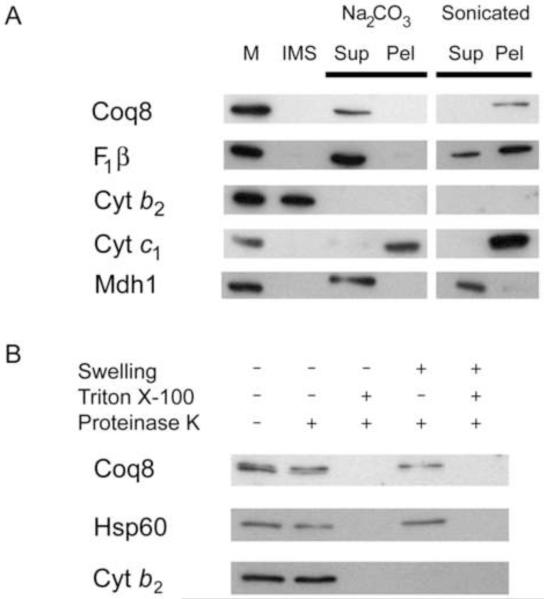
Coq8p is localized to the mitochondria [22] and a c-myc-tagged form of Coq8p is located within the mitochondrial matrix [34]. Recent analyses of yeast Coq8p have identified a membrane spanning region [47]. Although the Coq8-c-myc tagged polypeptide was shown to rescue growth on glycerol [34], Q content was not reported. Other tagged forms of Coq polypeptides have been observed to rescue growth on glycerol, but the Q content is not restored to wild-type [14], and in some cases the tagged form of the polypeptide was found to be mislocalized in the rescued yeast strains [48]. Thus it is important to localize the native form of the Coq8 polypeptide. We used antibodies recognizing Coq8p to determine the submitochondrial location of the untagged polypeptide. W303-1A yeast mitochondria were isolated and a hypotonic shock was performed to rupture the outer membrane, leaving the inner membranes intact, generating mitoplasts. Mitoplasts were sonicated to disrupt the remaining membranes, allowing separation of soluble matrix proteins and insoluble membrane associated proteins. Mitoplasts were also treated with alkaline buffer to extract proteins peripherally associated with the inner membrane [49]. Western blot analysis of these samples showed that Coq8p remained with the membrane components after sonication and was released into the supernatant after alkaline treatment, but not after sonication, indicating that Coq8p is a peripherally associated membrane protein (Fig. 1A). A proteinase K protection assay on purified mitochondria, or mitochondria subjected to hypotonic buffer to release intermembrane space components, identified Coq8p as located on the matrix side of the inner membrane (Fig. 1B). The results show that Coq8p is a peripherally associated membrane protein located on the matrix side of the inner membrane.
Fig. 1.
Coq8p is a peripheral membrane protein, associated with the mitochondrial inner membrane on the matrix side. A. Coq8p is peripherally associated with mitochondria membranes. Nycodenz purified mitochondria (M) were hypotonically swelled to generate the intermembrane space (IMS) fraction and mitoplasts. Mitoplasts were then treated with 0.1 M Na2CO3 pH 11.5, or sonicated. The treated mitoplasts were centrifuged and the supernatant (Sup) and pellet (Pel) fractions analyzed. B. Coq8p is located on the matrix side of the inner membrane. Mitochondria and mitoplasts were treated with Proteinase K with and without 1% Triton X-100. The β subunit of F1-ATPase (F1β) is a peripheral membrane protein [83], cytochrome b2 (Cyt b2) is located in the intermembrane space [84], cytochrome c1 (Cyt c1) is an integral membrane protein [85], malate dehydrogenase (Mdh1) [86], and Hsp60p [87], are localized to the matrix.
3.2 Mutation of the catalytic lysine-216 to alanine in Coq8p disrupts Q biosynthesis
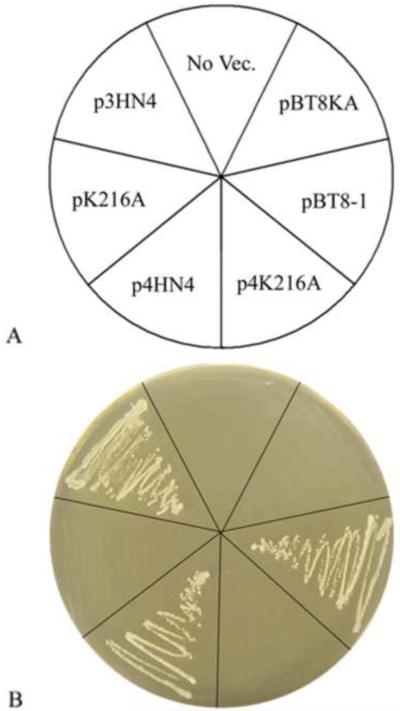
Amino acid sequence analyses of yeast Coq8p, human ADCK3, and homologues have identified protein kinase motifs [27-29]. The lysine present in motif II is believed to be involved in the transfer of protons during phosphorylation [50]. Previous studies have shown that mutation of this lysine to alanine in a variety of other protein kinases results in the loss of kinase function, but preserves structure [51]. To determine the role of this conserved lysine in Coq8p function, site directed mutagenesis was used to convert lysine-216 to alanine. The coq8Δ yeast strain, W303ΔABC1, was transformed with plasmids containing either wild-type or coq8-K216A and tested for growth on plate media containing glycerol as sole non-fermentable carbon source. No growth was detected after three to ten days of incubation of transformants expressing Coq8-K216A (Fig. 2). Identical results were obtained with another coq8Δ null mutant FYμABC1, in a distinct genetic background, (data not shown).
Fig. 2.
The Lysine216 in kinase subdomain II of Coq8 is required for rescue of glycerol growth in a yeast coq8 null mutant. The yeast coq8 null mutant (W303ΔABC1), was transformed with plasmids containing either wild-type COQ8 or coq8-K216A. p3HN4 (COQ8), and pK216A (coq8-K216A) are low-copy vectors; p4HN4 (COQ8), p4K216A (coq8-K216A), pBT8-1 (COQ8) and pBT8KA (coq8-K216A) are multi-copy vectors. Yeast transformants were incubated 3 days on YPG plate media at 30 °C.
3.3 Steady State Polypeptide Levels in coq8-K216A
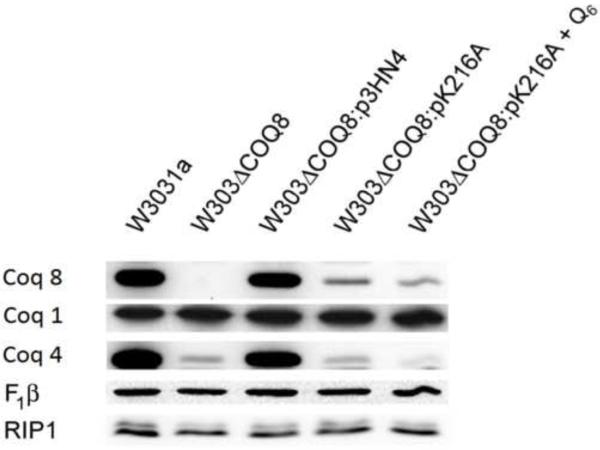
Previous work has shown that deletions of any of the COQ1-COQ9 genes results in a decrease in the steady state levels of several of the other Coq polypeptides [18]. For example, coq8 null mutants have decreased levels of Coq4, Coq6, Coq7, and Coq9 polypeptides [18], and the formation or integrity of the Q-biosynthetic complex is affected by the absence of Coq8p [34]. To determine the steady state level of the Coq8-K216A polypeptide, mitochondria from yeast expressing Coq8p-K216A were purified and steady state polypeptide levels were determined by Western blot. Polypeptide levels of Coq8p-K216A when expressed from a low copy plasmid are diminished relative to wild-type or the null mutant expressing wild-type Coq8 (W303ΔABC1:p3HN4) (Fig. 3), and steady state levels of Coq4p are also decreased. Thus, it is not possible to discern whether low Coq4p levels are due to the absence of kinase activity, or to the low level of the Coq8p-K216A.
Fig. 3.
The yeast Coq8-K216A polypeptide is not present at normal steady state levels. Yeast were grown in 1 liter of YPGal medium with 0.1% dextrose and harvested at an OD600nm of approximately 4. W303ΔABC1:pK216A + Q6 was grown in YPGal + 0.1% Dextrose media supplemented with 1.2 μM Q6. Purified mitochondria from W303-1A, W303ΔABC1 and W303ΔABC1 harboring the designated plasmids were subjected to Western blot analysis.
Previous studies have suggested that Q or a Q biosynthetic intermediate may be necessary for Coq polypeptide complex stability [52, 53]. For example, addition of Q6 to growth media has been shown to increase steady state levels of Coq3 and Coq4p polypeptides in coq1 and coq7 null mutants [52, 53], and addition of Q6 to yeast coq7 null mutant cultures restores assembly of a biosynthetic complex enabling synthesis of DMQ6, the penultimate intermediate in Q biosynthesis [33]. Addition of Q6 to cultures of coq8 null mutants has been shown to restore growth on media containing glycerol or ethanol as the sole nonfermentable carbon source [22]. However, addition of Q6 to cultures of W303ΔABC1:pK216A yeast did not rescue steady state Coq8 polypeptide levels. Similarly, addition of Q6 did not restore the Q-biosynthetic complex in coq8 null mutants [34]. Antibodies against the β subunit of the F1-ATPase and the Complex III Rieske iron-sulfur complex, Rip1, were used to verify equivalent sample loading, and indicate that disruption of the Coq-polypeptide complex has no effect on steady state levels of representative components in mitochondrial respiratory complexes. In summary, the very low steady state levels of the Coq8-K216A polypeptide produces the same limitations as the coq8 null mutant; thus, we sought to identify coq8 mutants that retained normal steady state levels of Coq8p.
3.4 Characterization of yeast coq8 point mutants with normal steady state level of Coq8p
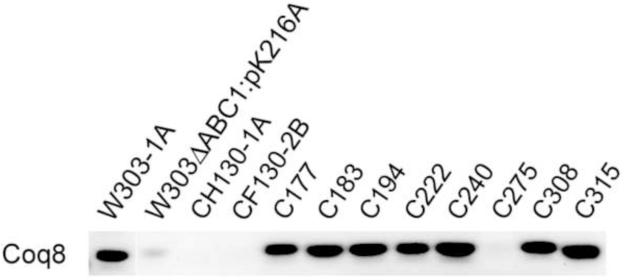
Eight different mutants in the G75/coq8 complementation group [54] whose coq8 gene defects had not previously been characterized were studied, with the goal of identifying mutants that retained steady state levels of Coq8p. These mutants were obtained via mutagenesis of the parental wild-type strain D273-10B/A1 [55, 56]. Each of the original coq8 mutants within this collection of strains (Table 1) was shown to be defective for growth on ethanol and/or glycerol as a carbon source, indicating a defect in respiration. The respiratory deficiency stems from recessive mutations in a nuclear gene, as they were complemented by a ρ0 strain with a normal complement of nuclear genes but lacking mitochondrial DNA (Table 1). Each strain was cultured and subjected to analysis by Western blot. Seven of the mutants expressed Coq8p polypeptide in amounts comparable to wild type, W303-1A (Fig. 4). The coq8 loci in each of these seven mutants were amplified by PCR and sequence analysis revealed that each mutant harbored a unique nucleotide substitution mutation resulting in amino acid substitution (Table 3). Four of the mutations affect residues within known kinase motifs (coq8-3, A197V in motif I; coq8-6, D229N in motif III; and coq8-8, D346N and coq8-4, N348Y, both in motif VIB; Fig. 5). The remaining three mutations result in glycine to aspartate substitution and are present in regions of unknown function (coq8-2, G89D; coq8-5, G130D; and coq8-7, G475D; Fig. 5).
Fig. 4.
Seven yeast coq8 mutant strains have normal steady state levels of the Coq8 polypeptide. The designated yeast strains were grown in YPD, lysed with 2% SDS and 425-600 μm glass beads (Sigma). Aliquots of whole cell lysates (corresponding to 0.1 OD600 of yeast cells) were separated by SDS-PAGE and then transferred to a PVDF membrane for western blot analyses with antibodies to Coq8p.
Table 3.
Amino Acid and Nucleotide Substitutions of coq8 Alleles
| Strain (abc1/coq8 allele) | Amino acid substitution (nucleic acid mutation)a |
|---|---|
| C130 (coq8-1) | Not determined |
| C177 (coq8-2) | G89D (G266A) |
| C183 (coq8-3) | A197V (C590T) |
| C194 (coq8-4) | N348Y (A1042T) |
| C222 (coq8-5) | G130D (G389A) |
| C240 (coq8-6) | D229N (G685A) |
| C275 (coq8-9) | Not determined |
| C308 (coq8-7) | G475D (G1424A) |
| C315 (coq8-8) | D346N (G1036A) |
Positions of mutations are relative to the ATG start codon of ABC1/COQ8.
Fig. 5.
Amino acid alignment of Abc1/Coq8p homologues. An alignment of Abc1/Coq8p and its homologues was created using DNASTAR Megalign. Clustal method was used with PAM250 residue weight table. Homologues are E. coli (GenBank™ accession number P27854), S. pombe (GenBank™ accession number CAA62818), A. thaliana (GenBank™ accession number AAC72875), C. elegans (GenBank™ accession number AAA62560), D. melanogaster (GenBank™ accession number NP_572836) and humans (ADCK1, ADCK2, ADCK3/CABC1, ADCK4 and ADCK5. GenBank™ accession numbers NP_065154, NP_443085, BAB91363, NP_079152 and NP_777582, respectively). S. cerevisiae coq8 mutant alleles and their mutations identified in this study (Table 3) are indicated on the alignment by arrows. Mutations detected in CABC1/ADCK3 in human patients [26, 27, 73] are indicated with triangles. Kinase subdomains I, II, III, VIB, VII and VIII, the G-x-G-x-x-G glycine rich region and the D-x-x-x-x-N catalytic loop are indicated. The invariant lysine of subdomain II is designated with an asterisk. A highly conserved motif in Coq8p with remarkable sequence identity to a region in the Bacillus subtilis protein RsbU is indicated by the hatched bar ( ). Numbers on the left indicate amino acid position relative to the start codon. Identical residues are shaded.
). Numbers on the left indicate amino acid position relative to the start codon. Identical residues are shaded.
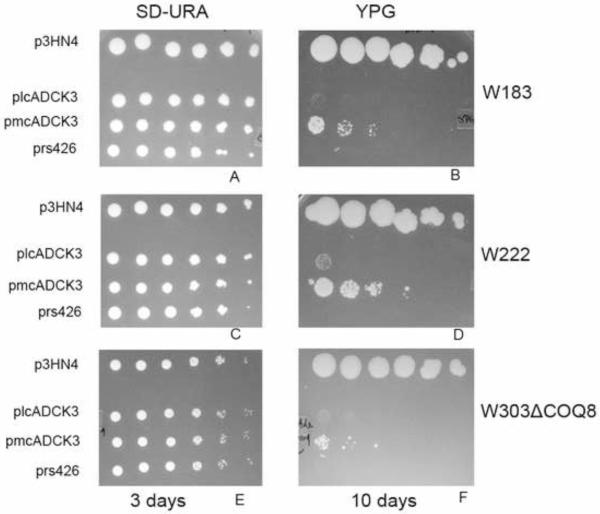
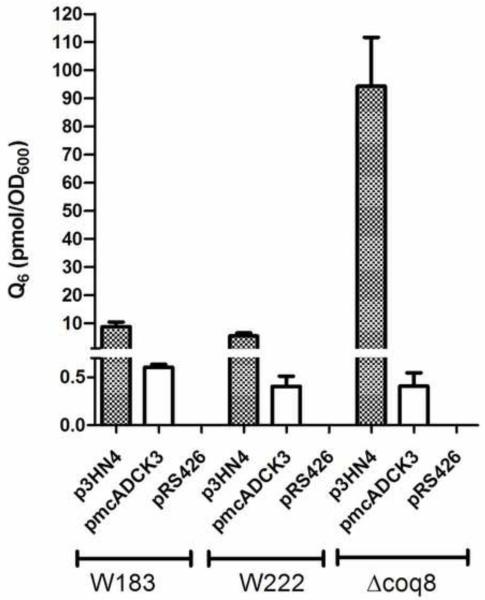
Derivatives of each of these seven coq8 point mutant yeast strains were prepared by mating to W303-1A, sporulation, and dissection of tetrads to yield respiratory defective mutants with designated auxotrophies (Table 1). Each of the coq8 mutant strains were rescued for growth on glycerol-medium following transformation with p3HN4 (low copy yeast COQ8), while transformation with pRS426 (multicopy yeast empty vector) failed to rescue growth. Two of the coq8 point mutants were chosen for further characterization: the W222 point mutant (coq8-5; G130D) was chosen because it mirrors a human ADCK3 mutation (G272D) present in a conserved region of unknown function and that is associated with disease and a lower level of Q10 content in skeletal muscle [26], and the W182 coq8 point mutant (coq8-3; A197V) was selected because it is located within conserved kinase motif I, and we anticipated that it would disrupt kinase activity of Coq8p. The rescue of W183 and W222, harboring the respective coq8-3 or coq8-5 allele, is shown (Fig. 6). To determine the effect of the mutations on Q6 content, lipids were extracted from wild-type and mutant yeast, and Q6 levels were measured with a QT4000 mass spectrometer coupled in-line with reversed phase HPLC. Q6 was not detected in W303ΔCOQ8, W183, or W222 harboring empty vector, pRS426. In contrast, Q6 levels were restored in W303ΔCOQ8, W183, and W222 mutant yeast harboring the p3HN4 plasmid (low copy yeast ABC1/COQ8) (Fig. 7). The rescue is less efficient in the point mutants (W183 and W222) than in the coq8 null mutant, perhaps due to interference by the mutant Coq8 polypeptide.
Fig. 6.
Expression of the human ADCK3 polypeptide rescues the growth of yeast coq8 mutants on non-fermentable carbon source media. Yeast coq8 point mutants W183 and W222, and coq8 null mutant (W303ΔABC1) were transformed with the designated plasmids: p3HN4, yeast low-copy COQ8; plcADCK3, low-copy ADCK3; pmcADCK3, multi-copy ADCK3; or pRS426, empty vector. Each strain was cultured overnight in SD–Ura selective media, the optical density (A600nm) was adjusted to 1.0, and 2 μl of 1:5 serial dilutions were spotted onto plate media, corresponding to a final A600nm of 1.0, 0.2, 0.04, 0.008, 0.0016, or 0.00032. A, C, & E, Growth is depicted on SD–Ura plates after three days of incubation at 30°C; B, D, & F, YPG plates after 10 days of incubation at 30°C.
Fig. 7.
Expression of the human ADCK3 polypeptide partially restores Q6 content in yeast coq8 mutants. Yeast cells were cultured in YPGal + 0.1% dextrose and collected at an optical density (A600nm) of about 1.0. The content of Q6 in yeast cell lipid extracts was determined by HPLC/MS-MS as described in Experimental Procedures. Yeast coq8 mutants were transformed with plasmids expressing yeast ABC1/COQ8 (low-copy, p3HN4), human CABC1/ADCK3 (multi-copy, pmcADCK3), or empty vector control (multi-copy, pRS426). Each bar represents a total of four measurements from two independent samples each with two injections. Standard deviations are represented with error bars. Q4 was used as internal standard.
3.5 Expression of the human ADCK3 polypeptide rescues yeast coq8 mutants
A human homolog ADCK3 shows 38% amino acid sequence identity with S. cerevisiae Coq8p. However, the human ADCK3 gene has not been shown to rescue the yeast coq8 mutant. To determine if ADCK3 might be an ortholog of yeast COQ8, its corresponding cDNA was subcloned into yeast expression vectors. Prior studies showed that rescue of yeast coq mutants by heterologous COQ genes required expression from multicopy vectors [46, 57]. In other cases, rescue by an E. coli homolog of Coq5p was observed for certain coq5 point mutants but not coq5 null mutants [58]. Neither low copy nor high copy plasmids expressing ADCK3 from the yeast CYC1 promoter (Table 2) were found to rescue yeast coq8 mutants [59]. It seemed possible that the human ADCK3 polypeptide may not be efficiently transported to the yeast mitochondrial matrix. To remedy this potential problem, we then tested plasmids expressing ADCK3 ORF with an amino-terminal yeast mitochondrial leader sequence (plcADCK3 and pmcADCK3). The resulting plasmids were transformed into a coq8 null mutant (W303ΔABC1) as well as yeast strains containing the coq8-3 (W183) and coq8-5 (W222) mutant alleles. To test rescue, yeast coq8 mutants harboring ADCK3 were plated on medium containing the non-fermentable carbon source, glycerol. Each of the coq8 mutants tested were rescued by pmcADCK3, and rescue was also observed after 10 days with plcADCK3 (Fig. 6). The pmcADCK3 plasmid partially restored Q6 content in yeast lipid extracts from transformed yeast (Fig. 7). Yeast abc1/coq8 mutants transformed with pmcADCK3 contained a small amount of Q6, while the same mutants transformed with the empty vector (pRS426) contained no detectable Q6.
The long incubation time needed to observe the rescue of the W222 coq8 mutant by low copy ADCK3, and the coq8 null mutant by multicopy ADCK3 probably reflects the time necessary to accumulate a sufficient amount of Q6. Yeast growth is observed in the spot containing only 20,000 cells (2 μl of yeast cells at A600nm = 1.0 or 1×107 cells/ml). Hence, it seems unlikely that spontaneous secondary mutations could account for the rescue on YPG, since they would not be expected to arise at such a high frequency. While there does not seem to be a strong correlation between the rescued growth in Fig. 6 and the Q6 content shown in Fig. 7, it seems likely that the content of Q6 as measured in YPGal liquid medium (Fig. 7) may not be indicative of the Q6 content in cells tested for growth on YPGlycerol. In fact, yeast coq7 mutants rescued with a heterologous gene and cultured on YPGal have been shown to have lower Q6 content as compared to the same cells cultured on YPG [53]. In summary, human ADCK3 is an ortholog of yeast Coq8p, since its expression rescues both growth on nonfermentable carbon source and Q6 biosynthesis in yeast coq8 mutants.
3.6 Coq8p is required for steady state levels of other Coq polypeptides
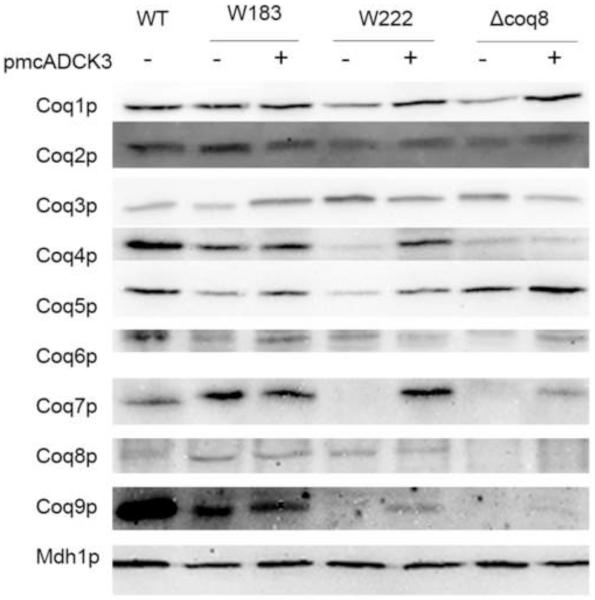
Previous studies have investigated the effects of deletions in the COQ genes on the steady state protein levels of Coq1p-Coq10p [18]. To examine whether coq8 point mutants destabilize other Coq polypeptides, mitochondria isolated from W183, W222, W303-1A (WT), and W303ΔABC1 were analyzed on immunoblots with antisera against the Coq1 – Coq9 proteins (Fig. 8). Mitochondria from W183 and W222 contained steady state levels of Coq8p, which was absent in W303ΔABC1. Steady state levels of Coq1p, Coq2p, Coq3p, Coq5p, and Coq6p were observed in each of the coq8 mutant strains. As reported by Tauche et al., [34], we observed that the presence of phosphatase inhibitors during mitochondria isolation preserved the steady state levels of Coq3p. Interestingly, W183 contained steady state levels of Coq4p and Coq7p, which were either dramatically decreased or absent in W222 and W303ΔCOQ8. All three coq8 mutant strains had dramatically decreased levels of Coq9p compared to wild type. Expression of human ADCK3 was able to restore steady state level of Coq4p in W222, and steady state level of Coq7p in both W222 and W303ΔCOQ8 strains. However, Coq9p level in the ADCK3 transformed yeast remained significantly decreased compared to wild-type yeast.
Fig. 8.
Expression of the human ADCK3 polypeptide in yeast coq8 mutants partially restores steady state levels of some of the yeast Coq polypeptides. Mitochondria were isolated from wild-type yeast (W303-1A) or from the indicated coq8 mutant yeast strains with or without pmcADCK3 (a multi-copy plasmid with ADCK3). Isolated mitochondria (20 μg protein) from the designated yeast strains were separated by 10% SDS-PAGE and then transferred to a PVDF membrane for immunoblotting with antibodies to one of the Coq polypeptides (Coq1-Coq9), or to malate dehydrogenase (Mdh1), as a loading control.
3.7 Coq3p, Coq5p, and Coq7p are putative substrates of Coq8p
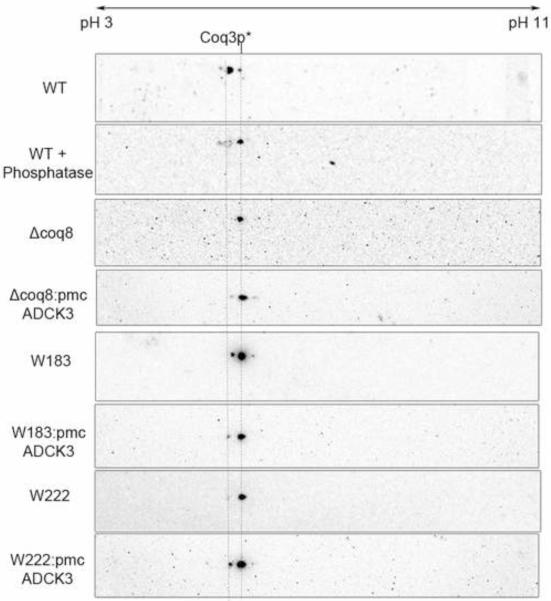
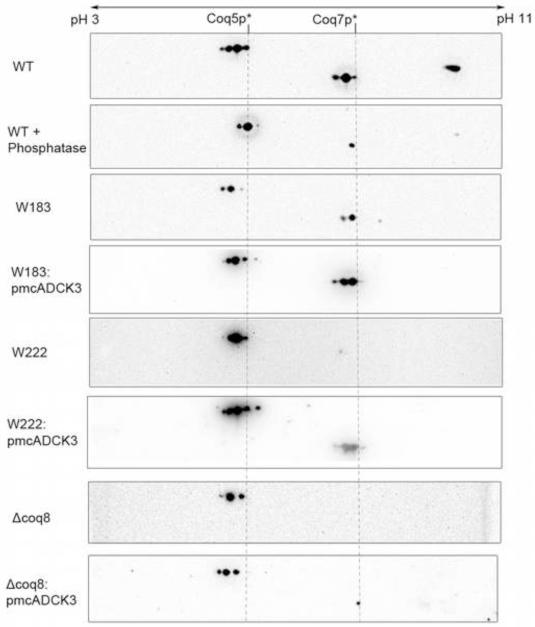
Phosphorylation of Coq3p has been shown to depend on the presence of Coq8p [34]. 2D-IEF/SDS-PAGE was performed to address whether other Coq polypeptides might also serve as substrates for Coq8p. Purified mitochondria from wild-type W303-1A and from coq8 mutants (W183, W222, and W303ΔCOQ8) were subjected to 2D-IEF/SDS-PAGE as described in Materials and Methods. Whereas previous studies made use of a tagged version of Coq3p [34], we utilized antibodies to Coq3p to detect the polypeptide in its natural state (Fig. 9). The detection of Coq3p revealed two spots in wild type in the pH range of 5~6, which is consistent with the predicted isoelectric point of 5.44 for Coq3p (Table 1S). The leftward, more acidic spot was no longer present in the phosphatase-treated wild-type mitochondria, identifying it as a phosphorylated isoform of Coq3p. These studies confirm the observation that W303ΔABC1 contained only the unphosphorylated isoform of Coq3p [34], and allowed us to use Coq3p as an internal marker in these analyses. Interestingly, W183 does contain a small fraction of phosphorylated Coq3p, suggesting this point mutation did not completely eliminate the kinase activity of Coq8p. On the other hand, W222 contained only the unphosphorylated isoform of Coq3p, with migration patterns similar to the coq8 null mutant. Expression of human ADCK3 was able to restore the phosphorylation state of Coq3p in both coq8 null mutant and W222 point mutant, although to different degrees.
Fig. 9.
Expression of the human ADCK3 polypeptide in yeast coq8 mutants partially restores the phosphorylation state of the yeast Coq3 polypeptide. Isolated mitochondria (200 μg protein) from wild-type yeast (W303-1A), or from the designated coq8 mutants, W183, W222, or W303ΔCOQ8, either without or with pmcADCK3 (as indicated), were separated by two-dimensional electrophoresis, with isoelectric focusing (IEF) in the first dimension and SDS-PAGE in the second dimension, followed by transfer to PVDF membranes for Western analysis. Where indicated, wild-type mitochondria were treated with phosphatase prior to rehydration and IEF. Coq3p was detected with anti-Coq3p antibody (1:1000 dilution). The two dashed lines indicate the positions of Coq3p isoforms. The pH range for IEF is indicated.
Detection of Coq5p and Coq7p with antibodies to these polypeptides revealed multiple spots for each (Fig. 10). For Coq5p, wild-type mitochondria contained at least four spots in the pH range of 5 to 6, consistent with the predicted isoelectric point of Coq5p (Table 1S). The two most acidic spots disappeared in wild-type mitochondria upon phosphatase treatment, indicating those two spots represent two different phosphorylation states of Coq5p. Interestingly, W183, W222, and W303ΔABC1 contained three spots, lacking the most acidic spot observed in wild type, indicating that Coq8p is responsible for the most acidic phosphorylated isoforms of Coq5p. This result suggests there may be another kinase working together with Coq8p on Coq5p, which is responsible for some Coq5p phosphorylation. The phosphorylation pattern of Coq5p is restored to wild type when ADCK3 was expressed in the point mutants W222 and W183. For Coq7p, three spots were observed in wild-type mitochondria in the pH range of 7 to 8, consistent with the predicted isoelectric point of Coq7p (Table 1S). The left two spots disappeared upon phosphatase treatment, indicating those two spots are phosphorylated isoforms of Coq7p. Coq7p is not detected in the coq8 null mutant (Fig. 10) since it is destabilized in the W303ΔABC1 strain (Fig. 8). In contrast, the W183 point mutant contained two spots, lacking the most acidic spot as compared to the wild type. However, it is difficult to discern whether a distinct kinase acts on Coq7p, or whether the A197V point mutation in Coq8p of W183 fails to completely eliminate the kinase activity. As shown in Fig. 8, W222 mitochondria do not contain stable levels of Coq7p. Expression of human ADCK3 is able to restore the phosphorylation pattern of Coq7p in both coq8 point mutants, but only the steady-state level of Coq7p is restored in coq8 null mutant (Figs. 8 and 10).
Fig. 10.
Expression of the human ADCK3 polypeptide in yeast coq8 mutants partially restores the phosphorylation state of the yeast Coq5 and Coq7 polypeptides. Isolated mitochondria (200 μg protein) from wild-type yeast (W303-1A), or from the designated coq8 mutants, W183, W222, or W303ΔCOQ8, either without or with pmcADCK3 (as indicated), were analyzed as described in Fig 7. Where indicated, wild-type mitochondria were treated with phosphatase prior to rehydration and IEF. Coq5p and Coq7 were detected with the designated antibodies (1:1000 dilution). The dashed lines designate the alignment and relative positions of the Coq5p and Coq7p isoforms The pH range for IEF is indicated.
4. Discussion
S. cerevisiae Coq8 polypeptide, and its human homologue ADCK3, have been shown to be required for biosynthesis of Q [22, 26, 27]. Yeast Coq8p is known to be required for the phosphorylation of the yeast Coq3 polypeptide, and this phosphorylation is important for the stability or formation of a multisubunit Coq polypeptide Q-biosynthetic complex [34]. In this work we have identified and utilized coq8 mutants that retain normal steady state levels of the Coq8 polypeptide and determined the effects of Coq8 point mutations on Q6 content, Coq polypeptide steady state levels, and the phosphorylated states of Coq3p, Coq5p, and Coq7p. The results suggest a profound functional conservation of kinase activity of human ADCK3, which when expressed in yeast coq8 mutant strains, rescues both Q6 synthesis and phosphorylation of yeast Coq3, Coq5 and Coq7 polypeptide substrates.
Initially, we introduced a K216A substitution within kinase motif II of Coq8p, because the corresponding mutation of this residue in other kinases resulted in correctly folded but kinase-inactive enzymes [51]. However, levels of the K216A-Coq8 polypeptide were profoundly decreased as compared to wild-type Coq8p. Consequently, steady state levels of the other Coq polypeptides were also decreased (for example Coq4p), as observed previously in coq8 null mutants [18]. For this reason we screened a collection of yeast coq8 mutants [54], with the goal of identifying coq8 mutant strains that retained normal steady state levels of the Coq8 polypeptide.
Seven distinct coq8 missense mutations were characterized in yeast mutants that produced a wild-type steady state level of Coq8p. Four of the seven yeast coq8 mutations affect residues that reside within known kinase subdomains (Fig. 5). A197V is located in subdomain I, which surrounds the bound ATP [51]. This subdomain in the majority of eukaryotic protein kinases contains the amino acid sequence G-x-G-x-x-G, present in many nucleotide-binding proteins. This motif helps form a β-strand-turn-β-strand structure that holds the α and β phosphate groups of ATP [51]. In Coq8p, this motif contains two alanine residues in place of the first two glycine residues (A-A-A-S-I-G). While the A197V (underlined) might cause steric hindrance and interfere with binding of phosphate moieties, our results indicate that the yeast W183 (coq8-3) mutant harboring A197V-Coq8p retained some kinase activity. Another mutation affected the aspartate in subdomain III (D229N), believed to help stabilize interactions between the invariant lysine of subdomain II and the phosphate groups of ATP [60, 61]. Disruption of the negative charge at that position would directly affect its ability to form a salt-bridge with the lysine. Two other mutations, D346N and N348Y, affected residues with kinase subdomain VIB, identified by the D-x-x-x-x-N motif that forms a catalytic loop. The invariant D residue of this loop is believed to be the catalytic base that accepts the proton from the amino acid substrate hydroxyl [51]. Thus D346N-Coq8p would be expected to slow or prevent phosphorylation. Distinct classes of eukaryotic protein kinases uniformly retain the invariant terminal D and N residues of motif VIB, but show variations of the “x” residues that mediate peptide substrate recognition [62-64]. For example, many serine and threonine kinases typically contain the sequence D-L-K-P-E-N, while tyrosine kinases contain D-L-R-A-A-N or D-L-A-A-R-N [65]. The sequence D-P-N-W-A-N constitutes another distinct motif that is highly conserved among the Coq8/ADCK3 homologues (Fig. 5). The yeast coq8 mutation resulting in N348Y (underlined) would likely disrupt the structure of this catalytic loop.
Three coq8 missense mutations (G89D, G130D, and G475D) are present in regions of unknown function. Both G89D and G130D are located near or in conserved regions. Patients harboring ADCK3 mutations resulting in R213W, G272V or G272D (corresponding to R77 and G130 in yeast Coq8, respectively) had deficiencies in muscle Q10 content [26]. Introduction of the human mutations into the corresponding positions of yeast Coq8 (R77WCoq8, G130VCoq8, or G130DCoq8) and expression in yeast coq8 null mutants, was shown to result in greatly decreased or absent Q6 content, respectively [26]. We confirm that the corresponding yeast coq8 mutant harboring G130D-Coq8p lacked Q6 and further determined that it failed to phosphorylate Coq3p and Coq7p, and was deficient in phosphorylation of Coq5p.
The mutation in coq8-7 (G475D) is near the sequence P-P-P-E-E-T-Y-S-L-H-R-K-x-x-G, identifying a remarkably conserved region among the Coq8 and human ADCK3 and ADCK4 homologues (Figure 5). A BLAST search with this sequence identifies a similar segment in the Bacillus subtilis protein, RsbU. This sequence is well conserved near the amino-terminus of RsbU homologues (Fig. 12). In crystal structures of RsbU, this region forms an α-helix and several conserved residues (including P44 and E45 in B. subtilis) are required for interaction with a neighboring α-helix [66]. This interaction stabilizes the RsbU polypeptide (a phosphatase), allowing it to form a homodimer necessary for binding of RsbU to RsbT (a kinase). The RsbU/RsbT interaction (termed partner switching) is controlled by serine phosphorylation and mediates stress response to environmental and nutritional signals in B. subtillis, and contributes to pathogenicity and enhanced intracellular growth of Staphylococcus aureus and Listeria monocytogenes [67, 68]. In Coq8p, this motif is located near the carboxyl-terminus. It is conceivable that in yeast Coq8 and human ADCK3, this region mediates a similar protein-protein interaction required for Q biosynthesis. Tauche et al. [34] determined that yeast Coq8p forms homomeric complexes. In fact, the rescue mediated by expression of wild-type yeast COQ8 was much less robust in yeast coq8 point mutants as compared to the coq8 null mutant (Fig. 7), suggesting that dysfunctional Coq8p present in the point mutants may interfere with functional interaction(s) of wild-type Coq8p, either with itself or with partner polypeptides.
Tauche et al. [34] discovered that Coq3p is a putative phosphorylation substrate of Coq8p using a tagged version of Coq3p. Here, we utilized antibodies to the Coq3 polypeptide to confirm this result, and extended the studies by examining Coq polypeptide steady state levels and phosphorylation status in wild-type, coq8 null and two of the coq8 point mutants that retained high steady state levels of the Coq8 polypeptide. The coq8 null mutant and W222 (coq8-5; G130D) were found to possess only the unphosphorylated isoform of Coq3p. However, W183 (coq8-3; A197V) retained phosphorylated Coq3p, suggesting that the A197V mutation did not eliminate the kinase activity of Coq8p, and allowed phosphorylation of Coq3p. W183 retained high steady state levels of Coq1 through Coq8 polypeptides, although levels of Coq9p were decreased relative to wild type. In contrast, W222 had dramatically lower levels of Coq4p and lacked Coq7p and Coq9p, a phenotype similar to the coq8 null mutant. One interpretation is that phosphorylated forms of Coq3p are required to preserve stability of Coq4, Coq7 and Coq9 polypeptides. In contrast, the W222 mutant lacked phosphorylated forms of Coq3p, contributing to profound destabilization of Coq4, Coq7 and Coq9 polypeptides. Alternatively, the mutation in W183 that allows for detectable steady state levels of Coq9p may in turn stabilize the other Coq polypeptides.
Coq7p and Coq5p were also found to be substrates of Coq8p. Wild-type yeast and the W183 coq8-3 mutant each contained normal steady state levels of Coq7p, and W183 retained one of the two phosphorylated Coq7 isoforms. In contrast, the coq8 null and W222 mutant both lacked steady state levels of Coq7p. We propose that the steady state level of Coq7p depends on Coq8 kinase activity, and that phosphorylation of Coq7p prevented Coq7p from destabilization. Coq7 has been previously identified as a regulated step in Q biosynthesis [33]. We further demonstrated that Coq5p also contained multiple phosphorylated isoforms. However, not all of the phosphorylation states of Coq5p were Coq8p dependent. Both the coq8 null mutant and the W222 coq8 point mutant lacked only the most acidic Coq5 spot in the 2D-IEF analyses. Three of the four Coq5 spots observed in wild-type mitochondria disappeared upon phosphatase treatment, suggesting that there is another kinase (or kinases) acting on Coq5p. Whether these phosphorylated isoforms of Coq5 are essential for Q synthesis still remains a question.
In these analyses we were not able to resolve phosphorylation states of several of the Coq polypeptides. Coq4p did not appear to be phosphorylated, since it migrated as one spot, and showed similar mobility with or without phosphatase treatment (data not shown). Coq2, Coq6, Coq8, Coq9 and Coq10 polypeptides were not detectable on 2D-IEF gels, even though they can be readily detected in isolated mitochondria by Western blot of SDS-PAGE gels. Although multiple spots of Coq1p were detected on 2D-IEF gels, no signal was detected following phosphatase treatment, thus the phosphorylation state of Coq1p is ambiguous (data not shown).
Steady state levels of Coq9p mirrored those of Coq7p, and thus it is tempting to speculate that Coq8 may also phosphorylate Coq9p. In fact, Coq9p has been detected in a yeast phosphoproteome analysis and is phosphorylated at T34 [69]. Currently, this is the only reported phosphorylation site in any yeast Coq polypeptide [70]. The amino terminal sequence of Coq9 residue is not conserved in metazoan Coq9 sequences, and it is interesting that T34 may be located within the predicted mitochondrial leader sequence of yeast Coq9p (Table 1S). Overexpression of COQ8 is known to suppress a nonsense (Q151STOP) coq9 mutant [18], and was attributed to a detectable presence of Coq9p due to read-through. Overexpression of COQ8 restores the steady state level of Coq4p in several coq null mutants, but not in the coq9 null mutant [71]. While experiments have not revealed a complex between Coq8p and Coq9p [30], it is certainly possible that phosphorylation of Coq9p by Coq8p might be necessary to form a Q biosynthetic complex and so preserve the otherwise unstable Coq polypeptides known to interact with Coq9, including Coq4p, Coq6p, and Coq7p. It is likely that such Coq8-mediated phosphorylation of Coq polypeptides might function to regulate Q6 synthesis, and thus also account for the COQ8 multicopy suppression of the coq10 mutant [31, 71].
Expression of the Coq8 human homolog, ADCK3, rescued the growth of yeast coq8 mutants on non-fermentable carbon source and enabled synthesis of Q6. Although ADCK3 (CABC1) has been localized to mitochondria [72], the rescue observed here required the addition of an amino-terminal yeast mitochondrial leader sequence to ADCK3 (pmcADCK3). In the yeast W222 and coq8 null mutants, pmcADCK3 rescued phosphorylation of Coq3p and Coq7p. The rescue afforded by pmcADCK3 was most dramatic in the W222 mutant because restoration of phosphorylated Coq3p and Coq7p coincided with significantly increased steady state levels of Coq7p, Coq4p, and Coq9p. ADCK3 also restored the phosphorylation state of Coq5p in W222. The effect of pmcADCK3 on the steady state level of Coq polypeptides was less significant in the coq8 null mutant. We speculate that the presence of the Coq8-G130D polypeptide may facilitate the phosphorylation or stabilization of yeast Coq polypeptides by human ADCK3. It is important to note that detection of phosphorylation by 2D-IEF and Western blot can detect whether or not a protein is phosphorylated, but does not indicate whether the polypeptide is correctly phosphorylated. While the rescue observed is dramatic, growth is much slower and the content of Q6 is much lower in the ADCK3 rescued mutants as compared to mutants rescued with yeast COQ8. ADCK3 shares 38% sequence identity with yeast Coq8p (Fig. 5), thus the activity of ADCK3 might not be ideal for the yeast substrates.
Patients harboring missense, deletion, or nonsense mutations in both copies of ADCK3 seem to have a distinct and milder presentation of symptoms as compared to patients with defects in other genes necessary for Q biosynthesis [26, 27, 73]. For example patients with severe mutations resulting in nonsense mediated decay of ADCK3 mRNA, develop progressive cerebellar ataxia, but indicate that ADCK3 itself is not essential for life [73]. In contrast, patients with PDSS1, PDSS2, COQ2, or COQ9 mutations generally exhibit a severe multisystemic infantile form of Q deficiency, and develop renal disease [74]. In the clinical setting it is important to identify Q-deficiencies in patients as early as possible, since treatment with oral Q10 can sometimes elicit dramatic improvement [74]. Partial rescue of kidney disease with oral Q10 supplements has been observed in the Pdss2 mouse model of kidney disease [75]. Yeast coq8 null mutants are indistinguishable from the other yeast coq mutants (coq1-coq7 and coq9); they lack Q, have severe respiratory deficiency, and exhibit similar sensitivity to oxidative stress [11, 15]. Therefore, it seems likely that one or more of the other human homologues of ADCK3 must also function in Q biosynthesis, perhaps supplying a partial overlap of function. Consistent with this idea is that fibroblasts cultured from patients with defects in ADCK3 have normal Q10 content [26]. The best candidate appears to be ADCK4, since it shares highest sequence identity. However, the function of ADCK4 in Q biosynthesis is not yet clear.
It is intriguing that plants contain numerous homologues of Coq8/ADCK3. Arabidopsis thaliana ABC1A is an ortholog of yeast Coq8p, and when expressed in yeast coq8(abc1) mutants, rescues the Q-deficient respiratory defect [76]. However, four distinct A. thaliana Coq8 homologues are present within plastoglobules, which are located within chloroplasts and appear to be important in lipid metabolism, including the synthesis and storage of other prenylated molecules such as plastoquinone, phylloquinone, and tocopherols [77]. It will be important to determine the function of plant Coq8(Abc1) homologues in the synthesis and regulation of these prenylated lipid molecules. Indeed, the stress sensitivity observed in A. thaliana strains harboring insertion mutations in the AtOSA1 gene (a Coq8 homologue) is consistent with a defect in synthesis of one of the prenylated quinone antioxidant lipids [47].
In summary, there is a striking conservation of Q biosynthesis from yeast to humans [11, 15]. This can now be extended to include the conserved function of yeast Coq8p and human ADCK3. Both yeast Coq8p and human ADCK3 appear to function as kinases recognizing conserved Coq polypeptide substrates, including Coq3p, Coq7p, and Coq5p. This phosphorylation is key to the formation or maintenance of a Q multisubunit complex [34] and may modulate biosynthesis of Q.
Research Highlights of Xie LX et al.
Yeast coq8 mutants were identified that maintain normal steady state levels of Coq8p
S. cerevisiae Coq8 polypeptide is required for phosphorylation of Coq3, Coq5 and Coq7
The human Coq8 homolog ADCK3 expressed in yeast coq8 mutants rescues Q biosynthesis
Human ADCK3 rescues phosphorylation of yeast Coq polypeptides in yeast coq8 mutants
The results indicate a profound conservation of Coq8/ADCK3 function in Q synthesis
Supplementary Material
Fig. 11.
The carboxyl terminal segment of Coq8 homologues and the amino terminal segment of RsbU homologues share a region of high identity. In RsbU the sequence designated by the black bar forms an alpha-helix, and side chains interact with a neighboring alpha-helix in a RsbU homodimer. Numbers indicate the amino acid position of the segment. The RsbU homologues shown are from Bacillus subtilis (GenBank accession number NP_388351), Bacillus lichenformis (GenBank accession number YP_077796), and Bacillus halodurans (GenBank accession number NP_241392). Shading indicates identical residues present in the majority of the sequences.
Acknowledgements
We are grateful to Dr. A. Tzagoloff for sending us the collection of yeast coq8 point mutants and for the cytochrome c1 antibodies. We thank Peter T. Lee for pBT8-1 and Drs. R. Saiki and S. Morvaridi for the gift of pRCM plasmid. We thank Dr. L. McAlister-Henn for malate dehydrogenase antibodies, Dr. B. Trumpower for antibodies to Rip1, and Dr. C. M. Koehler for antibodies to cytochrome b2, Atp2, and Hsp60. This work is supported by National Institutes of Health Grant GM45952 (to C.F.C.), and L.X.X. was supported by the Ruth L. Kirschstein National Service Award GM007185. The 2-D gel work was performed in the Proteomic Facility at the UCLA Molecular Instrumentation Center, which was established with a grant from the W. M. Keck Foundation. The LC-MS/MS determination of quinones was supported in part by Grant Number S10RR024605 from the National Center for Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center for Research Resources or the National Institutes of Health.
Footnotes
- 2D-IEF/SDS-PAGE
- two dimensional isoelectric focusing/sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Q
- coenzyme Q
- ORF
- open reading frame
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Ghisla S, Thorpe C. Acyl-CoA dehydrogenases. A mechanistic overview. Eur J Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- [2].Lenaz G, De Santis A. A survey of the function and specificity of ubiquione in the mitochondrial respiratory chain. In: Lenaz G, editor. Coenzyme Q. John Wiley & Sons; Chichester, U.K.: 1985. pp. 165–199. [Google Scholar]
- [3].Hildebrandt TM, Grieshaber MK. Three enzymatic activities catalyze the oxidation of sulfide to thiosulfate in mammalian and invertebrate mitochondria. Febs J. 2008;275:3352–3361. doi: 10.1111/j.1742-4658.2008.06482.x. [DOI] [PubMed] [Google Scholar]
- [4].Hill S, Hirano K, Shmanai VV, Marbois BN, Vidovic D, Bekish AV, Kay B, Tse V, Fine J, Clarke CF, Shchepinov MS. Isotope-reinforced polyunsaturated fatty acids protect yeast cells from oxidative stress. Free Radic Biol Med. 2010;50:130–138. doi: 10.1016/j.freeradbiomed.2010.10.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bentinger M, Brismar K, Dallner G. The antioxidant role of coenzyme Q. Mitochondrion. 2007;7S:S41–S50. doi: 10.1016/j.mito.2007.02.006. [DOI] [PubMed] [Google Scholar]
- [6].Papucci L, Schiavone N, Witort E, Donnini M, Lapucci A, Tempestini A, Formigli L, Zecchi-Orlandini S, Orlandini G, Carella G, Brancato R, Capaccioli S. Coenzyme Q10 prevents apoptosis by inhibiting mitochondrial depolarization independently of its free radical scavenging property. J Biol Chem. 2003;278:28220–28228. doi: 10.1074/jbc.M302297200. [DOI] [PubMed] [Google Scholar]
- [7].Walter L, Miyoshi H, Leverve X, Bernard P, Fontaine E. Regulation of the mitochondrial permeability transition pore by ubiquinone analogs. A progress report. Free Radic Res. 2002;36:405–412. doi: 10.1080/10715760290021252. [DOI] [PubMed] [Google Scholar]
- [8].Devun F, Walter L, Belliere J, Cottet-Rousselle C, Leverve X, Fontaine E. Ubiquinone analogs: a mitochondrial permeability transition pore-dependent pathway to selective cell death. PLoS One. 2010;5:e11792. doi: 10.1371/journal.pone.0011792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- [10].Azzolin L, von Stockum S, Basso E, Petronilli V, Forte MA, Bernardi P. The mitochondrial permeability transition from yeast to mammals. FEBS Lett. 2010;584:2504–2509. doi: 10.1016/j.febslet.2010.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kawamukai M. Biosynthesis and bioproduction of coenzyme Q10 by yeasts and other organisms. Biotechnol Appl Biochem. 2009;53:217–226. doi: 10.1042/BA20090035. [DOI] [PubMed] [Google Scholar]
- [12].Olson RE, Rudney H. Biosynthesis of ubiquinone. Vitam Horm. 1983;40:1–43. doi: 10.1016/s0083-6729(08)60431-8. [DOI] [PubMed] [Google Scholar]
- [13].Marbois B, Xie LX, Choi S, Hirano K, Hyman K, Clarke CF. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pierrel F, Hamelin O, Douki T, Kieffer-Jaquinod S, Muhlenhoff U, Ozeir M, Lill R, Fontecave M. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- [15].Tran UC, Clarke CF. Endogenous synthesis of coenzyme Q in eukaryotes. Mitochondrion. 2007;7S:S62–S71. doi: 10.1016/j.mito.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marbois B, Gin P, Faull KF, Poon WW, Lee PT, Strahan J, Shepherd JN, Clarke CF. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- [17].Marbois B, Gin P, Gulmezian M, Clarke CF. The yeast Coq4 polypeptide organizes a mitochondrial protein complex essential for coenzyme Q biosynthesis. Biochim Biophys Acta. 2009;1791:69–75. doi: 10.1016/j.bbalip.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hsieh EJ, Gin P, Gulmezian M, Tran UC, Saiki R, Marbois BN, Clarke CF. Saccharomyces cerevisiae Coq9 polypeptide is a subunit of the mitochondrial coenzyme Q biosynthetic complex. Arch Biochem Biophys. 2007;463:19–26. doi: 10.1016/j.abb.2007.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, Falk MJ. Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev Disabil Res Rev. 2010;16:200–218. doi: 10.1002/ddrr.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bousquet I, Dujardin G, Slonimski PP. ABC1, a novel yeast nuclear gene has a dual function in mitochondria: it suppresses a cytochrome b mRNA translation defect and is essential for the electron transfer in the bc1 complex. Embo J. 1991;10:2023–2031. doi: 10.1002/j.1460-2075.1991.tb07732.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Brasseur G, Tron G, Dujardin G, Slonimski PP, Brivet-Chevillotte P. The nuclear ABC1 gene is essential for the correct conformation and functioning of the cytochrome bc1 complex and the neighbouring complexes II and IV in the mitochondrial respiratory chain. Eur J Biochem. 1997;246:103–111. doi: 10.1111/j.1432-1033.1997.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- [22].Do TQ, Hsu AY, Jonassen T, Lee PT, Clarke CF. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- [23].Hsieh EJ, Dinoso JB, Clarke CF. A tRNA(TRP) gene mediates the suppression of cbs2-223 previously attributed to ABC1/COQ8. Biochem. Biophys. Res. Commun. 2004;317:648–653. doi: 10.1016/j.bbrc.2004.03.096. [DOI] [PubMed] [Google Scholar]
- [24].Macinga DR, Cook GM, Poole RK, Rather PN. Identification and characterization of aarF, a locus required for production of ubiquinone in Providencia stuartii and Escherichia coli and for expression of 2′-N-acetyltransferase in P. stuartii. J Bacteriol. 1998;180:128–135. doi: 10.1128/jb.180.1.128-135.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/jb.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mollet J, Delahodde A, Serre V, Chretien D, Schlemmer D, Lombes A, Boddaert N, Desguerre I, de Lonlay P, de Baulny HO, Munnich A, Rotig A. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am. J. Hum. Genet. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lagier-Tourenne C, Tazir M, Lopez LC, Quinzii CM, Assoum M, Drouot N, Busso C, Makri S, Ali-Pacha L, Benhassine T, Anheim M, Lynch DR, Thibault C, Plewniak F, Bianchetti L, Tranchant C, Poch O, DiMauro S, Mandel JL, Barros MH, Hirano M, Koenig M. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am. J. Hum. Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Leonard CJ, Aravind L, Koonin EV. Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res. 1998;8:1038–1047. doi: 10.1101/gr.8.10.1038. [DOI] [PubMed] [Google Scholar]
- [29].Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- [30].Johnson A, Gin P, Marbois BN, Hsieh EJ, Wu M, Barros MH, Clarke CF, Tzagoloff A. COQ9, a new gene required for the biosynthesis of coenzyme Q in Saccharomyces cerevisiae. J Biol Chem. 2005;280:31397–31404. doi: 10.1074/jbc.M503277200. [DOI] [PubMed] [Google Scholar]
- [31].Barros MH, Johnson A, Gin P, Marbois BN, Clarke CF, Tzagoloff A. The Saccharomyces cerevisiae COQ10 gene encodes a START domain protein required for function of coenzyme Q in respiration. J Biol Chem. 2005;280:42627–42635. doi: 10.1074/jbc.M510768200. [DOI] [PubMed] [Google Scholar]
- [32].Busso C, Tahara EB, Ogusucu R, Augusto O, Ferreira-Junior JR, Tzagoloff A, Kowaltowski AJ, Barros MH. Saccharomyces cerevisiae coq10 null mutants are responsive to antimycin A. Febs J. 2010 doi: 10.1111/j.1742-4658.2010.07862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Padilla S, Tran UC, Jimenez-Hidalgo M, Lopez-Martin JM, Martin-Montalvo A, Clarke CF, Navas P, Santos-Ocana C. Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell Mol Life Sci. 2009;66:173–186. doi: 10.1007/s00018-008-8547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Tauche A, Krause-Buchholz U, Rodel G. Ubiquinone biosynthesis in Saccharomyces cerevisiae: the molecular organization of O-methylase Coq3p depends on Abc1p/Coq8p. FEMS Yeast Res. 2008;8:1263–1275. doi: 10.1111/j.1567-1364.2008.00436.x. [DOI] [PubMed] [Google Scholar]
- [35].Sippel CJ, Goewert RR, Slachman FN, Olson RE. The regulation of ubiquinone-6 biosynthesis by Saccharomyces cerevisiae. J Biol Chem. 1983;258:1057–1061. [PubMed] [Google Scholar]
- [36].Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press; Plainview, NY: 2000. [Google Scholar]
- [37].Barkovich RJ, Shtanko A, Shepherd JA, Lee PT, Myles DC, Tzagoloff A, Clarke CF. Characterization of the COQ5 gene from Saccharomyces cerevisiae. Evidence for a C-methyltransferase in ubiquinone biosynthesis. J. Biol. Chem. 1997;272:9182–9188. doi: 10.1074/jbc.272.14.9182. [DOI] [PubMed] [Google Scholar]
- [38].Elble R. A simple and efficient procedure for transformation of yeasts. Biotechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- [39].Ackerman SH, Martin J, Tzagoloff A. Characterization of ATP11 and detection of the encoded protein in mitochondria of Saccharomyces cerevisiae. J Biol Chem. 1992;267:7386–7394. [PubMed] [Google Scholar]
- [40].Glerum DM, Koerner TJ, Tzagoloff A. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J. Biol. Chem. 1995;270:15585–15590. doi: 10.1074/jbc.270.26.15585. [DOI] [PubMed] [Google Scholar]
- [41].Hsu AY, Poon WW, Shepherd JA, Myles DC, Clarke CF. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- [42].Cormack B. Directed mutagenesis using the polymerase chain reaction. In: Chanda VB, editor. Current Protocols in Molecular Biology. Vol. 1. John Wiley & Sons; 1997. pp. 8.5.1–8.5.10.. [DOI] [PubMed] [Google Scholar]
- [43].Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- [44].Glick BS, Brandt A, Cunningham K, Muller S, Hallberg RL, Schatz G. Cytochromes c1 and b2 are sorted to the intermembrane space of yeast mitochondria by a stop-transfer mechanism. Cell. 1992;69:809–822. doi: 10.1016/0092-8674(92)90292-k. [DOI] [PubMed] [Google Scholar]
- [45].Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- [46].Jonassen T, Clarke CF. Isolation and functional expression of human COQ3, a gene encoding a methyltransferase required for ubiquinone biosynthesis. J. Biol. Chem. 2000;275:12381–12387. doi: 10.1074/jbc.275.17.12381. [DOI] [PubMed] [Google Scholar]
- [47].Jasinski M, Sudre D, Schansker G, Schellenberg M, Constant S, Martinoia E, Bovet L. AtOSA1, a member of the Abc1-like family, as a new factor in cadmium and oxidative stress response. Plant Physiol. 2008;147:719–731. doi: 10.1104/pp.107.110247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Belogrudov GI, Lee PT, Jonassen T, Hsu AY, Gin P, Clarke CF. Yeast COQ4 ncodes a mitochondrial protein required for coenzyme Q synthesis. Arch. Biochem. Biophys. 2001;392:48–58. doi: 10.1006/abbi.2001.2448. [DOI] [PubMed] [Google Scholar]
- [49].Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Kamps MP, Sefton BM. Neither arginine nor histidine can carry out the function of lysine-295 in the ATP-binding site of p60src. Mol Cell Biol. 1986;6:751–757. doi: 10.1128/mcb.6.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. Faseb J. 1995;9:576–596. [PubMed] [Google Scholar]
- [52].Gin P, Clarke CF. Genetic Evidence for a Multi-subunit Complex in Coenzyme Q Biosynthesis in Yeast and the Role of the Coq1 Hexaprenyl Diphosphate Synthase. J. Biol. Chem. 2005;280:2676–2681. doi: 10.1074/jbc.M411527200. [DOI] [PubMed] [Google Scholar]
- [53].Tran UC, Marbois B, Gin P, Gulmezian M, Jonassen T, Clarke CF. Complementation of Saccharomyces cerevisiae coq7 mutants by mitochondrial targeting of the Escherichia coli UbiF polypeptide: two functions of yeast Coq7 polypeptide in coenzyme Q biosynthesis. J Biol Chem. 2006;281:16401–16409. doi: 10.1074/jbc.M513267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol. Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Tzagoloff A, Akai A, Needleman RB. Assembly of the mitochondrial membrane system. Characterization of nuclear mutants of Saccharomyces cerevisiae with defects in mitochondrial ATPase and respiratory enzymes. J Biol Chem. 1975;250:8228–8235. [PubMed] [Google Scholar]
- [56].Tzagoloff A, Akai A, Needleman RB, Zulch G. Assembly of the mitochondrial membrane system. Cytoplasmic mutants of Saccharomyces cerevisiae with lesions in enzymes of the respiratory chain and in the mitochondrial ATPase. J Biol Chem. 1975;250:8236–8242. [PubMed] [Google Scholar]
- [57].Vajo Z, King LM, Jonassen T, Wilkin DJ, Ho N, Munnich A, Clarke CF, Francomano CA. Conservation of the Caenorhabditis elegans timing gene clk-1 from yeast to human: a gene required for ubiquinone biosynthesis with potential implications for aging. Mamm. Genome. 1999;10:1000–1004. doi: 10.1007/s003359901147. [DOI] [PubMed] [Google Scholar]
- [58].Baba SW, Belogrudov GI, Lee JC, Lee PT, Strahan J, Shepherd JN, Clarke CF. Yeast coq5 C-methyltransferase is required for stability of other polypeptides involved in coenzyme Q biosynthesis. J. Biol. Chem. 2004;279:10052–10059. doi: 10.1074/jbc.M313712200. [DOI] [PubMed] [Google Scholar]
- [59].Hsieh EJ. Chemistry and Biochemistry. Ph.D., University of California, Los Angeles; Los Angeles: 2006. Characterization of the Coenzyme Q Biosynthetic Genes ABC1/COQ8 and COQ9 in Yeast Saccharomyces cerevisiae; p. 126. [Google Scholar]
- [60].Facchin S, Lopreiato R, Stocchetto S, Arrigoni G, Cesaro L, Marin O, Carignani G, Pinna LA. Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem J. 2002;364:457–463. doi: 10.1042/BJ20011376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Spitaler M, Villunger A, Grunicke H, Uberall F. Unique structural and functional properties of the ATP-binding domain of atypical protein kinase C-iota. J Biol Chem. 2000;275:33289–33296. doi: 10.1074/jbc.M002742200. [DOI] [PubMed] [Google Scholar]
- [62].Taylor SS, Radzio-Andzelm E, Hunter T. How do protein kinases discriminate between serine/threonine and tyrosine? Structural insights from the insulin receptor protein-tyrosine kinase. Faseb J. 1995;9:1255–1266. doi: 10.1096/fasebj.9.13.7557015. [DOI] [PubMed] [Google Scholar]
- [63].Johnson LN, Lowe ED, Noble ME, Owen DJ. The Eleventh Datta Lecture. The structural basis for substrate recognition and control by protein kinases. FEBS Lett. 1998;430:1–11. doi: 10.1016/s0014-5793(98)00606-1. [DOI] [PubMed] [Google Scholar]
- [64].Hanks SK. Genomic analysis of the eukaryotic protein kinase superfamily: a perspective. Genome Biol. 2003;4:111. doi: 10.1186/gb-2003-4-5-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- [66].Delumeau O, Dutta S, Brigulla M, Kuhnke G, Hardwick SW, Volker U, Yudkin MD, Lewis RJ. Functional and structural characterization of RsbU, a stress signaling protein phosphatase 2C. J Biol Chem. 2004;279:40927–40937. doi: 10.1074/jbc.M405464200. [DOI] [PubMed] [Google Scholar]
- [67].Olivier AC, Lemaire S, Van Bambeke F, Tulkens PM, Oldfield E. Role of rsbU and staphyloxanthin in phagocytosis and intracellular growth of Staphylococcus aureus in human macrophages and endothelial cells. J Infect Dis. 2009;200:1367–1370. doi: 10.1086/606012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shin JH, Brody MS, Price CW. Physical and antibiotic stresses require activation of the RsbU phosphatase to induce the general stress response in Listeria monocytogenes. Microbiology. 2010;156:2660–2669. doi: 10.1099/mic.0.041202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Stark C, Su TC, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz BJ, Tyers M, Sadowski I. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford) 2010;2010 doi: 10.1093/database/bap026. bap026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zampol MA, Busso C, Gomes F, Ferreira-Junior JR, Tzagoloff A, Barros MH. Over-expression of COQ10 in Saccharomyces cerevisiae inhibits mitochondrial respiration. Biochem Biophys Res Commun. 2010;402:82–87. doi: 10.1016/j.bbrc.2010.09.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Iiizumi M, Arakawa H, Mori T, Ando A, Nakamura Y. Isolation of a novel gene, CABC1, encoding a mitochondrial protein that is highly homologous to Yeast activity of bc1 complex. Cancer Res. 2002;62:1246–1250. [PubMed] [Google Scholar]
- [73].Gerards M, van den Bosch B, Calis C, Schoonderwoerd K, van Engelen K, Tijssen M, de Coo R, van der Kooi A, Smeets H. Nonsense mutations in CABC1/ADCK3 cause progressive cerebellar ataxia and atrophy. Mitochondrion. 2010;10:510–515. doi: 10.1016/j.mito.2010.05.008. [DOI] [PubMed] [Google Scholar]
- [74].Quinzii CM, Hirano M. Coenzyme Q and mitochondrial disease. Dev Disabil Res Rev. 2010;16:183–188. doi: 10.1002/ddrr.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Saiki R, Lunceford AL, Shi Y, Marbois B, King R, Pachuski J, Kawamukai M, Gasser DL, Clarke CF. Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am J Physiol Renal Physiol. 2008;295:F1535–1544. doi: 10.1152/ajprenal.90445.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Cardazzo B, Hamel P, Sakamoto W, Wintz H, Dujardin G. Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene. 1998;221:117–125. doi: 10.1016/s0378-1119(98)00417-x. [DOI] [PubMed] [Google Scholar]
- [77].Ytterberg AJ, Peltier JB, van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].McEwen JE, Ko C, Kloeckner-Gruissem B, Poyton RO. Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J. Biol. Chem. 1986;261:11872–11879. [PubMed] [Google Scholar]
- [79].Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- [81].Hsu AY, Poon WW, Shepherd JA, Myles DC, Clarke CF. Complementation of coq3 mutant yeast by mitochondrial targeting of the Escherichia coli UbiG polypeptide: evidence that UbiG catalyzes both O-methylation steps in ubiquinone biosynthesis. Biochemistry. 1996;35:9797–9806. doi: 10.1021/bi9602932. [DOI] [PubMed] [Google Scholar]
- [82].Do TQ, Hsu AY, Jonassen T, Lee PT, Clarke CF. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- [83].Chen WJ, Douglas MG. Phosphodiester bond cleavage outside mitochondria is required for the completion of protein import into the mitochondrial matrix. Cell. 1987;49:651–658. doi: 10.1016/0092-8674(87)90541-1. [DOI] [PubMed] [Google Scholar]
- [84].Daum G, Bohni PC, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- [85].Ohashi A, Gibson J, Gregor I, Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- [86].Thompson LM, McAlister-Henn L. Dispensable presequence for cellular localization and function of mitochondrial malate dehydrogenase from Saccharomyces cerevisiae. J Biol Chem. 1989;264:12091–12096. [PubMed] [Google Scholar]
- [87].Reading DS, Hallberg RL, Myers AM. Characterization of the yeast HSP60 gene coding for a mitochondrial assembly factor. Nature. 1989;337:655–659. doi: 10.1038/337655a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.