Abstract
Therapies selectively targeting ischemic myocardium could be applied by intravenous injection. Here, we report an approach for ischemic tissue-selective targeting based on in vivo screening of random peptide sequences using phage display. We performed in vivo biopanning using a phage library in a rat model of ischemia-reperfusion and identified three peptide motifs, CSTSMLKAC, CKPGTSSYC, and CPDRSVNNC, that exhibited preferential binding to ischemic heart tissue compared to normal heart as well as other control organs. The CSTSMLKAC sequence was capable of mediating selective homing of phage to ischemic heart tissue. The CSTSMLKAC peptide was then made as a fusion protein with Sumo-mCherry and injected intravenously in a mouse model of myocardial ischemia-reperfusion injury; subsequently, bio-distribution of Sumo-mCherry-CSTSMLKAC was measured with quantitative ELISA. The targeting peptide led to a significant increase in homing to ischemic left ventricle compared to tissues from non-ischemic left ventricle, the right ventricle, lung, liver, spleen, skeletal muscle, and brain (all p<0.001). These results indicate that the peptide sequence CSTSMLKAC represents a novel molecular tool that may be useful in targeting ischemic tissue and delivering bioengineered proteins into the injured myocardium by systemic intravenous administration.
Keywords: phage display, ischemic-heart homing, peptide, ischemia-reperfusion, quantitative ELISA
Introduction
Myocardial infarction remains one of the leading causes of death worldwide, and patients with myocardial infarction benefit from revascularization and reperfusion therapies [1]. Despite the success of reperfusion therapy, the mortality and morbidity are still substantial, with 5 to 6% of patients having a subsequent cardiovascular event in the next 30 days [1]. Alternative therapeutic approaches are necessary to decrease heart failure and improve outcome after myocardial infarction.
Several different approaches including tissue engineering, cell therapy and protein therapy are potential treatments to limit cardiac damage and improve cardiac function after infarction. Although catheter-based and direct surgical procedures allow infusion of therapeutic moieties into coronary arteries or their direct injection into the myocardium, these modalities require invasive procedures. Furthermore, an approach that enabled systemic delivery to ischemic myocardium could benefit patients during the crucial early hours of infarction.
Targeted delivery is a method for the spatial and temporal release of therapeutically active molecules to specific locations or organs [2]. The concept of targeted delivery implies selective accumulation of drugs or proteins within the affected tissue after systemic administration of the carrier-bound particle with the minimal possible side effects on irrelevant organs [3]. Therapies that could be applied by intravenous injections with selectively homing to the ischemic region have the potential for protecting myocardium during infarction. For example, the ability to target drugs or proteins to the myocardium would allow ambulance-based treatment to initiate cardioprotective therapies before arrival of patients to the emergency room, long before catheterization.
Phage display is a proteomics technology that allows for selection of random peptide libraries displayed on the surface of genetically manipulated bacteriophage with various applications [4–12]. Since differential protein expression and distribution between the normal and ischemic myocardium have been reported [13–17], in vivo panning with phage display could be useful to identify peptides that exhibit preferential binding to ischemic heart tissue.
Here we report ischemic tissue-selective targeting based on in vivo screening of random peptide sequences using phage display. We identified peptides capable of mediating selective localization of phage particles to ischemic heart tissue. One of the peptides showed significant selectivity for ischemic myocardium in subsequent in vivo validation experiments. Furthermore, a fusion protein carrying the peptide was synthesized and injected intravenously in a murine model of myocardial ischemia-reperfusion injury, and this fusion protein homed to ischemic myocardium. Thus, ischemic myocardium-specific peptide sequences represent a novel molecular tool which may be useful in targeting ischemic myocardium.
Materials and Methods
Animals
Male Sprague-Dawley rats (body weight, 200–230 g; Harlan, Indianapolis, IN) were used for the in vivo screening of the phage library and to test the tissue specificity of synthetic peptides. Male FVB mice (9 weeks old, body weight, 25–28 g; Charles River, Wilmington, MA) were used to test the homing capability of recombinant proteins. All animal experiments were approved by the Harvard Medical School Standing Committee on Animals.
In vivo biopanning on ischemia/reperfusion rats
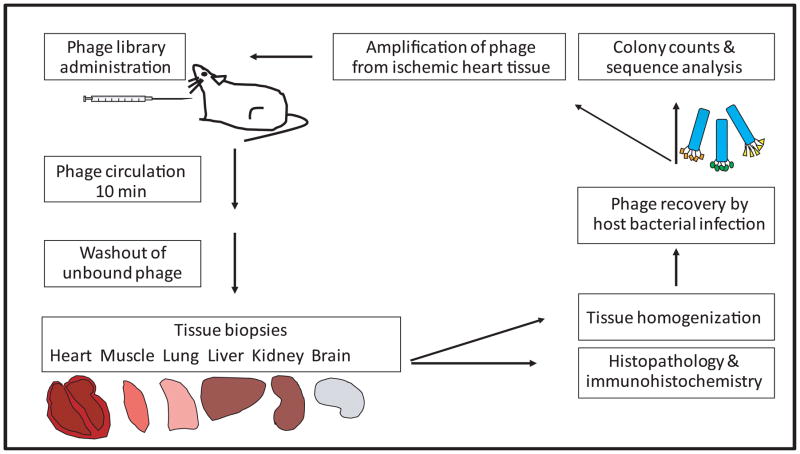
The Ph.D.-C7C phage library (New England Biolabs, Ipswich, MA) was used for the first panning. Ischemia/reperfusion injury was performed in rats. Briefly, a rat was anesthetized with pentobarbital (60 mg/kg), intubated, and mechanically ventilated with room air. The heart was exposed and the left coronary artery (LCA) was occluded below the appendage for 30 minutes. Following 10 minutes reperfusion, 1×1011 transducing units (TU) of phage particles were injected into left ventricular (LV) cavity from the LV apex with temporary occlusion of ascending aorta (2–3 seconds). The phage particles were allowed to circulate for 10 minutes, and then unbound phage were washed out with PBS injected from the LV apex. The 40 mg of ischemic region of LV, posterior wall of LV, right ventricle (RV), and the other organ tissues (lung, liver, kidney, skeletal muscle and brain) were excised (Figure 1). Each experiment for biopanning used 2 rats; one was injected with phage library or amplified phage for the next round, and the other was injected with insert-less wild-type M13KE phage (New England Biolabs) as a control.
Figure 1. Schematic description of in vivo phage display screen in a rat model of ischemia-reperfusion.
In the first round of biopanning, a phage library displaying random peptides is injected into a rat, and phage homing to various tissues are recovered for various assays. Phage recovered from ischemic heart tissue are amplified and used for the next round of biopanning. Three to four rounds of biopanning typically lead to enrichment of phage-peptide clones with high specificity of homing.
In vitro phage recovery and multiple consecutive rounds
The collected tissues were homogenized in DMEM (Sigma, St. Louis, MO) with protease inhibitor cocktail (Sigma) to generate single cell suspensions. The cell suspensions were centrifuged, the pellet was washed, and incubated with mid-log phase host bacteria, ER2738 (New England Biolabs) (rescued phage/ER2738). Serial dilutions of the rescued phage in mid-log phase ER2738 were mixed with the agarose-top (Bacto-Tryptone 10 g/L, yeast extract 5 g/L, NaCl 5 g/L, MgCl2 1 g/L, agarose 7 g/L), spread on duplicate LB/IPTG/Xgal plates (Trypton 10 g/L, Yeast extract 5 g/L, NaCl 10 g/L, agar 15 g/L, Xgal 20 mg/L, IPTG 0.1 μM), and incubated at 37°C. Blue-colored plaques on plates represented the relative binding units corresponding to tissue isolated from each organ. The phage rescued from the ischemic region of LV was amplified as the first-round phage. The second and the third rounds of biopanning were performed using the first-round phage and the second-round phage respectively. Each round had duplicate sets of two rats, with one rat injected with the test phage, and the other rat injected with insert-less phage as control. In each round, phage clones from blue-colored plaques on the plates of ischemic/non-ischemic LV and RV were randomly picked and stored in glycerol.
Ischemic homing phage amplification and titration
The rescued phage/ER2738 were amplified with early-log phase ER2738 in LB media, and then centrifuged to remove residual bacteria. The supernatant was mixed with PEG/NaCl (20% w/v polyethylene glycol-8000, 2.5 M NaCl in distilled water), incubated on ice overnight and centrifuged to collect the amplified phage pellet. Collected phage were rinsed with PBS, precipitated with PEG/NaCl, and resuspended in PBS for the next round. The titer of the amplified phage was measured according to the manufacturer’s manual of Ph.D.-C7C phage library.
Peptide sequences and amplification of phage clones
Phage clones recovered following the third selection round were sequenced for the peptide insert. 192 phage clones isolated from ischemic LV and 96 clones each obtained from non-ischemic LV and RV were selected for sequencing. To determine the amino acid sequences displayed by the selected phage, the insert in the integrated section of the phage was amplified by direct polymerase chain reaction (PCR) with the unique primer pairs; forward primer, 5′-TGTCGGCGCAACTATCGGTATCAA-3′, reverse primer, 5′-TAGCATTCCACAGACAGCCCTCTA-3′, and then DNA sequence was generated with the reverse primer by DNA sequencer ABI 310. The peptide sequences were translated from DNA sequence and analyzed for their frequency of occurrence. The peptide sequences with high frequencies were further analyzed for potential similarity/homology to known proteins by searching in online databases (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Testing fluorescein conjugated synthetic peptides in ischemia-reperfusion rats
The three most frequently appearing peptides recovered from ischemic myocardium, CSTSMLKAC, CKPGTSSYC, and CPDRSVNNC, were conjugated to a fluorescein tag and then tested for their homing capability to ischemic myocardium in vivo. Each peptide motif was tested in tandem with 3 controls: a modified peptide lacking the flanking cysteines (linear peptide, i.e. STSMLKA), a peptide made with a randomly scrambled sequence of the same amino acids (scrambled peptide, i.e. CSKTALSMC) and PBS. All peptides were dissolved in PBS at a concentration of 10 mg/mL and experimental steps were blinded such that all collection of data was performed without knowledge of sequence identity. Fluorescein-conjugated peptides were chemically synthesized, cyclized and purified by high-performance liquid chromatography with more than 95% purity and confirmed by mass spectrometry at Biopolymer Laboratory at Massachusetts Institute of Technology (Cambridge, MA) and NEO-Peptide (Cambridge, MA). Fluorescein-conjugated peptide 5 mg/kg body weight was injected intravenously into a rat with ischemia-reperfusion injury via a catheter placed in the right jugular vein during reperfusion. After allowing 10 min for peptide circulation, unbound peptides were washed out with PBS, and heart and other organ tissues (lung, liver, kidney, spleen, skeletal muscle and brain) were harvested and analyzed. Tissues were fixed in 4% paraformaldehyde and paraffin-embedded sections were counter-stained with Isolectin GS-IB4 antibody (Invitrogen, Carlsbad, CA) for endothelium and 4′,6′-diamidino-2-phenylindole (DAPI) for nuclei; images were acquired with an inverted epifluorescence microscope (model IX-70, Olympus) through an objective lens, LC Plan FI 20× (Olympus) equipped with a Roper CoolSnap HQ, photometrics camera (Roper Scientific, Tucson, AZ). Images were analyzed based on fluorescence intensity.
Western analysis and quantitative ELISA for detection of Sumo-mCherry-CSTSMLKAC
To test that the CSTSMLKAC peptide homes specifically to ischemic heart tissue with increased affinity even after conjugation to a large molecule complex, experiments were performed with fusion to two proteins, Sumo and mCherry. These protein domains were selected for conjugation to the peptide to facilitate biochemical detection by ELISA; the total molecular weight of the final complex was calculated to be ~40 kDa. Peptides were mutated at the 3′ end of mCherry cDNA, and then mCherry-peptides were cloned into pE-SUMO vector using the two restriction enzyme sites, XbaI and BsaI, according to the manufacturer instructions for SUMOpro Gene Fusion Technology (LifeSensors, Malvern, PA) (Figure 5A). Recombinant proteins were expressed in bacteria and purified with Ni-NTA affinity chromatography and size exclusion chromatography. Homing of recombinant SUMO-mCherry-CSTSMLKAC to ischemic heart tissue and control tissues was performed in tandem with 3 controls: Sumo-mCherry without any peptide conjugation, Sumo-mCherry conjugated to a modified peptide lacking the flanking cysteines (STSMLKA), and Sumo-mCherry conjugated to a peptide with a randomly scrambled sequence of the same amino acids. Sumo-mCherry-peptides were injected into mice subjected to ischemia-reperfusion injury (ischemia for 15 min and reperfusion for 5 min) in three independent experimental sets. Each experimental set (repeat) consisted of 5 mice such that one mouse received one of the following: Sumo-mCherry, Sumo-mCherry-CSTSMLKAC, Sumo-mCherry-STSMLKA, Sumo-mCherry-scrambled peptide, and PBS. All peptides were dissolved in PBS at a concentration of 10 mg/mL, and experimental steps were blinded by 2 individuals. Sumo-mCherry-peptide 1 mg was injected intravenously into a mouse with ischemia-reperfusion injury with the same procedure described previously. After 5 min of circulation time, 40 mg of heart tissue (ischemic LV, non-ischemic LV, and RV) and other organ tissues (lung, liver, kidney, spleen, skeletal muscle and brain) were excised and snap frozen. Frozen tissues were then homogenized in RIPA buffer on ice (PBS, SDS 0.1%, Nonidet P-40 1%, PMSF 0.6 mM, Sodium deoxycholate 0.5%, Sodium Orthovanadate 1 mM, protease inhibitor cocktail) (all from Sigma), sonicated and centrifuged to remove remnant debris. Tissue suspensions were analyzed by Western analysis and by quantitative ELISA. A 96 well Immulux HB flat bottom plate (Dynex Technologies, Chantilly, VA) was coated with anti-sumo antibody (rabbit, smt3 antibody, LifeSensors) for the capturing step, and the uncoated plastic surface was blocked with 2% bovine serum albumin (Sigma) in PBS. Fifty microliter of tissue suspensions were applied to coated plates, the plates were incubated for 2 hours at room temperature, and unbound proteins were washed four times with PBS-T containing 0.1% Tween 20. Signal detection with biotinylated mCherry antibody (rabbit, RFP antibody, Abcam, Cambridge, MA) followed with HRP-conjugated streptavidin (Thermo Scientific, Waltham, MA) and TMB substrate OptEIA™ (BD, Franklin Lakes, NJ). The colorimetric change was measured by the light absorbance at 540 nm by DTX800 (Beckman Coulter, Brea, CA). Each ELISA plate had a serial dilution of standard protein for derivation of a standard curve (0, 0.01, 0.1, 0.5, 1, 5, 10, 50, 100, and 500 ng/mL).
Figure 5. The CSTSMLKAC peptide conjugated with a large molecule showed specific affinity for mouse ischemic cardiac tissue.
(A) Construction of Sumo-mCherry conjugated with peptide CSTSMLKAC. Peptides were mutated at 3′ end of mCherry cDNA and mCherry-peptides were cloned into the pE-SUMO vector. The molecular size and isoelectric point (pI) were calculated 40 kDa and 5.8, respectively. (B) Western analysis in post-ischemic cardiac tissues with an anti-mCherry antibody showed that a greater amount of protein complex was delivered to the ischemic left ventricle (LV) by the CSTSMLKAC peptide compared to negative controls (Upper panel). Lane 1–5, ischemic heart tissue samples from mice injected with PBS (Lane 1), Sumo-mCherry (vehicle) (Lane 2), Sumo-mCherry-STSMLKA (Lane 3), Sumo-mCherry-CSTSMLKAC (Lane 4), and Sumo-mCherry-scrambled peptide (Lane 5). Coomassie blue staining of the gel (lower panel) showed that each well was loaded with a comparable amount. (C) Quantitative ELISA analysis showed that significantly more Sumo-mCherry was delivered in mouse cardiac tissues by the CSTSMLKAC peptide compared with the negative controls: vehicle, STSMLKA, and the scrambled peptide (all p<0.05). In all groups except STSMLKA, there were significant differences between the ischemic and non-ischemic LV. +p<0.05, and ++p<0.0001. N=6 per group in three independent mice.
Statistical analysis
All replicate data are expressed as mean ± SEM. Significant differences among groups were assessed using one-way ANOVA followed by Bonferroni’s test or Newman-Keuls post hoc analysis, and significant differences between groups were assessed using unpaired t-test. Statistically significant differences were confirmed at p-value smaller than 0.05.
Results
Enrichment of phage particles with preferential binding to ischemic heart tissue after three rounds of biopanning
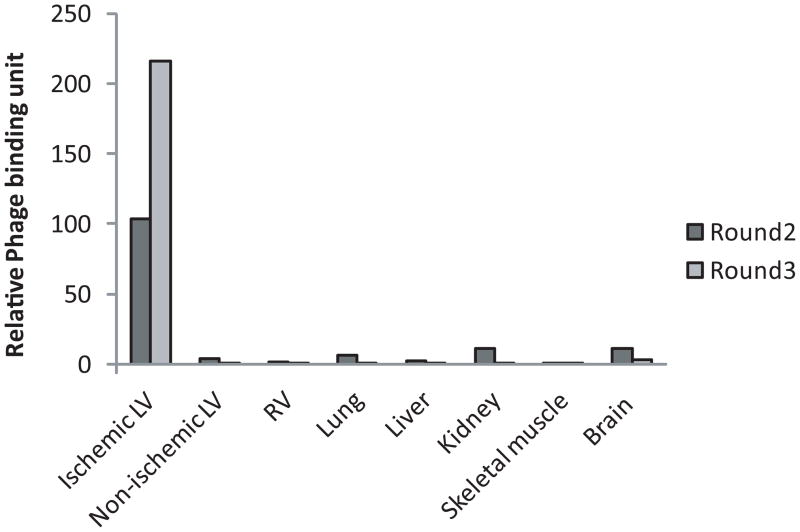
Following the first round of biopanning with the Ph.D.-C7C random phage library in a rat model of ischemia-reperfusion, the relative phage binding units corresponding to various tissues were as follows: 25.4 in ischemic LV, 1870 in non-ischemic LV, 8.5 in RV, 10.9 in lung, 1.5 in liver, 26.9 in kidney, 1.2 in skeletal muscle, and 52.4 in brain tissue. The second round of bio-panning performed using as input amplified phage recovered from ischemic LV tissue in the first round showed a modest increase in relative phage binding to ischemic LV (with 25.4 relative binding units on Round 1, and 103 on Round 2). The third round performed using amplified phage recovered from ischemic LV on the second round resulted in significant enrichment in phage displaying preferential binding to ischemic heart tissue compared to normal heart and to the other control tissues with relative phage binding units 216 on Round 3 in ischemic LV and less than 3 in all other organ tissues (Figure 2). Following the third round, we identified and analyzed the amino acid sequences displayed by the phage clones rescued from the ischemic region of LV, non-ischemic region of LV, and RV. In the ischemic LV, 79 of 180 clones had the sequence CSTSMLKAC (occurrence rate 44%), 20 clones had CKPGTSSYC (11%) and 18 clones had CPDRSVNNC (10%). In non-ischemic LV, 14 of 84 sequenced clones had CWEPMNHLC (17%), 13 clones had CPFASYLHC (16%), 9 clones had CENVWYPRC (11%). In RV, 13 of 89 sequenced clones had CQSTMSTLC (15%), 9 clones had CPTSFLTDC (10%), and 7 clones had CYDLRSHQC (8%). Using a BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and considering a minimum of tri-peptide sequence matching, we compared the nine peptide motifs that were enriched in the heart tissue (either in LV or RV), to known proteins with special attention to those with bio-functional relevance to cardiac ischemia. Based on sequence homology, we identified a number of strong matching candidates. Examples of candidate proteins potentially mimicked by the heart-homing peptides are listed in Table 1. Three peptide sequences CSTSMLKAC, CKPGTSSYC, and CPDRSVNNC identified from ischemic heart tissue with highest occurrence rate have some homologies with proteins that may be involved in ischemic heart diseases, and we then tested their homing capability to ischemic myocardium in vivo.
Figure 2. In vivo biopanning resulted in successful enrichment for ischemic heart homing phage.
Three rounds of panning using the Ph.D.-C7C random phage library in a rat model of ischemia-reperfusion yielded strong enrichment for peptide motifs that exhibited preferential binding to ischemic heart tissue compared to the normal heart and other control tissues. Phage binding units were normalized between panning rounds relative to the binding units of the M13KE control insert-less phage in various tissues.
Table 1.
Sequences of high frequency peptides recovered from heart tissues following the third biopanning round in an ischemia-reperfusion rat model and matching motifs in candidate proteins mimicked by the phage inserts.
| Peptide Motifs (Frequency) | Matching Motifs in Candidate Protein Mimics | Accession Number (UniProtKB/Swiss-Prot) | |
|---|---|---|---|
| Ischemic Region of LV | C-STSM5LKA-C (44%) | 34834- STSKMLKA -34841 (Titin) | Q8WZ42 |
| 531- TSMLKA -536 (OPA-1) | Q2TA68 | ||
| 716-MLKA-719 (Dynamin-1 like protein, DRP-1) | O00429 | ||
| C-KPGTSSY-C (11%) | 889- CKPGTTGRYC -898 (Laminin, alpha 2) | P24043 | |
| 705- GTSSYC -710 (Brain-specific angiogenesis inhibitor 2) | O60241 | ||
| C-PDRSVNN-C (10%) | 747- DRSVENC -753 (Talin 2) | Q9Y4G6 | |
| 3869- CPDRTV -3874 (Dynein) | Q96JB1.2 | ||
| Non-ischemic Region of LV | C-WEPMNHL-C (17%) | 698- PMNHL -702 (Oxoglutarate dehydrogenase) | Q02218 |
| 961- PMTHLC -966 (Death-associated protein kinase-1) | P53355.5 | ||
| C-PFASYLH-C (16%) | 1375- PFASYL -1380 (Centlein) | Q9NXG0 | |
| 264- PFASYGLH -271 (Neutral sphingomyelin phosphodiesterase) | Q9NXE4 | ||
| C-ENVWYPR-C (11%) | 192- NIWYP -196 (Purinoceptor 4) | Q99571 | |
| 315- WFPRC (Inhibitor of apoptosis protein-1) | Q13489 | ||
| RV | C-QSTMSTL-C (15%) | 352- QSTMSTL -358 (Epidermal growth factor receptor 5) | Q86T13 |
| 132- QSTMS -136 (Tensin 4) | Q81ZW8 | ||
| C-PTSFLTD-C (10%) | 1001- CPTGFLGTDC -1010 (Multiple EGF-like domain-6) | O75095.3 | |
| 65- CPTSWVTD -72 (Cytochrome c oxidase subunit Vib polypeptide 1) | P14854.2 | ||
| C-YDLRSHQ-C (8%) | 324- CYDLRS -329 (ADAMTS-like1) | Q8N6G6 | |
| 510- YDLRSQ -515 (Schlafen family member 12) | Q8IYM2 | ||
CSTSMLKAC peptide homes to ischemic myocardium
Three peptide sequences identified in the in vivo screen were validated for their homing potential to ischemic heart tissue using fluorescein-conjugated synthetic peptides. Peptides were observed as green fluorescence signal in the examined tissue, and we assessed green fluorescence intensity relative to background fluorescence. CSTSMLKAC in the ischemic LV showed relative fluorescence intensity of 1.60±0.059, which was significantly higher than that measured for control peptides (1.19±0.031, p<0.001 for linear peptide, 1.32±0.048, p<0.01 for scrambled, and 1.38±0.054, p<0.001 for PBS only); moreover, it was significantly higher compared to CSTSMLKAC in the normal LV, with relative fluorescence intensity of 1.12±0.013 (p=0.001) (Figure 3A). The other two peptide motifs did not show any statistical differences compared with the negative controls in the examined ischemic heart tissue or in comparison to the normal LV (Figures 3B and 3C). These results indicate that among the three enriched peptide motifs that were originally identified in the in vivo screen, the CSTSMLKAC peptide was the only peptide with preferential homing to ischemic cardiac tissue. Figure 4A showed that the CSTSMLKAC peptide is localized in the ischemic portion (anterior wall) of the myocardium, compared to the non-ischemic region (posterior wall). However, it is possible that the peptide can localize to not only cardiomyocytes but also other cell types, such as vascular smooth muscle cells. To address this, aortic tissues were harvested following myocardial ischemia-reperfusion injury. No significant accumulation of the peptide was detected in either endothelial or smooth muscle cells in the aortic tissue (Figure 4C and F). These results suggest that fluorescein-tagged CSTSMLKAC peptide is primarily concentrated in cardiomyocytes in the ischemic myocardium but not by cells in non-ischemic LV or in non-coronary blood vessels.
Figure 3. The CSTSMLKAC peptide homes to ischemic myocardium.
Peptide sequences CSTSMLKAC, CKPGTSSYC, and CPDRSVNNC were tested for their homing capability to ischemic left ventricle (LV) as synthetic peptides conjugated with fluorescein in vivo. Peptides were injected into a rat heart with ischemia-reperfusion. The CSTSMLKAC peptide showed significantly higher intensity in the ischemic LV compared to the negative controls: linear form of the peptide (***p<0.001), scrambled peptide with the same amino acids of peptide (**p<0.01), and PBS (*p<0.05) (A). The other two peptide motifs did not show any significant differences compared to the negative controls and the non-ischemic tissue (B and C).
Figure 4. Fluorescein-conjugated CSTSMLKAC concentrated in cardiomyocytes in ischemia-reperfusion injured myocardium.
Following ischemia-reperfusion injury, rats were treated with 1 mg of either fluorescein-CSTSMLKAC (A, B and C) or fluorescein-scrambled peptide (D, E and F) by intravenous injections. Hearts and aortic tissues were harvested and fixed 10 minutes after injection. Histological sections were stained with Isolectin-IB4 to stain endothelium (red) and DAPI to stain nuclei (blue). Panel A and D show the anterior wall of left ventricle (LV) of the ischemic region, and B and E show the posterior wall of LV of the non-ischemic region. Fluorescein-CSTSMLKAC (green), but not scrambled peptide, was concentrated in cardiomyocytes in the injured myocardium, while fluorescein-CSTSMLKAC did not concentrate in cells in aortic tissues (C and F). Scale bars represent 50 μm.
To determine temporal effects of CSTSMLKAC peptide on cardiac function, rats were subjected to ischemia/reperfusion injury, and hemodynamic and echocardiographic parameters were measured 24 hours after systemic administration of fluorescein-tagged CSTSMLKAC, scrambled peptide, or PBS (n=4 each). There were no significant differences in heart rate, end-systolic pressure, end-diastolic pressure, stroke volume, ejection fraction, cardiac output, cardiac index by catheterization, or % fractional shortening by echocardiography between the homing peptide group and other groups (Table 2). All groups had similar LV volumes and wall thickness. These results suggest that the administration of the homing peptide did not induce any significant impairment in LV contractile function.
Table 2.
Hemodynamic and echocardiographic assessment of systemic administration of fluorescein- CSTSMLKAC on heart function in rats after ischemia/reperfusion injury.
| PBS | Homing peptide | Scrambled peptide | |
|---|---|---|---|
| Hemodynamic measurement | |||
| Heart rate [beats/min] | 444.8±13.14 | 454.75±13.88 | 430.75±20.59 |
| End-systolic volume [μl] | 309±33.9 | 280.76±15.91 | 260.91±10.99 |
| End-diastolic volume [μl] | 406.1±42.53 | 393.56±25.62 | 362.72±14.82 |
| End-systolic pressure [mmHg] | 107.5±9.2 | 119±3.16 | 120.1±3.48 |
| End-diastolic pressure [mmHg] | 3.87±0.35 | 3.96±0.67 | 3.08±0.22 |
| Stroke volume [μl] | 97.15±14.58 | 112.82±13.61 | 101.81±13.74 |
| Ejection fraction [%] | 23.79±2.79 | 28.44±2.09 | 27.89±3.01 |
| Cardiac output [μl/min] | 42640±5533 | 51430±6708 | 44530±7633 |
| Cardiac index [ml/min per 100 g] | 20.38±2.71 | 19.87±2.54 | 20.96±4.48 |
| Echocardiography | |||
| Anterior wall thickness [mm] | 2.01±0.06 | 2.00±0.04 | 2.08±0.08 |
| Posterior wall thickness [mm] | 1.81±0.09 | 1.73±0.10 | 1.83±0.06 |
| End-systolic dimension [mm] | 3.24±0.20 | 2.49±0.51 | 2.93±0.18 |
| End-diastolic dimension [mm] | 5.36±0.10 | 5.33±0.36 | 5.16±0.23 |
| % Fractional shortening | 39.39±4.35 | 54.05±6.72 | 43.20±3.22 |
| Left ventricle mass [g] | 655±30.24 | 635±84.31 | 646±55.48 |
| LVM/BW | 3.13±0.10 | 2.41±0.23 | 2.94±0.32 |
| Body weight [g] | 209.3±7.36 | 260.25±13.14 | 224±22.01 |
Specific delivery of Sumo-mCherry to ischemic cardiac tissue in mice by the CSTSMLKAC peptide
The molecular mass of fluorescein is 332 Da and total mass of fluorescein-CSTSMLKAC is 1.2 kDa. To test whether the CSTSMLKAC peptide motif can deliver a protein moiety - with a much greater molecular weight - to ischemic cardiac tissue, we designed, expressed and purified a recombinant protein comprised of Sumo-mCherry conjugated to CSTSMLKAC peptide with a total molecular weight of 40 kDa. As shown in Figure 5A, the recombinant protein was easily detected with commercially available antibodies. To quantify Sumo-mCherry in tissue homogenates, we developed a sandwich ELISA using anti-sumo antibody for the capturing step and biotinylated anti-mCherry antibody for detection of analytes. On each ELISA plate, we ran a standard curve with an R-squared greater than 0.99 in the range from 0.01 to 5 ng/mL. To minimize the amount of recombinant protein used, and to determine if the peptide homing strategy was effective in other species, we performed these experiments in mice.
We analyzed post-ischemic cardiac tissue samples using Western analysis by probing with an anti-mCherry antibody (rabbit anti-RFP antibody, Abcam) (Figure 5B). PBS was loaded as a null control to show non-specific detection of proteins in post-ischemic cardiac tissue. Western analysis showed clear homing of the protein by the CSTSMLKAC motif. ELISA with the same tissue samples was performed to quantitate the homing of Sumo-mCherry in the ischemic LV and the non-ischemic LV (Figure 5C). The unmodified peptide conjugated to Sumo-mCherry showed a statistically significant increase in affinity for post-ischemic mouse cardiac tissue compared to the three controls (dark bars, Figure 5C). The final protein concentration of the peptide-complex was quantified as 4.0±1.4 μg/g tissue (N=6). This result precludes the possibility that the Sumo-mCherry complex itself is responsible for the increased affinity and also shows sequence specificity requiring the flanking cysteines for the homing property of our peptide. Compared to the non-ischemic tissue from the same mouse heart, our unmodified peptide showed the greatest specificity towards the post-ischemic tissue, increasing the affinity by 11.4-fold with p<0.05 (Figure 5C). Interestingly, Sumo-mCherry itself (without peptide conjugation) showed a statistically increased affinity to post-ischemic tissue by 2.7-fold with a p<0.0001 by unpaired t-test (Figure 5C), possibly due to the increased vascular permeability, edema and/or the decreased vascular clearance from the post-ischemic area resulting in trapping of large molecules. The scrambled amino acid sequence also showed an increased affinity by 3.4-fold for post-ischemic tissue with statistically significant with p<0.01, suggesting that some local amino acid content may provide some homing capability even without the specific sequence.
To further characterize the specificity and the bio-distribution of Sumo-mCherry-CSTSMLKAC in mice, we homogenized tissues after intravenous administration of the peptide-complex and analyzed them using quantitative ELISA (Figure 6). We conducted the experiments on three mice and aggregated average data from duplicate wells from the same ELISA plate. We found a significantly increased amount of peptide-Sumo-mCherry complex (4.0±1.4 μg/g of tissue) in the post-ischemic LV compared to tissues from non-ischemic LV, RV, lung, liver, spleen, skeletal muscle, and brain, (all p<0.001). The amount of protein complex found in the kidney was the second highest at 1.99 ±0.13 μg/g of tissue, possibly due to renal clearance or glomerular entrapment of the complex.
Figure 6. Biodistribution of Sumo-mCherry-CSTSMLKAC was measured with quantitative ELISA after intravenous injection in mice.
The concentration of peptide-Sumo-mCherry complex (4.0 ±1.4 μg/g of tissue) were significantly increased in the ischemic left ventricle (LV) compared to other organ tissues (non-ischemic LV, right ventricle (RV), lung, liver, spleen, skeletal muscle, and brain) with a p<0.001 (indicated with *). N=6 per group in three independent mice.
Discussion
Our results demonstrate that a peptide motif identified by a phage display screen can target proteins selectively to ischemic myocardium. Intravenous administration of the fusion protein carried by the peptide could selectively home to the ischemic myocardium relative to the non-ischemic myocardium. The significance of these experiments is the potential for a new method that can deliver bioengineered proteins to the heart by systemic intravenous administration for stimulating cardiac repair during and after myocardial ischemia.
To discover tissue-specific peptide binding sequences, we used phage display for high-throughput screening of protein interactions, and it was applied in vivo “biopanning”. In vivo biopanning comprises a multiple-round phage display screen in a living animal [18]. The peptide-ligands can serve as “peptidomimetic” leads for the development of targeted therapeutics, imaging agents, and diagnostic markers in many diseases [19–22]. Zhang and colleagues employed in vivo phage display to identify a number of peptides with selective binding properties to normal heart endothelium [23]. Their peptide sequences identified by phage-display technology could specifically target cardiac endothelium. The heart specificity of a specific sequence, CRPPR, was 300-fold higher compared to control phage, suggesting that the CRPPR sequence is effective for cardiac targeting. Thus, these peptide sequences provide a new method for protein delivery to the myocardium. One of the differences between their study and ours is that their phage display approach was to target toward normal hearts, while we targeted ischemic myocardium [23]. In addition, they employed a method in which, prior to in vivo biopanning with the selected phage pool, the peptides were screened by ex vivo panning in cultured endothelial cells for phage libraries. In the present study, we hypothesized that cardiac injury might lead to more preferential targeting to the injured myocardium, perhaps by allowing more binding sites for peptides to be exposed in ischemic cells. We therefore performed in vivo bio-panning to avoid the potential loss of peptide candidates.
Our results demonstrate that this screen yielded a new peptide that was capable of preferentially binding to the ischemic myocardium. Although the precise mechanism by which this peptide binds to the ischemic myocardium remains unclear, it might mimic natural ligands that bind to receptors exposed by the ischemic injury. A cytoskeletal protein titin was identified as a protein with sequence similarity to the peptide CSTSMLKAC. Previous reports suggest that ischemia promotes the binding of alpha-B crystalline to the I-band portion of cardiac titin in cardiomyocytes, thereby stabilizing titin against denaturation by ischemic damage [24]. This raised the hypothesis that the mechanism by which CSTSMLKAC is concentrated in ischemic cardiomyocytes may be mediated by the direct interaction of CSTSMLKAC with alpha-B crystalline. To test this hypothesis, we evaluated binding of CSTSMLKAC and alpha-B crystalline using the Surface Plasmon Resonance assay. Biotinylated CSTSMLKAC was hybridized to streptavidin coated chips and mounted in a Biacore 2000 device. As shown in Supplemental Figure 1, a significant interaction between CSTSMLKAC and alpha-B crystalline was not demonstrated with this technique. While this biochemical experiment does not exclude the possibility that ischemia induces the binding of the homing peptide with alpha-B crystalline in vivo, further studies are necessary to address the precise mechanism by which the peptide is concentrated by ischemic myocardium.
This approach could be used to identify receptors for the ischemia-homing peptides and their functional relevance. For instance, we identified proteins mimicked by these peptide motifs as; Optic Atrophy-1 (OPA-1), and Dynamin-1 like protein (DLP-1). OPA-1 is a protein that is essential for normal mitochondrial fusion. It has been reported that ischemia markedly decreases OPA-1 levels in the heart and this leads to further apoptosis [25]. DLP-1 is also required protein for mitochondrial fission [26]. This suggests importance of proteins involved in mitochondrial fusion and fission in ischemia-reperfusion injury. Future identification of the receptor(s) targeted by the ischemia homing peptide may provide new insights into the molecular mechanisms underlying ischemic injury.
Short peptide sequences can be incorporated into fusion proteins, which would theoretically lead to increased targeting of these moieties to the heart. Therefore, we expressed and purified the Sumo and mCherry expression proteins with the targeting peptide, which were used for detection in vitro and in vivo. The results imply the possibility that this approach can be used for other fusion proteins to target injured myocardium. While this study describes a proof-of-concept with multiple in vivo validations, a thorough examination of a new fusion of peptide and a therapeutic drug will be necessary for proving the benefits of therapeutic targeting. As an example, cyclosporine is widely used as an immunosuppressive agent and significantly reduces infarct size and improves LV function after myocardial ischemia [27, 28], but systemic use of cyclosporine has several potentially detrimental effects such as renal and hepatic toxicity and hypertension. The conjugation of cyclophilin inhibitory molecules or other drugs with the targeting peptides could represent a new therapeutic strategy for the treatment of myocardial infarction.
Before use in humans, multiple issues must be considered. The long-term toxicity of this peptide and specificity in all species is unknown. For instance, any delivered protein can be immunogenic, and fusion proteins with added epitopes may lead to antibody responses. There is potential for any peptide sequence to elicit an immunological response, or an unexpected toxicity. Furthermore, inter-species differences could influence pharmacokinetics and receptor interactions. Figure 6 shows that Sumo-mCherry conjugated CSTSMLKAC accumulated in kidney as well as ischemic injured myocardium, which may raise a limitation in human use. Therefore, we determined whether molecular size has any impact on its distribution in vivo. Our results are consistent with the physiological principle that small peptides and larger proteins are handled differently in the kidney (Supplemental Figure 2). In this study, we did not determine the half-life of the peptide itself in the circulation as this would not reflect the pharmacokinetics of fusion proteins with the peptide sequence. Furthermore, it is important to consider that N-terminus or C-terminus fusion of peptide sequences could alter not only pharmacokinetics but receptor dynamics and immunological properties of potential therapeutic proteins. Thus, further studies are necessary to address these issues. In addition, our experiments primarily used internal jugular injections to mimic venous delivery, but more peripheral delivery such as limb injection could be more relevant to intravenous limb injection in humans.
In conclusion, we showed that a phage display-based approach resulted in identification of a new peptide that can preferentially target ischemic myocardium. Peptides discovered in this manner could be used for specific targeting of a fusion protein to the ischemic region of the myocardium by intravenous injection. This approach could lead to a novel approach to cardiac repair: systemic administration of bioengineered proteins can be delivered to a targeted injured tissue.
Supplementary Material
Acknowledgments
This work was supported by American Heart Association Post-doctoral Fellowships (to SK and DEJ), a fellowship of the Koeln Fortune Program/Faculty of Medicine, University of Cologne, Cologne, Germany (to SL), and an NIH grant HL092930 (to RTL).
Footnotes
Conflict of Interest: Richard T. Lee is a founder of and consultant for Provasculon.
Disclosures
Richard T. Lee is a founder of and consultant for Provasculon.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keeley EC, Boura JA, Grines CL. Primary angioplasty versus intravenous thrombolytic therapy for acute myocardial infarction: a quantitative review of 23 randomised trials. Lancet. 2003 Jan 4;361(9351):13–20. doi: 10.1016/S0140-6736(03)12113-7. [DOI] [PubMed] [Google Scholar]
- 2.Putnam D. Drug delivery: the heart of the matter. Nat Mater. 2008 Nov;7(11):836–7. doi: 10.1038/nmat2309. [DOI] [PubMed] [Google Scholar]
- 3.Ye X, Yang D. Recent advances in biological strategies for targeted drug delivery. Cardiovasc Hematol Disord Drug Targets. 2009 Sep;9(3):206–21. doi: 10.2174/187152909789007025. [DOI] [PubMed] [Google Scholar]
- 4.Paschke M. Phage display systems and their applications. Appl Microbiol Biotechnol. 2006 Mar;70(1):2–11. doi: 10.1007/s00253-005-0270-9. [DOI] [PubMed] [Google Scholar]
- 5.Sergeeva A, Kolonin MG, Molldrem JJ, Pasqualini R, Arap W. Display technologies: application for the discovery of drug and gene delivery agents. Adv Drug Deliv Rev. 2006 Dec 30;58(15):1622–54. doi: 10.1016/j.addr.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem Rev. 2005 Nov;105(11):4056–72. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 7.Koivunen E, Arap W, Rajotte D, Lahdenranta J, Pasqualini R. Identification of receptor ligands with phage display peptide libraries. J Nucl Med. 1999 May;40(5):883–8. [PubMed] [Google Scholar]
- 8.Mintz PJ, Kim J, Do KA, Wang X, Zinner RG, Cristofanilli M, et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003 Jan;21(1):57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 9.Pini A, Giuliani A, Ricci C, Runci Y, Bracci L. Strategies for the construction and use of peptide and antibody libraries displayed on phages. Curr Protein Pept Sci. 2004 Dec;5(6):487–96. doi: 10.2174/1389203043379323. [DOI] [PubMed] [Google Scholar]
- 10.Adda CG, Anders RF, Tilley L, Foley M. Random sequence libraries displayed on phage: identification of biologically important molecules. Comb Chem High Throughput Screen. 2002 Feb;5(1):1–14. doi: 10.2174/1386207023330561. [DOI] [PubMed] [Google Scholar]
- 11.Samoylova TI, Smith BF. Elucidation of muscle-binding peptides by phage display screening. Muscle Nerve. 1999 Apr;22(4):460–6. doi: 10.1002/(sici)1097-4598(199904)22:4<460::aid-mus6>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 12.Kolonin MG, Saha PK, Chan L, Pasqualini R, Arap W. Reversal of obesity by targeted ablation of adipose tissue. Nat Med. 2004 Jun;10(6):625–32. doi: 10.1038/nm1048. [DOI] [PubMed] [Google Scholar]
- 13.Sakai J, Ishikawa H, Kojima S, Satoh H, Yamamoto S, Kanaoka M. Proteomic analysis of rat heart in ischemia and ischemia-reperfusion using fluorescence two-dimensional difference gel electrophoresis. Proteomics. 2003 Jul;3(7):1318–24. doi: 10.1002/pmic.200300432. [DOI] [PubMed] [Google Scholar]
- 14.White MY, Cordwell SJ, McCarron HC, Prasan AM, Craft G, Hambly BD, et al. Proteomics of ischemia/reperfusion injury in rabbit myocardium reveals alterations to proteins of essential functional systems. Proteomics. 2005 Apr;5(5):1395–410. doi: 10.1002/pmic.200400995. [DOI] [PubMed] [Google Scholar]
- 15.De Celle T, Vanrobaeys F, Lijnen P, Blankesteijn WM, Heeneman S, Van Beeumen J, et al. Alterations in mouse cardiac proteome after in vivo myocardial infarction: permanent ischaemia versus ischaemia-reperfusion. Exp Physiol. 2005 Jul;90(4):593–606. doi: 10.1113/expphysiol.2005.030296. [DOI] [PubMed] [Google Scholar]
- 16.Kim N, Lee Y, Kim H, Joo H, Youm JB, Park WS, et al. Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics. 2006 Feb;6(4):1237–49. doi: 10.1002/pmic.200500291. [DOI] [PubMed] [Google Scholar]
- 17.Sakai J, Ishikawa H, Satoh H, Yamamoto S, Kojima S, Kanaoka M. Two-dimensional differential gel electrophoresis of rat heart proteins in ischemia and ischemia-reperfusion. Methods Mol Biol. 2007;357:33–43. doi: 10.1385/1-59745-214-9:33. [DOI] [PubMed] [Google Scholar]
- 18.Pasqualini R, Ruoslahti E. Organ targeting in vivo using phage display peptide libraries. Nature. 1996 Mar 28;380(6572):364–6. doi: 10.1038/380364a0. [DOI] [PubMed] [Google Scholar]
- 19.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998 Jan 16;279(5349):377–80. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 20.Rhyner C, Weichel M, Fluckiger S, Hemmann S, Kleber-Janke T, Crameri R. Cloning allergens via phage display. Methods. 2004 Mar;32(3):212–8. doi: 10.1016/j.ymeth.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Hajitou A, Pasqualini R, Arap W. Vascular targeting: recent advances and therapeutic perspectives. Trends Cardiovasc Med. 2006 Apr;16(3):80–8. doi: 10.1016/j.tcm.2006.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zurita AJ, Troncoso P, Cardo-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004 Jan 15;64(2):435–9. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L, Hoffman JA, Ruoslahti E. Molecular profiling of heart endothelial cells. Circulation. 2005 Sep 13;112(11):1601–11. doi: 10.1161/CIRCULATIONAHA.104.529537. [DOI] [PubMed] [Google Scholar]
- 24.Golenhofen N, Arbeiter A, Koob R, Drenckhahn D. Ischemia-induced association of the stress protein alpha B-crystallin with I-band portion of cardiac titin. J Mol Cell Cardiol. 2002 Mar;34(3):309–19. doi: 10.1006/jmcc.2001.1513. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial OPA1, apoptosis, and heart failure. Cardiovasc Res. 2009 Oct 1;84(1):91–9. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hom J, Sheu SS. Morphological dynamics of mitochondria--a special emphasis on cardiac muscle cells. J Mol Cell Cardiol. 2009 Jun;46(6):811–20. doi: 10.1016/j.yjmcc.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mewton N, Croisille P, Gahide G, Rioufol G, Bonnefoy E, Sanchez I, et al. Effect of cyclosporine on left ventricular remodeling after reperfused myocardial infarction. J Am Coll Cardiol. 2010 Mar 23;55(12):1200–5. doi: 10.1016/j.jacc.2009.10.052. [DOI] [PubMed] [Google Scholar]
- 28.Piot C, Croisille P, Staat P, Thibault H, Rioufol G, Mewton N, et al. Effect of cyclosporine on reperfusion injury in acute myocardial infarction. N Engl J Med. 2008 Jul 31;359(5):473–81. doi: 10.1056/NEJMoa071142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.