Abstract
Virus capsid assembly requires recruiting and organizing multiple copies of protein subunits to form a closed shell for genome packaging that leads to infectivity. Many viruses encode scaffolding proteins to shift the equilibrium towards particle formation by promoting inter-subunit interactions and stabilizing assembly intermediates. Bacteriophage HK97 lacks an explicit scaffolding protein, but the capsid protein (gp5) contains a scaffold-like N-terminal segment termed the delta domain. When gp5 is expressed in E. coli, the delta domain guides 420 copies of the subunit into a procapsid with T=7 laevo icosahedral symmetry named Prohead-I. Prohead-I can be disassembled and reassembled under mild conditions and it cannot mature further. When the virally encoded protease (gp4) is co-expressed with gp5, it is incorporated into the capsid and digests the delta domain followed by auto-proteolysis to produce the meta-stable Prohead-II. Prohead-I+P was isolated by co-expressing gp5 and an inactive mutant of gp4. Prohead-I and Prohead-I+P were compared by biochemical methods, revealing that the inactive protease stabilized the capsid against disassembly by chemical or physical stress. The crystal structure of Prohead-I+P was determined at 5.2Å resolution and distortions were observed in the subunit tertiary structures similar to those observed previously in Prohead-II. Prohead-I+P differed from Prohead-II due to the presence of the delta domain and the resulting repositioning the N-arms, explaining why Prohead-I can be reversibly dissociated and cannot mature. Low-resolution X-ray data enhanced the density of the relatively dynamic delta domains, revealing their quaternary arrangement and suggesting how they drive proper assembly.
Keywords: Virus crystallography, HK97 bacteriophage, virus assembly, virus maturation, capsid dynamics, viral protease, scaffold protein
Introduction
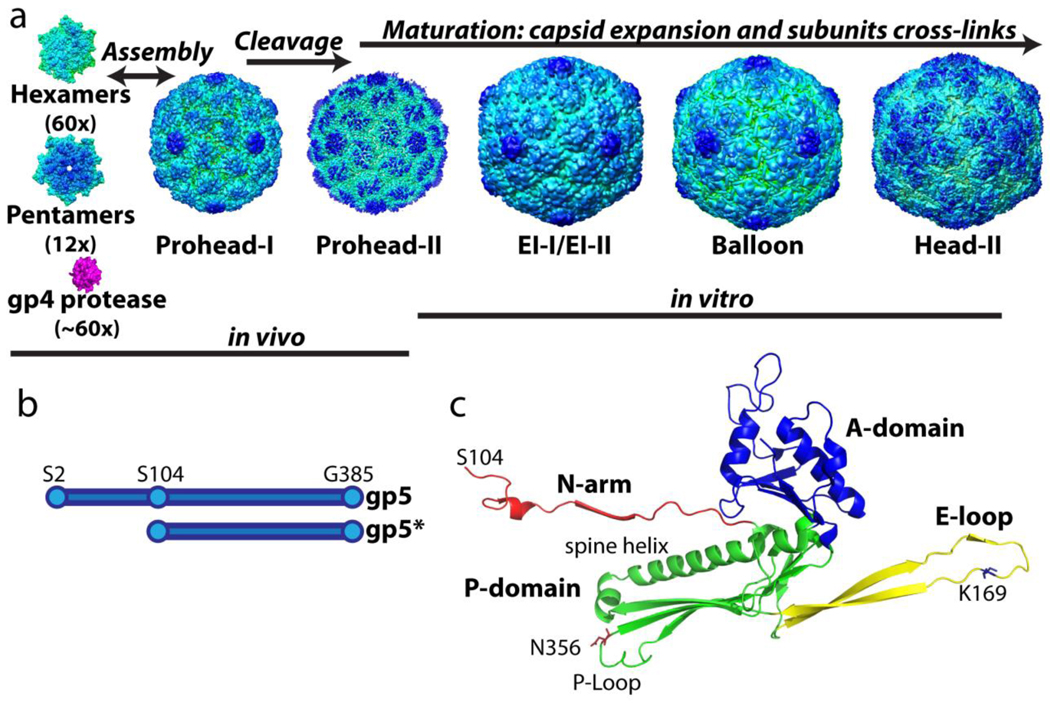
Virus capsid maturation is the process by which a non-infectious, initial assembly product gains stability and infectivity through a series of particle transformations, often involving chemical and physical reorganization of the initial constituents. The process is required in most complex viruses because initial assembly generally involves weak, moderately reversible, interactions (~2–4kBT) to allow self-correcting and annealing of the assembling subunits1; 2. Subsequently these weak interactions must be transformed into robust associations to protect the packaged viral genome and deliver it to the host cell. Bacteriophage HK97 is among the most accessible systems for such studies, since maturation can be performed in vitro with virus-like-particles (VLPs) prepared by expressing the appropriate genes in E. coli3. HK97 is a double stranded DNA bacteriophage that employs a sophisticated and efficient assembly and maturation pathway. The N-terminal delta domain (residues 2–103) of the gp5 major capsid protein is responsible for facilitating and guiding proper formation of the first assembly product called Prohead-I (Figure 1A). The delta domain maintains solubility of the assembly intermediates by preventing the capsid subunits from inappropriately aggregating and prevents premature capsid expansion4. The Prohead-I particle can be disassembled readily to capsomers (hexamers and pentamers) and these properly reassemble in vitro under mild conditions5. Maturation of Prohead-I can only occur following removal of the delta domains.
Figure 1.
The assembly and maturation pathway of HK97 virus-like-particles. (A) The hexamers and pentamers, composed of full length gp5 (residue 2–385 shown in B,) co-assemble with the viral protease gp4 to form Prohead-I. Once the assembly is complete, the protease cleaves off the delta domain (residue 2–103) followed by auto-proteolysis to form Prohead-II composed of gp5*. Capsid assembly and delta domain proteolysis take place in the E. coli expression system. When Prohead-II is purified by previously described methods, it can be matured in vitro under a variety of conditions8; 37. The final mature capsid, Head-II, contains catenated networks (“chain-mail”) formed by isopeptide covalent bonds between subunits. (C) The crystal structure of the Head-II capsid subunit. The two domains and two extensions are color coded. Cross-links join lysine 169 on the E-loop of one subunit to asparagine 356 on the P-domain of the neighboring subunit. The delta domain, which is absent in Prohead-II and Head-II, is connected to the N-arm in capsomers and in Prohead-I.
During assembly approximately 60 copies of the HK97-encoded serine protease (gp4) are incorporated into the Prohead-I particle6. The encapsidated proteases are believed to be activated through dimerization, a process known to activate the herpesvirus protease7, that then proteolyze the delta domains at multiple sites. The digested polypeptide fragments of the delta domains and the auto-proteolyzed gp4 apparently exit through ~10Å pores between capsomer junctions as the capsid matures to Prohead-II. Prohead-II, formed by gp5* subunits (containing residues 104–385) (Figure 1B), is meta-stable and primed to mature through the intermediates shown in figure 1A to the fully expanded and cross-linked mature capsid, Head-II8; 9; 10. Although the protease is not required for Prohead-I formation, it is essential for producing Prohead-II5. Prohead-II can be matured in vitro using a variety of chemical and physical perturbations8; 11. During maturation the capsid subunits undergo multiple tertiary and quaternary conformational changes and the capsid diameter expands from 560 to 660Å. These conformational changes allow lysine 169 and asparagine 365 of neighboring capsomer subunits to form an auto-catalytic isopeptide linkage. The mature particle is fully cross-linked by these residues into a unitary chain-mail structure that can withstand the high internal pressure of the packaged DNA chromosome12.
The HK97 Prohead-II, expansion intermediates, and the Head-II maturation product have been characterized8; 13; 14. However, there is limited structural knowledge of Prohead-I. Prohead-I is interesting as it is the starting point for the maturational processing. Here we report the 5.2Å and 10Å X-ray crystal structures of Prohead-I+P, the recombinant particle assembled with a mutant, inactive protease that is closely similar to authentic Prohead-I, just prior to proteolysis. The structure revealed that there is subunit tertiary structure distortion correlated with the strength of quaternary interactions at three different classes of trimer contacts. The 10Å structure illuminates the role of the delta domain in capsid assembly, stabilization, and inhibition of maturation. Biophysical studies of Prohead-I with and without an inactive protease demonstrate that stabilization of the capsid is conferred by the packaged protease suggesting that its presence provides significant free energy favoring assembly.
HK97 is believed to share common ancestry with the herpes virus family based on similarities in subunit structure and maturation pathway15; 16; 17. A homology model of the viral protease, demonstrating structural similarity to the protease in herpes virus was previously established18. Close similarity between the two proteases suggests that both viruses utilize similar mechanisms to remove accessory proteins for regulating assembly. The homology model allows us to have a preliminary understanding of the biochemical and structural features of the HK97 protease.
Results
Prohead-I+P is composed of full length capsid protein (gp5) and viral protease (gp4)
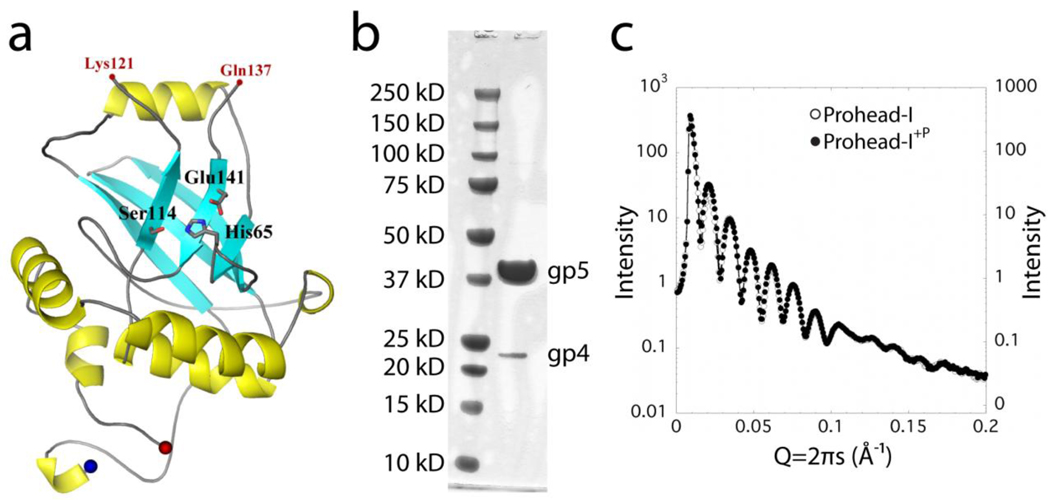
The authentic Prohead-I particle is a transient species that has a short life time in virus maturation and cannot be isolated in a pure form from the expression system. To determine the structure of the Prohead-I-protease complex, an inactive protease construct was made by mutating histidine 65 at the active site of the serine protease to an alanine. When this mutant protease was co-expressed with gp5, Prohead-I like particles referred to as Prohead-I+P were purified. These particles show no gp5 cleavage and contain the mutant gp4 protease (Figure 2B). Based on the gel densitometry, there were 60–120 copies of packaged protease, in good agreement with previously reported estimates5. Empty Prohead-I particles are made by expressing only gp5. The subunits spontaneously assemble into particles with a size and shape indistinguishable from Prohead-I+P, which was confirmed by essentially identical small angle x-ray scattering (SAXS) curves (Figure 2C).
Figure 2.
Prohead-I+P encapsidates enzymatically inactive gp4 protease. (A) The proposed model of gp4 protease generated using Herpes protease homology. The active site of the serine protease is shown in stick representation. (B) SDS-PAGE of Prohead-I with inactive protease. Prohead-I gp5 is at 42 kDa and the gp4 protease is at 24 kDa. Gp4 is incorporated into the Prohead-I capsid. (C) Overlapping curves from Prohead-I with and without protease in their SAXS patterns indicate that both forms have virtually identical particle shape and overall structure at this resolution.
A HK97 gp4 protease homology model was made using a computational algorithm described in Materials and Methods. The core of the protease appears similar to HSV-1 protease, also a serine protease. The active site residues (serine, histidine, and glutamate) are highlighted in figure 2A.
Disassembly of Prohead-I and Prohead-I+P
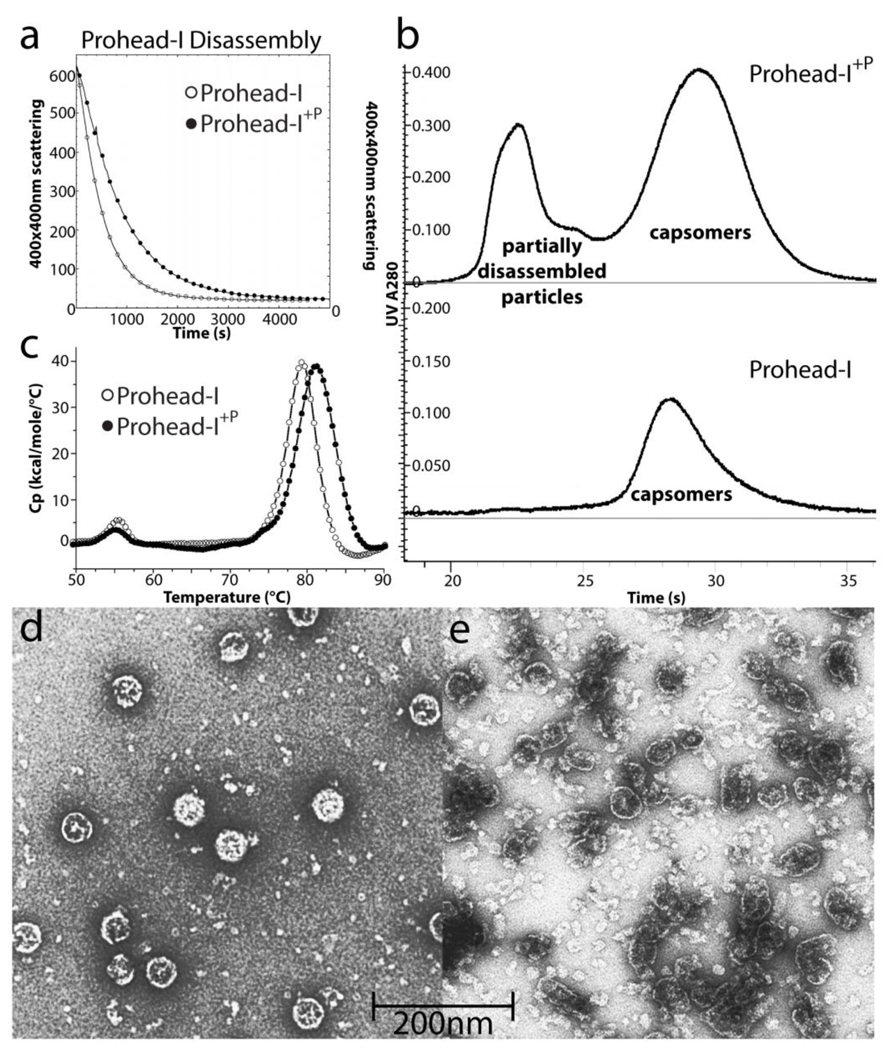
We compared Prohead-I and Prohead-I+P employing a previously established protocol that disassembles Prohead-I into capsomers.5 The disassembly process was monitored by light scattering, using light at 400 nm which is scattered strongly by assembled particles and weakly by dissociated capsomers (Figure 3A). Prohead-I disassembled more rapidly than Prohead-I+P. Moreover, the final state of disassembled Prohead-I was more homogeneous than Prohead-I+P after 12 hours incubation in the disassembly buffer (Figure 3B). Electron micrographs of negatively stained particles were recorded with the early elution fraction of Prohead-I+P from size exclusion chromatography showing that it was not completely disassembled.
Figure 3.
Biochemical comparisons of Prohead-I with and without protease. (A) Disassembly kinetics of Prohead-I and Prohead-I+P monitored by light scattering at 400nm. By incorporating inactive protease, Prohead-I capsid disassociated significantly slower than without protease, indicating capsid stabilization by protease-delta domain interactions. (B) Both forms of Prohead-I were loaded into Superdex 200 size exclusion column after disassembly overnight. Prohead-I+P remains partially disassembled while Prohead-I was completely disassociated after overnight in disassembly buffer. (C) The thermogram of both forms of Prohead-I analyzed by Differential Scanning Calorimetry. Prohead-I undergoes a major thermal event at 79°C while Prohead-I+P undergoes a similar transition at 82°C. The recorded thermal event corresponds to particle disassembly and protein denaturation since there is no additional peak up to 110°C (not shown). Three degrees of thermal stabilization is contributed by interacting with the protease. (D) Negative stain electron microscopy of intact Prohead-I+P and (E) partially assembled particles eluted from the size exclusion column show that subjecting these protease-containing particles to conditions that completely dissociate wild type Prohead-I causes severe damage to the particles but does not fully disassociate them.
Prohead-I+P and Prohead-I were further compared for their thermo-stability by Differential Scanning Calorimetry (DSC). Figure 3C shows that Prohead-I undergoes a major thermally-induced transition at 79.3°C ± 0.5°C that corresponds to particle disassembly and protein denaturation, whereas Prohead-I+P undergoes a comparable transition at 82.3°C ± 0.5°C (Figure 3C). Our results for Prohead-I agree well with previous DSC measurements19.
These data show that the protease stabilizes Prohead-I+P against both chemical and thermal stress, an observation consistent with its interactions with the interior surface of the capsid.
Crystallization of Prohead-I and Prohead-I+P
Crystallization studies of Prohead-I+P and Prohead-I were initiated to establish a structural basis for their properties. Isomorphous crystals diffracting to 7.5 Å resolution were obtained for both constructs, confirming that the two particle types have identical dimensions to that resolution. To obtain crystals that diffracted to higher resolution, a screen specific for virus crystallization was developed (see Materials and Methods) that covered a broad range of pH and salt conditions previously used in successful viral capsid crystallization studies20. Prohead-I was unstable and readily disassociated in crystallization buffers containing high salt or pH values above 9 as confirmed by a native agarose gel. Prohead-I+P was more stable than Prohead-I under all crystallization conditions and during cryo-stabilization. The best crystals were obtained with Prohead-I+P and they diffracted to 5.2Å resolution. These crystals were isomorphous with those described above (Table 1).
Table 1.
Crystallographic statistics.
| High Resolution Data | Low Resolution Data | |
|---|---|---|
| Space Group | I23 | I23 |
| Cell Dimension (Å) | a=b=c=560.120 | a=b=c=560.730 |
| Dmin-Dmax (Å) | 45-5.2 | 100-10 |
| # of reflections | 3,472,359 | 48,522 |
| Unique reflections | 86,130 | 15,010 |
| <I/σ(I)> | 11.4 (1.6) | 4 (1.5) |
| Completeness % | 77.5 (55.5) | 93.9 (92.5) |
| Rmerge % | 7.7 (26.1) | 25.2 (57.2) |
| Rave % | 16.3 (45.8) | 22.7 (36.9) |
| CCave % | 96.4 (43.5) | 86.3 (64.1) |
| Rcrys / Rfree %/ | 46.1/46.7 | n/a |
Values in parentheses are for the outer shell resolution bin (5.29-5.20) for the high resolution data and (10.36-10.00) for the low resolution data. Rmerge=(∑h ∑i(Ihi−<Ih>)/∑h ∑iIh) × 100. <Ih> is the mean of the Ihi observations of reflection h. <I/σ(I)> is the average signal-to-noise ratio of observed intensity (I).
CCave=∑((|FO|−<|FO|>)(|FC|−<|FC|>)/(∑(|FO|−<|FO|>)2∑(|FC|−<|FC|>)2)1/2. Rcryst=∑h|FO−FC|/∑hFO, FO is observed structure factor and FC is calculated structure factor.
Rave=∑h|FO−FC|/∑hFO, FO is observed structure factor and FC is calculated from a 5-fold NCS-averaged electron density map.
The structure determination of Prohead-I+P is described in Materials and Methods. Briefly, the structure was determined by molecular replacement with Prohead-II as the starting model. Phase extension was initiated at 45Å resolution, to eliminate model bias, and extended to 5.2 Å resolution in shells, corresponding to single steps in the reciprocal lattice, by applying non crystallographic 5-fold symmetry and solvent leveling. The final phase-refined electron density map at 5.2 Å resolution clearly revealed the gross tertiary structure of the capsid protein and quaternary interactions. A polyalanine model, based on this density and informed by known previous crystal structures of the subunit, was constructed. While there was clear similarity to the Prohead-II subunit tertiary structure, there were many features, described below, that were different, confirming the unique structure and role of Prohead-I+P in HK97.
Structural Features of Prohead-I+P
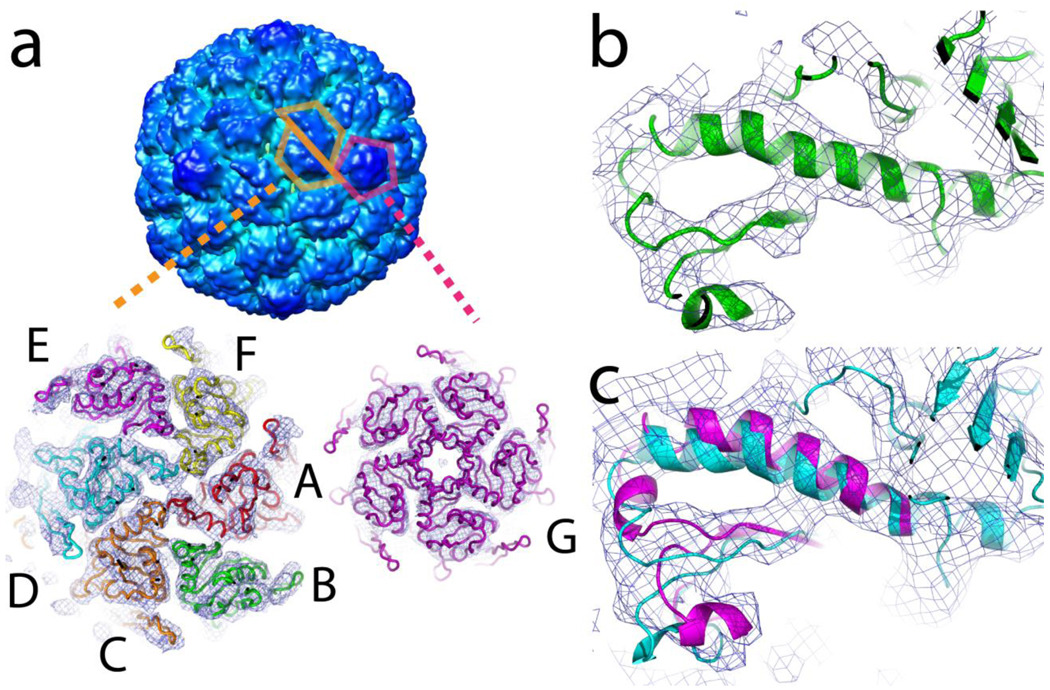
A signature feature of Prohead capsids, readily observable in moderate resolution cryoEM image reconstructions4, is the pseudo 2-fold symmetry of the hexamers that relates two asymmetric trimers, one composed of subunits A, F, and E and the other by subunits B, C, and D (Figure 4A). The skewed hexamers become 6-fold symmetric in the first expansion intermediate9. Virtually exact 6-fold symmetry is maintained in all subsequent particles 13 including the end point Head-II10. The pentamers, following the global icosahedral symmetry, are symmetric throughout the maturation process.
Figure 4.
Skewed hexamers and bent helices. (A) The hexamers of Prohead-I+P exhibit a skewed morphology with psudo-2-fold symmetry that relates subunits A, F, and E, to D, C, and B respectively. After proteolysis of the delta domains by the gp4 protease, the symmetry of the skewed hexamers remains the same. Following capsid expansion, the hexamers become 6-fold symmetric. The pentons, however, maintain 5 fold symmetry throughout the maturation pathway. (B) The atomic coordinates of Prohead-II were fitted as a rigid unit into the Prohead-I+P electron density map at 5.2Å resolution. The spine helix of subunit B shown in green exhibits the most bending in Prohead-I and is closely similar to that observed in Prohead-II. This helix straightens as the capsid expands during maturation. (C) The spine helix in subunit E of Prohead-I+P shown in magenta required remodeling to fit the Prohead-I+P electron density map and exhibits less distortion than in Prohead-II (shown in cyan). This behavior correlates to the broken Class 2, quasi 3-fold interaction described in Figure 6. All densities were contoured at 1.5 σ.
The HK97 subunit has two principal domains, termed axial (A) and peripheral (P) (Figure 1C)10. The A domain is a compact α/β structure that interacts with neighboring subunits in the same capsomer, around the corresponding 5-fold or quasi-6-fold symmetry axis at the center of each capsomer. The P domain mediates inter-capsomer contacts at the icosahedral and quasi 3-fold axes where the capsomers join together. The P-domain consists of the spine helix (residue 200–239) as its core, with the P (peripheral)-loop and E (extended)-loop lying at opposite ends of the P domain, on the perimeter of the capsomer. It is the P-loop and immediately surrounding residues that make the initial 3-fold contacts that are found in Prohead-I+P and Prohead-II and are discussed in more detail below. In the Head-II model, the N-arm begins at residue 104 (the first residue after the site of gp4 protease cleavage), and forms an intra-capsomer beta structure with a neighboring subunit. The core structures of the seven subunits in the icosahedral asymmetric unit of Head-II are closely similar with only small differences found in the E-loops and the N-arms. The subunit secondary and tertiary structures show no unusual characteristics.
In contrast to the conformational uniformity among capsid proteins in Head-II, the Prohead-I+P structure revealed that the subunits in the icosahedral asymmetric unit are significantly different from each other. The spine helices are bent and unwound to varying degrees among the seven subunits. There is a striking correlation between the degree of helix bending and the location of the subunit in the skewed hexamer. The B and E subunits, related by the pseudo 2-fold symmetry, are furthest from this axis. The helix in the B subunit is bent the most. Although the E and B subunits are virtually equivalent distances from the pseudo 2-fold axis, the E subunit is only moderately bent and is comparable to the helices in the C and F subunits. The A and D subunits are positioned closest to the pseudo 2-fold symmetry axis and have spine helices that are bent the least. The C and F helices have intermediate bending and these subunits are at intermediate distances from the pseudo 2-fold axis. The G subunits associated with the pentamers have moderate helix bending (Figure 4B). The degree of helix bending also correlates closely with a twist that occurs about beta strands in the P domain. It is likely that the bent spine helices are accommodating this atypical beta structure.
Prohead-II subunits21, with one exception, exhibit the same bending and unwinding distortion of the spine helices observed in Prohead-I+P. The E and B subunits in Prohead-II have closely similar distortions, whereas in Prohead-I+P, as described above, these two subunits have different degrees of distortion. The B subunit is very similar to its counterpart in Prohead-II, but the E subunit shows little distortion (Figure 4C). As described below, this can be attributed to differences in the quasi 3-fold subunit interactions that occur in Prohead-I+P and Prohead-II.
The N-arm and the delta domain
Residues 104–130, referred to as the N-arm, undergo large positional changes with maturation and are covalently attached to the delta domain in Prohead-I+P particles. Strong density is observed for the Prohead-I+P N-arms leading to the interior surface of the capsid and connecting with density attributed to the delta domains within the particle (Figure 5A). Loss of the delta domain in Prohead-II increased the mobility of the N-arm and density was only visible beginning at residue 119 in this intermediate21.
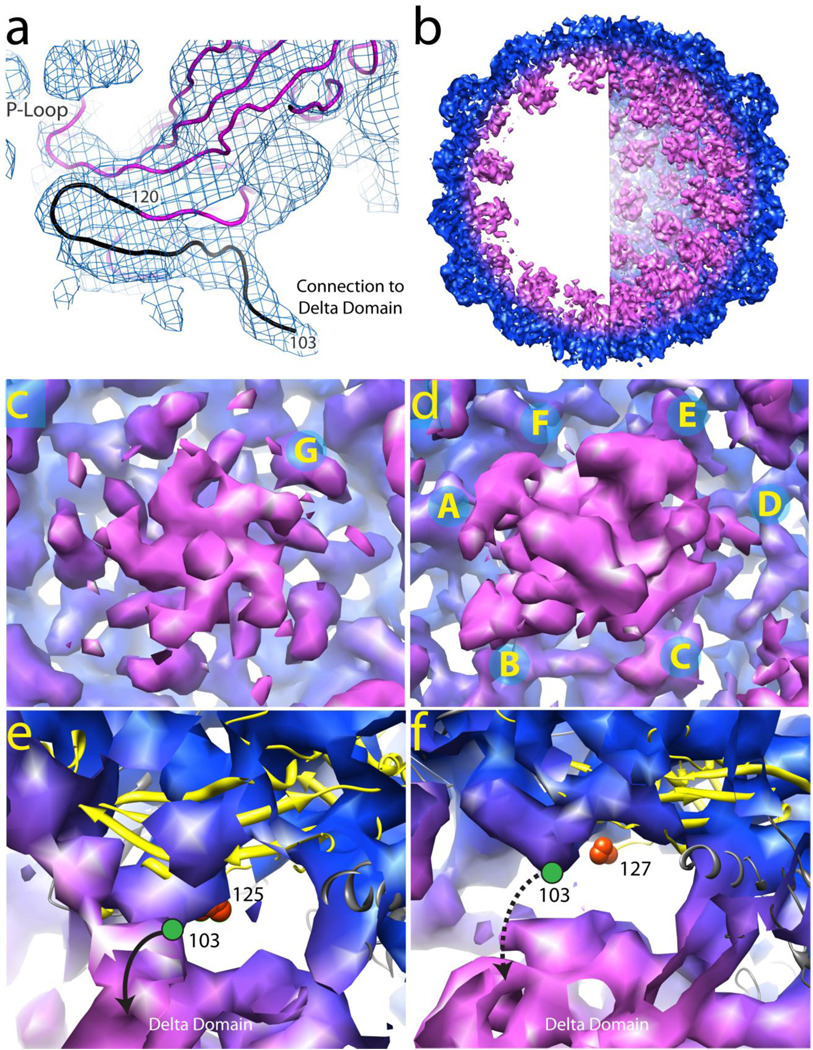
Figure 5.
The density associated with the N-arm at 5.2Å resolution and the delta domain at 10Å resolution in Prohead-I+P. (A) The density of the N-arm and the connection to the delta domain is displayed at 5.2Å resolution. The model shown in magenta is based on the E subunit polyalanine coordinates derived from the Prohead-II crystal structure and refined into the Prohead-I+P density map. Residue 120 is the last observed residue in Prohead-II. The additional density observed in Prohead-I+P is sufficient and properly positioned to account for ~17 residues (residues 103–119 hypothetically, in black) proposed that connects to the delta domain. (B) A 65 Å thick cross section slice of Prohead-I+P 10 Å electron density is shown on the left and a hemisphere of the particle density is shown on the right. Regions in magenta are not present in Prohead-II and correspond to the delta domain and possibly part of the protease. (C) Delta domain density (magenta) at the particle pentamers (Subunits G) viewed from the interior. (D) Delta domain density in magenta at the hexamers exhibiting approximate tetrameric symmetry. Subunits A, B, and D, E show continuous density to the delta domain, but subunits C and F do not. (E) Subunit B ribbon model highlighted in yellow. The last modeled residue in Prohead-II, 125, is shown as an orange sphere. The C-terminus of the delta domain, residue 103 is shown as a green circle in its probable location. We proposed, as indicted in the figure, that this N-arm is connected to the density associated with the delta domain. Subunits A, B, D, E, and G display similar features (data not shown). (F) Subunit F ribbon model highlighted in yellow. The last modeled residue in Prohead-II, 127, is shown as an orange sphere. The C-terminus of the delta domain, residue 103 is shown as a green circle in its probable location. This N-arm does not connect to the delta domain indicated as a dashed line. Similarly, subunit C also does not connect to the delta domain (data not shown). All densities were contoured at 1.5 σ.
The interior delta domain density was weak and lacked continuity, presumably due to either dynamic or spatial disorder. A data set including virtually all of the unique reflections between 100 and 10Å resolution was collected in an effort to overcome this problem and define an average volume occupied by the delta domains. Phase extension was initiated with data between 100 and 30Å resolution and extended in shells of constant volume by applying non crystallographic 5-fold symmetry and solvent leveling to a final resolution of 10Å. This map displayed defined electron density for the delta domains in both the hexamers and pentamers (Figure 5B, 5C, 5D), however, the volume defined at the hexamers was inadequate to accommodate six complete delta domains. Indeed, the shape of the delta domain density in the hexamers is roughly tetrameric with an approximate 4-fold axis coincident with the pseudo 2-fold axis (Figure 5D). The density connectivity strongly suggests that the visible delta domains come from the A, B, D, and E subunits in the hexamer. A convincing connection to the delta domain is also observable in the G subunits of the pentamer (Figure 5C and 5E). In all of these subunits the connectivity to the delta domains extends from the density assigned as the N-arm in the 5.2Å map. In contrast, density in subunits C and F, in the 10Å map, terminates at the same point that they did in the 5.2Å map, consistent with high mobility for the delta domains associated with these subunits (Figure 5F).
The X-ray data were collected from Prohead-I+P crystals, however, density attributable to the protease was not well defined or may not be resolved from the delta domains at this resolution. The SAXS data did not resolve any differences in the two forms of Prohead-I, which together with the crystal data suggest that the protease may form transient interactions with the delta domains or be spatially disordered. Crystals of empty Prohead-I were too poor to allow a structure comparison between the two forms. A difference map between the two Prohead-I forms computed with cryoEM reconstructions may be informative about interactions between the delta domains and the protease molecules.
Broken 3-fold Interactions
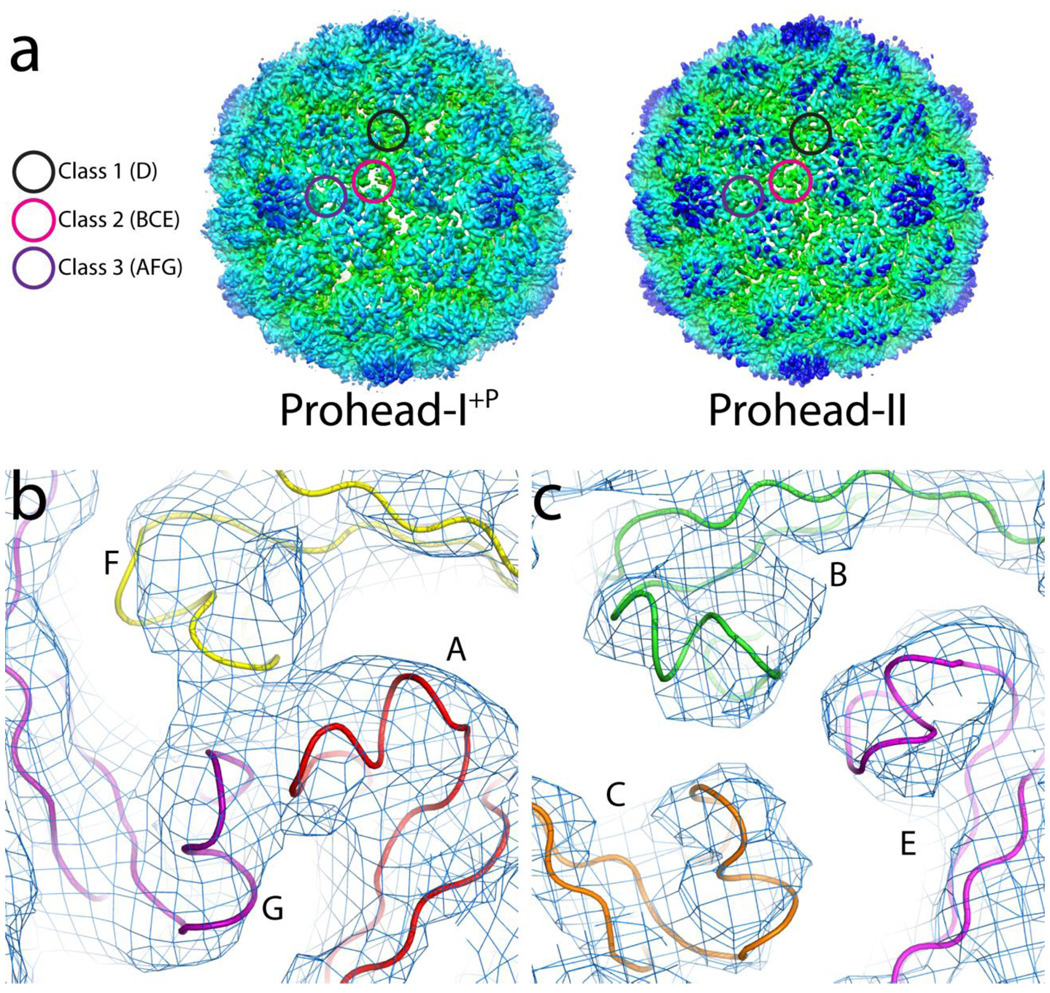
Prohead-I particles, particularly in the absence of encapsidated protease, are readily disassembled and efficiently reassembled under moderate conditions5. In contrast, conditions have not been found where Prohead-II could be disassembled and reassembled- such attempts invariably drive Prohead-II down the maturation pathway shown in figure 1. Significant changes in the 3-fold associations of capsomers in Prohead-I+P and Prohead-II suggest a structural origin for this difference in particle behavior. Interactions near the 3-fold axes in Prohead-II are formed by strong associations of P-loops at three classes of 3-fold interactions: Class 1: 20 icosahedral 3-fold contacts of hexamer-only junctions between D subunits, Class 2: 60 quasi-3-fold contacts of hexamer-only junctions with subunits B, C, and E, and Class 3: 60 quasi-3-fold contacts at junctions of two hexamers and one pentamer with subunits A, F, and G (Figure 6A). These strong 3-fold interactions play a major role in capsid stabilization and maturation, with structural and biochemical data demonstrating that they remain virtually identical in Prohead-II to Head-II14; 21.
Figure 6.
3-fold interactions. (A) Electron density of Prohead-I+P and Prohead-II, each calculated at 5.2Å resolution and contoured at 1.5 σ. The Class 1 and 3 trimers are similar in Prohead-I+P and Prohead-II, but Class 2 trimers are strikingly different, since they form pores in Prohead-I+P and sealed interactions in Prohead-II. Sealed, Class 3 trimer contacts in Prohead-I+P are shown in (B) whereas the porous, Class 2 trimers are shown in (C).
The 3-fold contacts of the Class 2 type are strikingly different in the crystal structures of Prohead-I+P and Prohead-II (Figure 6A). Subunits B, C, and E in Prohead-I+P show no contact at all between P-loops (Figure 6A and C), while the densities at the 3-fold interactions of Class 1 and 3 are consistent with P-loop contacts (Figure 6B). Poor P-loop contacts at Class 2 3-folds in Prohead-I+P correlates with reduced distortion of the spine helix in subunit E. As described above, the E subunit in Prohead-II shows a greater bend in the spine helix and additional twist in the beta sheets than it does in Prohead-I. This additional distortion appears to be a direct result of the stronger P-loop interactions in Prohead-II, where all 3-fold contacts display equivalent, closely packed density consistent with strong P-loop interactions and with additional stability confirmed by DSC19.
The P-loop density (or lack of it) observed in Prohead-I+P provides an explanation for the reversible formation of the initial assembly product22. Prohead-I fits the description of a self-assembling system (i.e., weak reversible interactions that permit subunit annealing and self-correction), but its stability is inadequate to ensure viability in the dilute and potentially harsh environments where the mature virion survives. The 60 Class 2 interactions (BCE subunits), where there is no electron density or P-loop contact, account for 43% of the 140 3-fold interactions in the particle. Without these P-loop interactions the capsid stability is compromised and we suggest that this explains why this capsid can disassemble in vitro. Furthermore, lack of these interactions, together with the delta domain-tethered N-arms, blocks maturation due to a deficient number of critical pivot points for subunit rearrangements21;14. Thus the P-loop interactions determine assembly, stability, and the timing of the transition from Prohead-I+P to Prohead-II. The delta domains ultimately regulate all of this since their removal results in strong, equivalent P-loop trimer intractions in Prohead-II.
Discussion
Origin of quaternary and tertiary structure distortions
Major signatures of the Prohead-I+P structure are the skewed hexamers and the distorted tertiary structure of the capsid protein, neither of which are observed in Head-II. Previous studies have shown that both of these distortions are present in unassembled subunit capsomers derived either from disassembly of Prohead-I or mutations near the P-loops resulting in capsomers that cannot assemble23. We posit that interactions of delta domains create these distortions. The assembly of Prohead-I is a reversible process in which particle formation is highly favored within the cell, but the equilibrium can be readily shifted in vitro to generate free capsomers. Prohead-I is essentially an organizational intermediate in which the capsomers are properly positioned, but weakly associated. Delta domains are critical to maintain the weak interactions and reversible character of Prohead-I.
The X-ray model shows that Prohead-I+P has visibly weaker inter-capsomer interactions at the Class 2 3-fold contacts when compared to Prohead-II and we conclude that this destabilizes the entire particle relative to Prohead-II. The N-arm positions in Prohead I+P directly contribute to this destabilization. Density for residues 104–124 (N-arm) differs significantly between the Prohead-I+P and Prohead-II structures. Prohead-I+P displays strong density for these residues in all the subunits, whereas there is virtually no density corresponding to the N-arms in Prohead-II. The N-arm densities are immediately adjacent to the region near the P-loops and are stabilized by the internal delta domains to which they are still attached. We infer that this stabilization directly affects the P-loops and their quaternary structure interactions. Thus, delta domains impose their effect indirectly by constraining the conformation of residues that follow them. Removal of the delta domain by proteolysis releases the N-arms, allowing them to be dynamic and reducing their influence on the P-loop. P-loops can then form much more stable interactions with each other at the trimers, making Prohead-II particles meta-stable and irreversible with respect to capsomer association.
In addition to the tertiary structure distortion, the delta domain arrangement induces and locks the skewed hexamer conformation. Only after their removal by protease do the hexamers become symmetric at ambient temperatures. Strong delta domain density on the pseudo 2-fold (quasi 6-fold) axis suggests its involvement in causing hexamer skew. This role was further demonstrated in studies showing that the hexamers have 6-fold symmetry and the delta domain density is disordered when Prohead-I is heated to 60°C. After heated samples are cooled to room temperature, the hexamers return to skewed conformation and the delta domain density is restored4. This strong evidence, that the skewed conformation is the result of the delta domain interactions, is consistent with the structural results reported here.
The second role of the delta domain suggested by our data is to inhibit maturation. Though the N-arm is dynamic in Prohead-II, in later intermediates it is engaged in extensive, quaternary structure interactions. It contributes to a three stranded beta sheet in the mature particle. This sheet is formed from two strands in the E-loop from one subunit and the N-arm from another subunit. As this sheet is on the outside of the particle, it could never form in the presence of the delta domain.
The role of the protease in assembly and maturation
Prohead-I assembles effectively in the expression system with or without protease. Previously, the role of protease was assigned only to its catalytic activity associated with digestion of the delta domains and the generation of Prohead-II. Here we showed that protease stabilizes Prohead-I employing both DSC (Fig. 3C; Prohead-I is ~3° more stable than Prohead-I−P) and chemical disassembly experiments (Figure 3B and D). (Prohead-I+P does not fully disassemble under the mild conditions whereas Prohead-I does). We surmise that presence of the protease favors Prohead-I assembly within the cell. Indeed, the presence of the inactive protease was critical for the production of crystals that diffracted beyond 7Å resolution, implying there are specific interactions with the capsid that may reduce particle flexibility and enhance stability.
Combining biochemical, biophysical, and structural methods, we determined the structure of the first icosahedral particle in the maturation of HK97 (Figure 7). The Prohead-I+P structure revealed the mechanism for organizing subunits into the proper quaternary structure, utilizing weak to moderate interactions that facilitate annealing and error correction. The Prohead-I+P particle is thus poised for the series of events associated with maturation, requiring only the activity of the protease to transform the reversible assembly state of Prohead-I+P to the metastable state of Prohead-II that then cascades down an energy landscape to the mature virion.
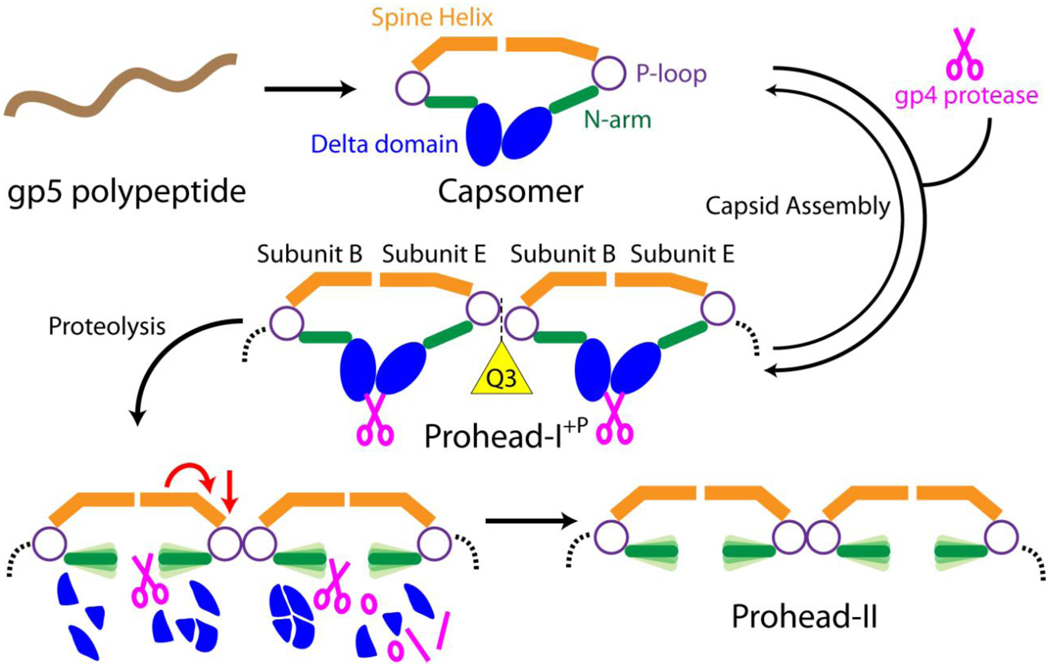
Figure 7.
A schematic illustration that depicts subunit interactions and transitions during maturation to Prohead-II. The gp5 polypeptide is expressed and immediately folds into capsomers in E. coli. Incorporating the gp4 protease, the delta domains promote inter-capsomer interactions to drive formation of Prohead-I+P. In this particle, the delta domain serves as a lock to prevent maturation by demobilizing the N-arms. As a result, the Class 2 quasi 3-fold contacts, composed of subunits B, E (shown in the figure) and C (not shown in the figure), are open whereas other trimer contacts are sealed. Protease removes the delta domain, allowing the N-arms to gain flexibility, and the spine helix in subunit E undergoes additional bending to completely close the quasi 3-fold contact. The transformed particle, Prohead-II, is a meta-stable intermediate that is energetically positioned for spontaneous maturation when electrostatically triggered.
Materials and Methods
Cloning and Protein Expression
Prohead-I with inactive protease is produced by co-expressing H65A gp4 and wild type gp5. Mutagenesis was performed using standard quick change PCR in a Perkin Elmer thermocycler (Waltham, MA) with forward primer 5’-GCGATGTTTTTCAACGCCAAGACGTGGGAGCTGCCG and its corresponding reverse complement synthesized by Integrated DNA Technologies (San Diego, CA). DNA sequencing of the resulting plasmid constructs was performed by Eton Biosciences (San Diego, CA).
Prohead-I with and without inactive protease were expressed in BL21(DE3) pLysS cells carrying pT7-Hd2.9:4− vector. One liter of LB growth medium containing 0.50 µg/ml ampicillin and 0.25 µg/ml chloramphenicol was inoculated with an overnight culture, and when an optical density of 0.35–0.45 was reached (measured at a wavelength of 550 nm) the culture was induced with 0.4 mM IPTG and incubated overnight at 28°C. Cells were collected by centrifugation at 4640 × g and were resuspended in 45 ml of ice cold lysis buffer (0.05 M Tris HCl pH 8.0, 0.005 M EDTA, 0.01 M DTT). Cells were lysed by addition of 0.2 % (v/v) Triton X100. The cell suspension was cycled three times between warming to 22 – 24°C and cooling on ice to facilitate complete lysis. The lysate was heated to 22–24 °C, and MgSO4 at a final concentration of 7.5 mM and DNase I (20 ug/ml) were added to decrease lysate viscosity. Cell debris was removed by centrifugation at 15300 × g.
Capsid particles were precipitated by adding 0.5 M monosodium glutamate (MSG) and 6% (w/w) PEG 8000 and stirring on ice for 1 hour. Following centrifugation at 10000 × g, the precipitate was resuspended by adding Prohead-I buffer (0.02 M Tris HCl, pH 7.5, 0.05 M MSG) and particles were pelleted by ultracentrifugation in the Beckman Ti50.2 rotor at 35K rpm for 2 hours at 4°C. The pellet was resuspended overnight in Prohead-I buffer. Particles were further purified over a 10–30% glycerol gradient by ultracentrifugation in an SW28 rotor at 27K rpm, 1.5 hours at 4°C, and the fraction containing particles, visible by its light scattering, was removed by side-puncture of the tube with an 18 gauge needle and syringe. The particles were loaded onto an anion exchange column (Poros PI, Applied Biosystems, Foster City, CA) for final purification. Eluted particles were buffer exchanged by two cycles of ultracentrifugation and stored in Prohead-I buffer. SDS-PAGE confirmed pure particles containing inactive proteases shown in figure 2B.
SAXS Analysis
Solution X-ray scattering data were recorded at the SSRL Beam Line 4-2. A bent cylinder mirror focused the beam onto the detector position, and a pair of Si(111) crystals was used as a monochromator. X-ray wavelength was 1.38 Å in these experiments. SPEAR operating parameters, beamline configuration, and detector instrumentation and calibration methods were similar to those used in previously reported experiments.24 The sample-to-detector distance was 2.5 m. Radial averaging, intensity scaling, and background subtraction were performed by MarParse 25.
A polycarbonate sample cell with mica windows was used for measurements. Sample temperature was held at 20°C. SAXS measurements were taken for Prohead-I and Prohead-I+P particles and for matching blank solutions containing the Prohead-I buffer. Measurements were gathered at particle concentrations ranging from 2–10 mg/ml. No concentration dependent effects were observed. The data shown are for particles at 5 mg/ml.
Disassembly Assay
The capsid sample, either Prohead-I with or without protease, was diluted in 100 mM CHES, pH 9.5, and 40% glucose. The final protein concentration was less than 5 mg/ml. The kinetics of capsid dissociation was measured with a Varian Eclipse fluorometer (Walnut Creek, CA), measuring scattering at 400 nm as a function of time. After overnight dissociation, the capsid sample in Prohead-I buffer was loaded onto a Superdex 200 size exclusion column with a Biorad Duoflow flow pressure liquid chromatography and the chromatogram was analyzed.
Differential Scanning Calorimetry
The MicroCal (Piscataway, NJ) VP-DSC calorimeter was used for calorimetry experiments. Prohead-I with or without the protease was diluted to 0.5 mg/ml in degassed 20 mM potassium phosphate, pH 7.5, 20 mM potassium chloride. The temperature was incrementally increased at a rate of 90 degrees per hour from 10°C to 110°C. The data were analyzed with the graphic software Origin version 5.0 provided by MicroCal. Tm was measured as the centroid of the peak in the isotherm. ΔH is defined to be the area under the peak.
Crystallization and Data Collection
Crystals of Prohead-I+P were grown in 200 mM sodium acetate, pH 4.6, 300 mM calcium chloride, 4% w/w PEG 4000, 15% w/v glycerol, 100 mM sarcosine, and 6% n-propanol. They were crystallized using the hanging drop method at 20°C. The reservoir contained 200mM sodium acetate, pH 4.6, 75mM calcium chloride, 4% w/w PEG 4000, 3.75% w/v glycerol, 25 mM sarcosine, and 1.5% n-propanol. Crystals started to appear within 2 weeks and were fully grown to 400–700 µm in about 1 month.
Crystals were transferred sequentially to 10%, 20%, and 30% glycerol solutions containing half concentrations of the crystallization buffer components, except for PEG 4000, which was kept same as the original concentration. Soaking time was 30 seconds in each buffer, following which the crystals were flash frozen in liquid nitrogen for storage or data collection.
Diffraction data was collected at the BL11-1 beamline at the Stanford Synchrotron Radiation Laboratory, Menlo Park, CA. Crystals were exposed to 0.99 Å wavelength (12658.079 eV) synchrotron radiation for 20 seconds with 0.25 degrees of oscillation. 250 frames of diffraction images were collected. The low resolution data of Prohead-I+P were collected at GM/CA CAT, sector 23-IDD beamline at the Advanced Photon Source, in Argonne, IL. Crystals were exposed to 0.775 Å wavelength (15997 eV) synchrotron radiation for 5 seconds with 99% attenuation and 0.25 degrees of oscillation. 760 frames of diffraction images were collected.
To obtain the low resolution data, the detector was positioned 600 mm from the crystal at the APS GM/CA CAT beamline, with an in-line helium cone.
Data processing and Electron Density Map calculation
Diffraction images were indexed, integrated, and scaled using the HKL2000 package26. The crystal space group was determined to be I23 with 2 particles in the unit cell. The orientation was determined by the 5-fold self-rotation function in the GLRF program27. The Prohead-II atomic model (PDB code: 3E8K) was used to determine the initial phases13; 28. Phases were improved and extended by multiple cycles of 5-fold real-space, non-crystallographic symmetry (NCS) averaging and solvent flattening with the RAVE package29. After several cycles of 5-fold, non-crystallographic symmetry (NCS) averaging and solvent flatteing to remove model bias, the polypeptide backbone density became clearly interpretable. The Prohead-I+P model was built by fitting each Prohead-II subunit as a rigid body into the Prohead-I+P density. The Prohead-I+P model was further refined using Crystallography and NMR System (CNS) rigid body refinement30 of each subunit from the structure factors calculated from the NCS averaged map. Coot was used to further refine the pseudo-atomic model with real space refinement31.
Viral protease (gp4) modeling based on homology
Structure prediction of the HK97 gp4 sequence with the FFAS03 server produced a number of herpesvirus proteases with high confidence32; 33. These alignments were manually refined using the Swiss-PdbViewer and the Swiss-Model server to produce predictions with improved energetics33; 34; 35; 36.
Acknowledgements
This work was funded by NIH grant 2R01-AI40101 to J.E. Johnson. We thank Drs. Steven Edgcomb and Jamie Williamson for guidance and training with DSC data collection and analysis. We also appreciate user supports from APS GM/CA CAT and SSRL BL11-1 X-ray facilities.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with accession number RCSB ID code rcsb062097 and PDB ID code 3P8Q.
References
- 1.Katen S, Zlotnick A. The thermodynamics of virus capsid assembly. Methods Enzymol. 2009;455:395–417. doi: 10.1016/S0076-6879(08)04214-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caspar DL. Movement and self-control in protein assemblies. Quasi-equivalence revisited. Biophys J. 1980;32:103–138. doi: 10.1016/S0006-3495(80)84929-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duda RL, Martincic K, Hendrix RW. Genetic basis of bacteriophage HK97 prohead assembly. J Mol Biol. 1995;247:636–647. doi: 10.1006/jmbi.1994.0169. [DOI] [PubMed] [Google Scholar]
- 4.Conway JF, Cheng N, Ross PD, Hendrix RW, Duda RL, Steven AC. A thermally induced phase transition in a viral capsid transforms the hexamers, leaving the pentamers unchanged. J Struct Biol. 2007;158:224–232. doi: 10.1016/j.jsb.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Z, Hendrix RW. Assembly in vitro of bacteriophage HK97 proheads. J Mol Biol. 1995;253:74–85. doi: 10.1006/jmbi.1995.0537. [DOI] [PubMed] [Google Scholar]
- 6.Hendrix RW, Duda RL. Bacteriophage HK97 head assembly: a protein ballet. Adv Virus Res. 1998;50:235–288. doi: 10.1016/s0065-3527(08)60810-6. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt U, Darke PL. Dimerization and activation of the herpes simplex virus type 1 protease. J Biol Chem. 1997;272:7732–7735. doi: 10.1074/jbc.272.12.7732. [DOI] [PubMed] [Google Scholar]
- 8.Gan L, Conway JF, Firek BA, Cheng N, Hendrix RW, Steven AC, Johnson JE, Duda RL. Control of crosslinking by quaternary structure changes during bacteriophage HK97 maturation. Mol Cell. 2004;14:559–569. doi: 10.1016/j.molcel.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J Mol Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 10.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 11.Lee KK, Gan L, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Evidence that a local refolding event triggers maturation of HK97 bacteriophage capsid. J Mol Biol. 2007;340:419–433. doi: 10.1016/j.jmb.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 12.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 13.Gan L, Speir JA, Conway JF, Lander G, Cheng N, Firek BA, Hendrix RW, Duda RL, Liljas L, Johnson JE. Capsid conformational sampling in HK97 maturation visualized by X-ray crystallography and cryo-EM. Structure. 2006;14:1655–1665. doi: 10.1016/j.str.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Lee KK, Gan L, Tsuruta H, Moyer C, Conway JF, Duda RL, Hendrix RW, Steven AC, Johnson JE. Virus capsid expansion driven by the capture of mobile surface loops. Structure. 2008;16:1491–1502. doi: 10.1016/j.str.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belnap DM, Steven AC. 'Deja vu all over again': the similar structures of bacteriophage PRD1 and adenovirus. Trends Microbiol. 2000;8:91–93. doi: 10.1016/s0966-842x(00)01704-2. [DOI] [PubMed] [Google Scholar]
- 17.Hendrix RW. Evolution: the long evolutionary reach of viruses. Curr Biol. 1999;9:R914–R917. doi: 10.1016/s0960-9822(00)80103-7. [DOI] [PubMed] [Google Scholar]
- 18.Cheng H, Shen N, Pei J, Grishin NV. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 2004;13:2260–2269. doi: 10.1110/ps.04726004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ross PD, Conway JF, Cheng N, Dierkes L, Firek BA, Hendrix RW, Steven AC, Duda RL. A free energy cascade with locks drives assembly and maturation of bacteriophage HK97 capsid. J Mol Biol. 2006;364:512–525. doi: 10.1016/j.jmb.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL, 3rd, Reddy VS. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gertsman I, Gan L, Guttman M, Lee K, Speir JA, Duda RL, Hendrix RW, Komives EA, Johnson JE. An unexpected twist in viral capsid maturation. Nature. 2009;458:646–650. doi: 10.1038/nature07686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duda RL, Martincic K, Xie Z, Hendrix RW. Bacteriophage HK97 head assembly. FEMS Microbiol Rev. 1995;17:41–46. doi: 10.1111/j.1574-6976.1995.tb00186.x. [DOI] [PubMed] [Google Scholar]
- 23.Gertsman I, Fu CY, Huang R, Komives E, Johnson JE. Critical salt bridges guide capsid assembly, stability, and maturation behavior in Bacteriophage HK97. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.M000039-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KK, Tsuruta H, Hendrix RW, Duda RL, Johnson JE. Cooperative reorganization of a 420 subunit virus capsid. J Mol Biol. 2005;352:723–735. doi: 10.1016/j.jmb.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Smolsky IL, Liu P, Niebuhr M, Ito K, Weiss TM, Tsuruta H. Biological small-angle X-ray scattering facility at the Stanford Synchrotron Radiation Laboratory. J Appl Cryst. 2007;40:s453–s458. [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 27.Tong L, Rossmann MG. Rotation function calculations with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 28.Rossmann MG. The molecular replacement method. Acta Crystallogr A. 1990;46(Pt 2):73–82. doi: 10.1107/s0108767389009815. [DOI] [PubMed] [Google Scholar]
- 29.Kleywegt GJ, Jones TA. From first map to final model. Warrington, United Kingdom: SERC Daresbury Laboratory; 1994. pp. 59–66. [Google Scholar]
- 30.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallographica Section D-Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile--profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 34.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 35.Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Res. 2009;37:D387–D392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duda RL, Hempel J, Michel H, Shabanowitz J, Hunt D, Hendrix RW. Structural transitions during bacteriophage HK97 head assembly. J Mol Biol. 1995;247:618–635. doi: 10.1006/jmbi.1995.0168. [DOI] [PubMed] [Google Scholar]