Abstract
Molecular recognition of RNA structure is key to innate immunity. The protein kinase PKR differentiates self from non-self by recognition of molecular patterns in RNA. Certain biological RNAs induce autophosphorylation of PKR, activating it to phosphorylate eukaryotic initiation factor 2α (eIF2α), which leads to inhibition of translation. Additional biological RNAs inhibit PKR, while still others have no effect. The aim of this article is to develop a cohesive framework for understanding and predicting PKR function in the context of diverse RNA structure. We present effects of recently characterized viral and cellular RNAs on regulation of PKR, as well as siRNAs. A central conclusion is that assembly of accessible long double-stranded RNA (dsRNA) elements within the context of biological RNAs plays a key role in regulation of PKR kinase. Strategies for forming such elements in biology include RNA dimerization, formation of symmetrical helical defects, A-form dsRNA mimicry, and coaxial stacking of helices.
Introduction
Numerous remarkable roles for RNA in biology have been uncovered [1]. RNA is central to translation; it can function as an enzyme (ribozyme) and genetic switch (riboswitch); and small RNAs play key roles in regulating genes. Many of these discoveries have been transformative to our understanding of life processes [2].
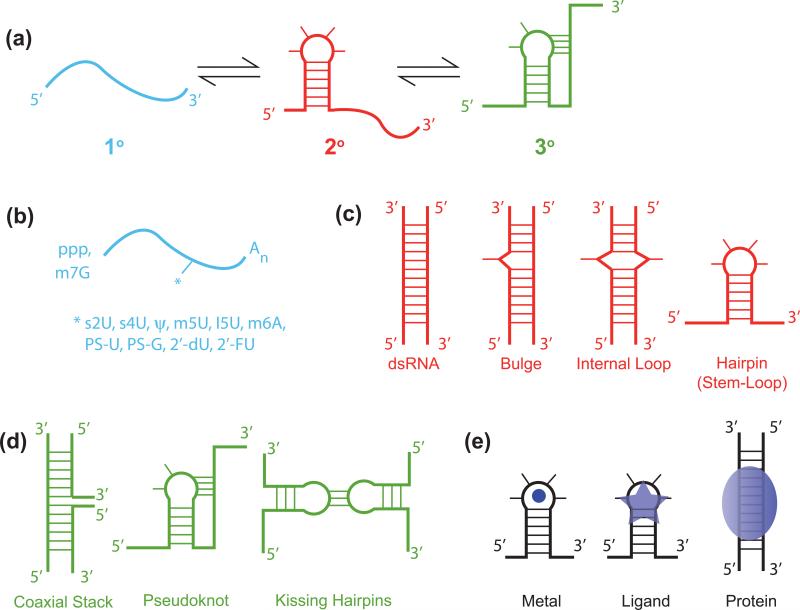
A central reason why RNA plays crucial roles in biology is that it embodies both diverse structural and decodable sequence information. The folding of RNA has been described as hierarchical [3], in which primary structure forms as the RNA is being transcribed, followed by folding of secondary structure, and then tertiary structure, as the nascent secondary structural elements assemble (Figure 1a).
Figure 1.
Hierarchy of RNA folding. (a) Two-step folding pathway of a pseudoknot RNA, involving primary structure (blue) forming secondary structure (red), here a 5’-proximal hairpin, followed by tertiary structure (red), here interaction of the 3’-tail with the hairpin loop. (b) Primary structural elements of RNA (blue), with certain 5’-end, 3’-end, and internal modifications provided. (c) Secondary structural elements of RNA (red), with perfect dsRNA, imperfections on one strand to give a bulge, on both strands to give an internal loop, or a stem-loop provided. (d) Tertiary structure elements of RNA (green), with coaxial stacking of helices, pseudoknot, and kissing hairpin loops depicted. (e) Binding of various species to RNA, with metal ion, ligand, and protein shown.
There is great diversity present in each element of the hierarchy: Primary structure embodies different sequence and length, as well as modifications at the ends and internally (Figure 1b). Secondary structure has as its basis the A-form helix, but is highly diverse owing to assorted imperfections (defects) present in most helices such as bulges, hairpin loops, and internal loops (Figure 1c). Tertiary structures are compact and often (but not always) globular forms of RNA that bring together helices and are highly diverse (Figure 1d). Adding even further to this complexity, the fold and interactions of RNA are dynamic as well: RNA folds as it is being transcribed, and it interacts with ions, metabolites, proteins, and other RNAs (Figure 1e) [4].
Innate immunity is the initial immune response to invasion by pathogens [5]. Many proteins are involved in this process, including toll-like receptors (TLRs), retinoic acid-inducible gene 1 (RIG-I), and the RNA-activated protein kinase (PKR). One key function of these proteins is distinguishing self from non-self through so-called pathogen-associated molecular patterns, or ‘PAMPs’ [6]. Given RNA's diversity in sequence and structure, it comes as no surprise to find that nature has chosen RNA for many key PAMPs. Specific sequences and structures present in pathogenic RNA allow the innate immune system to distinguish between cellular RNAs and RNAs from viruses and foreign organisms [7].
This review focuses on the RNA-based activation of PKR and how RNAs can serve as PAMPs. The last few years have witnessed increased understanding of PKR interaction with RNAs of diverse structure. We begin with an overview of PKR structure and its well-known interaction with dsRNA. We then describe recent contributions within the context of the RNA folding hierarchy, proceeding from primary to tertiary structure and ending with siRNAs and a brief comparison to other RNA-based regulating proteins of innate immunity. Our central goal is to develop a cohesive framework for understanding and predicting PKR function in the context of RNA structure.
Structure and function of PKR
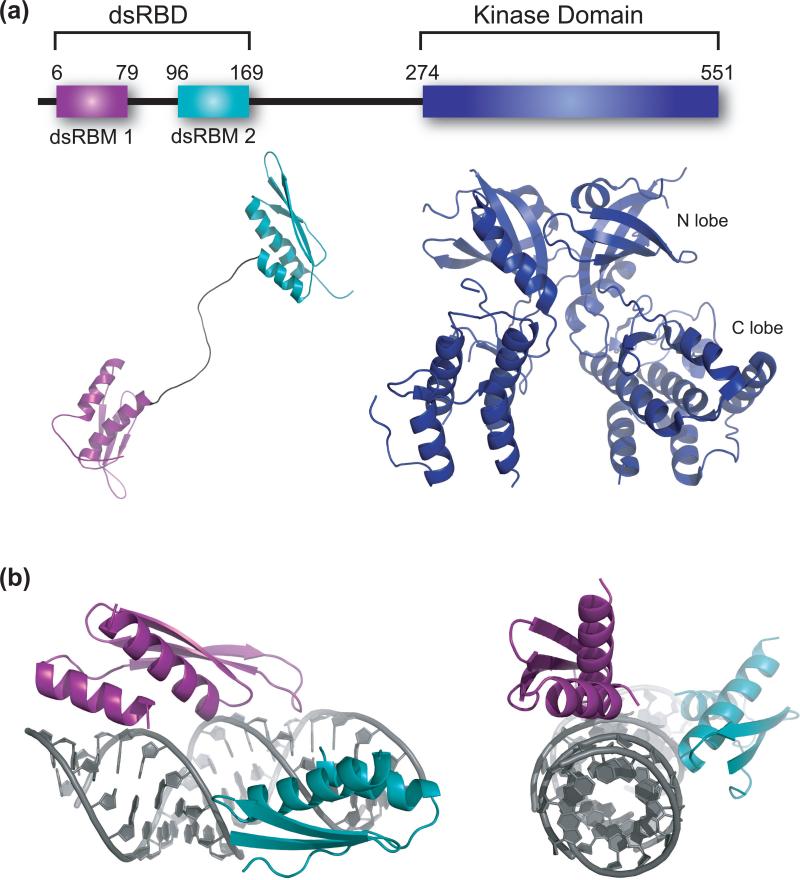
The structural biology of PKR is best viewed as a work in progress. PKR is a 551 amino acid protein that consists of two functional domains: an N-terminal dsRNA binding domain (dsRBD) that comprises two dsRNA binding motifs (dsRBMs) spaced by a flexible 20 amino acid linker,1 and a C-terminal kinase domain that contains the major sites for phosphorylation (Figure 2a) [8,9]. The dsRBM is a common motif that occurs in all kingdoms of life and is present in a number of notable proteins beyond PKR, including dicer, drosha, and adenosine deaminases that act on RNA (ADARs) [10]. The dsRBM typically recognizes dsRNA non-sequence specifically via minor groove interactions, and several reports indicate interactions with the bases [11,12]. Available structural biology of PKR includes an NMR structure of the dsRBD solved without RNA present [13], and a crystal structure for the kinase domain complexed with eIF2α substrate [14]. The NMR structure reveals the typical αβββα architecture for each dsRBM [13], while the X-ray structure indicates a smaller, mostly β-sheet N-terminal lobe (N-lobe) with a larger, stable, largely helical C-terminal lobe (C-lobe) (Figure 2a). The N-lobe of the kinase domain is involved in dimerization of PKR, whereas the αG helix from the C-lobe acts as a substrate-docking motif [14]. Low-resolution structural models of full length latent (inactive) PKR have been constructed by small angle X-ray scattering (SAXS) and reveal that PKR has intrinsically disordered regions, which may become ordered upon RNA binding; interestingly, data from this method are not fully consistent with the autoinhibition model previously proposed for PKR (described below) in which the latent protein is locked into closed conformation, as described below [15]*.
Figure 2.
Structural biology of PKR. (a) Structures of the two domains of PKR. PKR is a 551 amino acid protein that contains an N-terminal dsRNA binding domain (dsRBD) that is comprised of two dsRNA binding motifs (dsRBMs) spaced by a flexible 20 amino acid linker depicted below the dsRBMs, and a C-terminal kinase domain with small and large N- and C-terminal domains. Available are an NMR structure of the dsRBD (pdbid 1QU6) and a crystal structure of the kinase domain complexed with eIF2α substrate (eIF2α omitted here). The kinase crystallizes as a dimer (pdbid 2A1A). (b) Structure of a dsRBM from X. laevis rbpa bound to dsRNA (pdbid 1DI2) [16]. The dsRNA is 10 bp in length, and two helices are shown stacked end-to-end. Each dsRBM occupies one face of the dsRNA, and packing occurs along different faces of the dsRNA. Shown are side-on and end-on views.
At present there are no RNA-bound structures of PKR, probably because the non-sequence specific nature of RNA binding and the disordered region between the dsRBD and the kinase domain leads to heterogeneous states. However, a few structures of other dsRBMs bound to dsRNA have been solved; see for example [16,17]. In general, the dsRBM binds into the wide accessible minor groove of dsRNA, and multiple dsRBMs can pack along the length of the helix. Shown in Figure 2b is packing of two dsRBMs on ~20 bp of dsRNA. Packing of four dsRBMs on 33 bp of dsRNA, which is the minimum activating length, can be modeled similarly.
The function of PKR in biology is quite diverse. A number of excellent reviews of PKR function are available [8,18-20], and only a very brief overview is presented here. In general, activation of latent PKR requires dimerization and autophosphorylation, which occurs upon recognition of sufficiently long dsRNA, such as from intermediates generated during viral replication. In general, 33 bp are needed for minimal activation, with longer dsRNAs activating to a greater extent. Shorter dsRNA, 15-30 bp, inhibits PKR through competitive binding [21,22]. The protein activator PACT and the polyanion heparin can also activate PKR [20], and PKR can even autophosphorylate in the absence of activator if its concentration is high enough [23]. The activated dimer of PKR goes on to phosphorylate its cellular substrate eIF2α on Ser51 leading to translational arrest [8,19]. This process provides essential antiviral and antiproliferative capabilities for the host cell. More recently it was found that phosphorylation of three tyrosine residues on PKR, in addition to multiple serine/threonine phosphorylating sites, is required for full-scale activation of the kinase [24].
In addition to the antiviral functions, PKR has been implicated in modulating cell-signalling pathways to alter numerous cellular responses [19]. In addition, several diseases, such as Huntington, Parkinson and Alzheimer's, have been linked to PKR regulation [20]. A recent report suggested that p53-mediated tumor suppression can be attributed to p53's induction of PKR under genotoxic conditions [25], while another recent study indicated that PKR regulates insulin action and metabolism in response to nutrient signals and endoplasmic reticulum stress [26].
RNA primary structure-based regulation of PKR
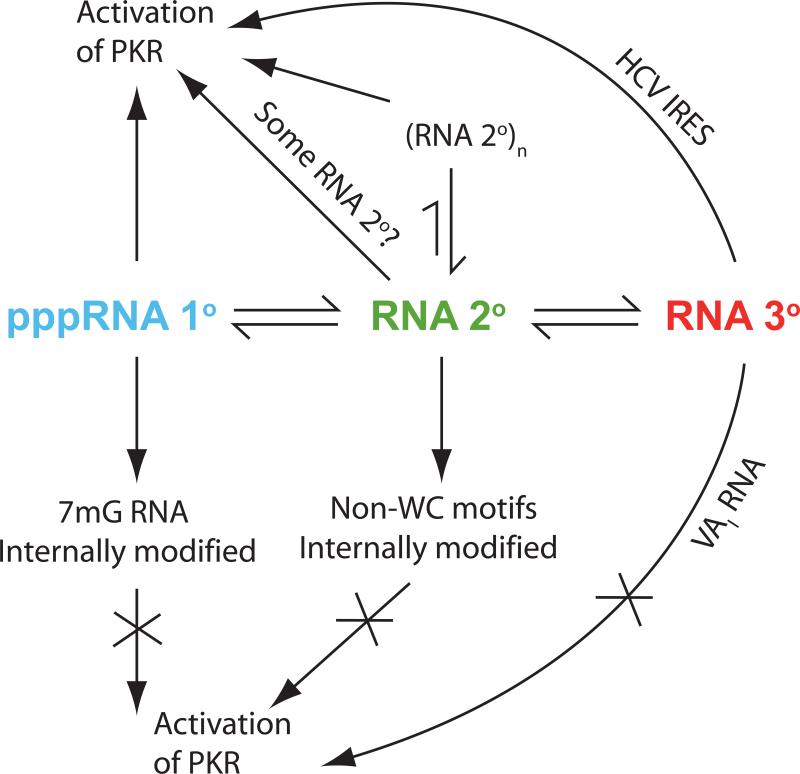
As presented in the Introduction, the folding of RNA is largely hierarchical (Figure 1), and high information content exists at each level in the folding hierarchy. The next three sections consider the three levels of RNA folding. The interplay of these RNA elements with regulation of PKR function is summarized in Figure 3.
Figure 3.
Interplay between the hierarchy of RNA folding and the activation of PKR. The same overall two-step RNA folding pathway as present in Figure 1 is shown, with coloring maintained. At each of the three states along the pathway, the potential for regulation of PKR by RNA exists. At the primary structural level, a 5’-triphosphate helps largely single-stranded RNA activate PKR, whereas a native 7mG cap and internal chemical modifications are incompatible with activation. At the secondary structural level, aggregates of RNA, depicted as ‘(RNA 2°)n’, and certain RNA secondary structures activate PKR, while internal chemically modified RNAs and certain non-Watson-Crick motifs do not. Additionally, certain RNA tertiary structures, such as that of IFN-γ mRNA (see Figure 4b), appear to activate PKR, while others, such as that of the VAI RNA, do not. See text for details and references.
Early studies used perfect dsRNAs such as poly I:C and T7 transcribed dsRNAs of various lengths to characterize PKR activation [19]. More recent studies reveal additional activation by RNAs that are non-perfectly double stranded [27,28]. At the primary RNA sequence level, ssRNAs with a small stem-loop and an imperfect 16 base-paired dsRNA with 10-15 nt single-strand tails (so-called ‘ss-dsRNA’) have been shown to activate PKR in a 5’-triphosphate dependent manner [27,28]. This 5’-triphosphate functional group of ssRNA is key in PKR activation, as 5’-diphosphate, -monophosphte, -hydroxyl and 7mG cap-containing ssRNAs do not activate PKR [28]. Most endogenous cytoplasmic RNAs contain 5’-monophophate or 7mG cap, generated through RNA processing, whereas bacterial and some viral RNAs contain 5’-triphosphate; the 5’-triphosphate functionality thus constitutes a PAMP for PKR. In contrast, activation of PKR by dsRNA does not require a 5’-triphosphate, indicating that PKR uses different strategies for recognition of ssRNA and dsRNA. The 5’-triphosphate serves as a PAMP for PKR in recognition of the viral RNA from influenza B virus as well [29]**. Additional experiments have demonstrated that internal nucleoside modifications in 5’-triphosphate ssRNA abrogate PKR activation [30]*, indicating that these may also serve in distinguishing self from non-self.
RNA secondary structure-based regulation of PKR
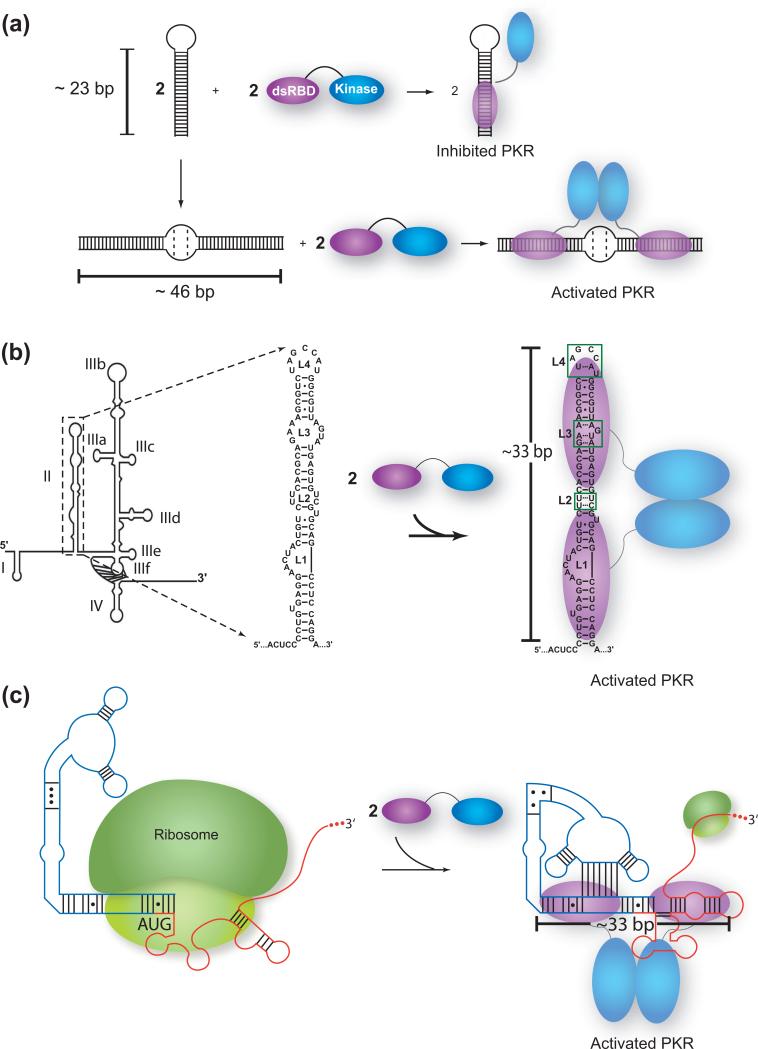
Long stretches of double-stranded RNA (≥33 bp) activate PKR potently, and have been proposed as the major activators of PKR in vivo. The molecular mechanism behind dsRNA-based activation of PKR has been studied extensively. Several models have been advanced, including an autoinhibition model, in which dsRNA binding to the dsRBD releases PKR from an inactive conformation, and a dimerization model, in which dsRNA binding serves to promote kinase dimerization [8]. Recently, analytical ultracentrifugation (AUC) has been employed to investigate the length dependence and stoichiometry of PKR binding to dsRNA [22,31]. These studies have demonstrated that dsRBM1 functions primarily in recognition of shorter dsRNA sequences (<20 bp), while both dsRBMs participate in recognition of longer dsRNAs, which are capable of activating PKR. Additionally, AUC studies have shown that the minimum requirement of ~33 bp for activation of PKR correlates with the ability to bind two PKR monomers. These data are consistent with a model in which long dsRNA functions to bring two PKR monomers into close proximity, which promotes dimerization and thus activation of the kinase domains.
In addition to dsRNA, PKR has been found to be activated by a variety of viral and cellular RNAs, which typically contain various secondary structure imperfections. One such viral RNA is the human immunodeficiency virus transactivation-response region (HIV TAR), a 23 bp hairpin RNA interrupted by three bulges that can exist as a dimer (Figure 4a) [32]. There has been longstanding discrepancy about the role of HIV TAR RNA in regulation of PKR; recent evidence, however, strongly supports that a dimeric form of TAR activates PKR [33,34]. In this study, monomers and dimers of TAR were isolated by native gel electrophoresis and studied both structurally and functionally. In particular, it was found that two TAR hairpin monomers refold to form an extended duplex with two asymmetric bulges, which effectively doubles the number of base pairs from ~23 bp in TAR monomer to ~46 bp in TAR dimer. It was found that monomer inhibited PKR, while dimer activated it, consistent with the known dependence of PKR function on dsRNA length. Thus, in this case, RNA dimerization promoted PKR dimerization and activation. In addition, this study showed that RNA dimers with fewer asymmetrical secondary structure defects were more potent activators of PKR, suggesting that such defects function as antideterminants of PKR binding.
Figure 4.
Activation of PKR by complex RNAs. (a) HIV TAR RNA undergoes dimerization, which effectively doubles its length from 23 to 46 bp. The 23 bp TAR monomer inhibits PKR, whereas the 46 bp dimer activates PKR. The former binds PKR as a monomer, whereas the latter binds it as a dimer. (b) HCV IRES containing four domains, with domain II highlighted. Domain II contains base paired and loop segments, both of which contribute to activation and provide the equivalent of ~33 bp of dsRNA. This element has been shown to mimic A-form dsRNA, including the loop interactions boxed in green [36]. (c) Pseudoknot from the IFN-γ mRNA. The 124 nt 5’-untranslated region is blue, and the coding sequence is red. This domain forms an extended structure with several base pairing elements that also leads to the equivalent of ~33 bp of dsRNA [50]. In both panels, a dimer of PKR appears to assemble onto the ~33 bp region, leading to an activated state. Panel (b) is adapted with permission from [63].
The IRES of HCV has been reported to regulate PKR [35-37]. A strategy by which dsRNAs with imperfections can activate PKR is through structural mimicry of perfect A-form dsRNA, as recently demonstrated in activation of PKR by domain II of hepatitis C virus internal ribosome entry site (HCV IRES) RNA [36]*. The IRES element of HCV has a complex secondary structure with four distinct structural domains containing multiple symmetric and asymmetric bulges, internal loops, and a pseudoknot (Figure 4b). Despite these complicated structural elements, several domains of HCV IRES RNA have been reported as activators of PKR, including domains III-IV, which contains several multi-helix junctions and a pseudoknot, and domain II, a shorter hairpin with several internal loops and bulges [36-38]. Given both the presence of imperfections and the limited number of canonical base pairs (< 33 bp), activation of PKR by domain II in particular is surprising. Footprinting and mutational analysis suggest that PKR binds and is potently activated by domain II RNA because the overall topology of its symmetrical loop regions is primarily A-form [36]*. Non-Watson-Crick interactions in the loops of domain II maintain an overall A-form helical backbone geometry and contribute to an activating total of ~33 bp. Mimicry of A-form dsRNA by symmetrical loops may serve as a general mechanism for PKR activation by RNAs with multiple helical imperfections.
Regulation of PKR by RNA secondary structure is also typified by abrogation of PKR dimerization and activation through binding of inhibitory RNAs, such as those encoded by adenovirus (VAI) and Epstein-Barr virus (EBERI). Both RNAs bind PKR with similar affinity as activating RNAs, but prevent PKR dimerization and subsequent autophosphorylation [39]. VAI and EBERI have roughly similar structures with three distinct domains: an apical stem-loop, a central domain, and a terminal stem. In the case of VAI, the apical stem-loop has been identified as the PKR dsRBD binding site, and the three-way junction within the central domain is the determinant for PKR inhibition [40]*; this domain includes elements of tertiary structure, which will be discussed in the next section. The terminal stem is completely dispensable for inhibition [41]*. The VAI apical stem-loop consists of ~18 canonical and non-canonical base pairs, which is sufficient for binding one PKR monomer but not long enough to promote PKR dimerization. Interestingly, the apical stem-loop domain of VAI exists as a population of two conformations, one of which potently inhibits PKR, and the other of which displays markedly decreased inhibition activity [42]*. Possible benefits of these functionally distinct structures for either the virus or the host have yet to be determined.
Although the function of PKR is typically to serve as a sensor of non-self RNA, certain cellular RNAs activate PKR. Previous work by Davis et al. and Nussbaum et al. identified the 3’-UTRs (untranslated region) of several highly structured cytoskeletal mRNAs as activators of PKR [43,44]. Interestingly, PKR activation by cytoskeletal 3’-UTRs is predicted to play a role in the tumor-related activities of these sequences. Similar to previously discussed viral RNAs, these cellular RNAs contain long helical stretches interrupted by bulges, internal loops, and branch points. Also, an element of the 3’-UTR of tumor necrosis factor α mRNA (TNF-α) has also been shown to activate PKR [45]. Control of exogenous gene expression by PKR is attenuated by full-length ADAR1 as well as its dsRBMs alone, suggesting that PKR and ADAR1 compete for binding to the same RNAs [46,47]. Whether this effect carries over to cellular RNAs is unclear at present [48].
RNA tertiary structure-based regulation of PKR
Tertiary structure has the potential to activate or inhibit PKR. Given PKR's penchant for dsRNA, one simple idea is that if the tertiary structure is globular, activation is unlikely, but that if it is extended, activation is possible. The 5’-UTR of the cellular mRNA for interferon-gamma (IFN-γ) fits this model (Figure 4c). As part of the interferon-mediated antiviral response, PKR participates in a negative feedback loop whereby IFN-γ regulates its own translation via competition between the ribosome and PKR for binding to IFN-γ mRNA [49,50]. If the level of PKR in the cell is low, the ribosome binds to IFN-γ mRNA to promote interferon synthesis. Upon clearing the ribosome, the 5’-UTR refolds to generate an RNA structure containing a pseudoknot, which is capable of activating PKR. Four adjoining short helices within IFN-γ mRNA coaxially stack within the pseudoknot to cumulate to an activating total of ~33 bp. Thus, in addition to RNA oligomerization by HIV TAR and A-form structural mimicry by HCV domain II, the amalgamation of secondary and tertiary features in IFN-γ mRNA demonstrates another means by which the hierarchical nature of RNA folding can generate RNA structures capable of activating PKR.
Finally, a role for RNA tertiary structure in PKR activation lies in the VAI viral RNA. It was recently determined that Mg2+, which is often required for stabilization of RNA tertiary structure, is required for correct folding of the VAI central domain and leads to binding of just one PKR. This helps explain the well-established inhibitory role of this RNA [51]*. Melting profiles and compensatory base pair modifications suggested a possible role of RNA tertiary structure in PKR inhibition by VAI RNA [40]. It has been suggested that, while the terminal stem of VAI may function to stabilize this tertiary structure, in the absence of the terminal stem, PKR binding to VAI may stabilize tertiary structure [41], although the exact nature of this tertiary structure has not been fully characterized [52].
siRNA-based regulation of PKR
There exist conflicting results on the activation of PKR by small interfering RNAs (siRNA). siRNA are short, 19-27 bp, dsRNAs that mediate RNA interference. Several groups have reported that siRNAs of 19-21 bp do not activate PKR, supporting the aforementioned requirement of 33 bp dsRNA for PKR activation [28,53]. In particular, Kim et al. [53] designed long siRNAs of 27 bp to enhance RNAi potency and efficiency and showed that they do not activate PKR, while we found that 21 bp double-stranded siRNAs do not activate PKR [27,28].
In contrast to these observations, activation of PKR by siRNA containing just 19 to 21 bp has been reported [54,55]. A proposed model for PKR activation by these shorter dsRNAs suggested that a PKR dimer assembled on one siRNA to phosphorylate a PKR dimer bound to a different siRNA [55].
Activation of PKR has a strong dependence on ionic strength, and lower salt conditions favor short RNA binding [22]. Also blunt end siRNAs activated PKR less potently than sticky ends [55]. Thus, experimental conditions, sequence, and helix termini of siRNA may play crucial roles in determining which siRNAs activate PKR [55]. Lastly, other studies indicate that activation of PKR by siRNAs is more efficient in vitro than in vivo [56], while others indicated that in vivo effects may be indirect [57]. More work is needed to sort out these discrepancies.
Comparison of PKR to other RNA-based regulating proteins in innate immunity
RIG-I and Toll-like receptors (TLR 3, 7 and 8) are additional sensors in innate immunity that recognize patterns associated with non-self RNAs. Indeed, dsRNA and 5’-triphosphate groups, which PKR recognizes as mentioned, can also be recognized by RIG-I [28,58,59]. Moreover, several natural nucleoside modifications in RNA can negate the 5’-triphosphate and dsRNA dependent activation of PKR and RIG-I [30,58,60]. Indeed, in vitro transcribed pseudouridine-containing mRNA translates better than unmodified mRNA owing to diminishing PKR activation [61]*. Regarding TLRs, they are similarly affected: TLR3 is regulated by similar nucleoside modifications in dsRNA [62], while TLR 7 and 8 are regulated by such modifications in ssRNA. Remarkably, PKR, RIG-I, and TLRs are not sequence homologues, supporting unique molecular recognition strategies by each and suggesting convergent evolution.
Conclusions and outlook
The RNA-activated protein kinase PKR is activated by much more than long perfect RNA helices. Recent studies indicate that biological RNAs activate PKR by diverse strategies and to varying extents: dimerization of RNA, inclusion of symmetrical defects, mimicry of A-form dsRNA, and coaxial stacking of helices. A common theme is assembly of accessible double-stranded elements that reach the activating length of ~33 bp. Covalent modifications to the 5’–end and internal regions of RNA can either activate or inhibit the kinase. Much remains to be understood about the link between RNA structure and extent of PKR activation, including roles of RNA tertiary structure, RNA aggregation, and co-transcriptional folding. Additional cellular and viral RNAs that regulate PKR surely await discovery, and high-resolution structures of PKR bound to dsRNA and complex biological RNAs are needed. Moreover, ways in which various RNAs alter the fraction and extent of PKR phosphorylation are unknown. Such future advances would help to further define the RNA features that allow PKR to perform its essential functions in innate immunity.
Acknowledgements
We thank Pete Beal, Jim Cole, Rick Russell and Scott Showalter for helpful comments on the manuscript, and National Institutes of Health grant R01-58709 for support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
For submission to Current Opinion in Structural Biology
This nomenclature is the convention used in the PKR field. However, more generally speaking, ‘dsRBM’ refers to the sequence motif, while ‘dsRBD’ refers to an independently folding domain.
References and recommended reading
Papers of particular interest, published within the last two years, have been highlighted as:
• of special interest
• • of outstanding interest
- 1.Atkins JF, Gesteland RF, Cech TR, editors. RNA Worlds: From Life's Origins to Diversity in Gene Regulation. Cold Spring Harbor Laboratory Press; 2010. [Google Scholar]
- 2.Nilsen TW. RNA 1997-2007: a remarkable decade of discovery. Mol Cell. 2007;28:715–720. doi: 10.1016/j.molcel.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 3.Tinoco I, Jr., Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Woodson SA. Compact Intermediates in RNA Folding. Annu Rev Biophys. 2010;39:61–77. doi: 10.1146/annurev.biophys.093008.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: Recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 7.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol Med. 2006;12:167–176. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Cole JL. Activation of PKR: an open and shut case? Trends Biochem Sci. 2007;32:57–62. doi: 10.1016/j.tibs.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadler AJ. Orchestration of the activation of protein kinase R by the RNA-binding motif. J Interferon Cytokine Res. 2010;30:195–204. doi: 10.1089/jir.2010.0005. [DOI] [PubMed] [Google Scholar]
- 10.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 11.Wu H, Henras A, Chanfreau G, Feigon J. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc Natl Acad Sci U S A. 2004;101:8307–8312. doi: 10.1073/pnas.0402627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stefl R, Oberstrass FC, Hood JL, Jourdan M, Zimmermann M, Skrisovska L, Maris C, Peng L, Hofr C, Emeson RB, et al. The solution structure of the ADAR2 dsRBM-RNA complex reveals a sequence-specific readout of the minor groove. Cell. 2010;143:225–237. doi: 10.1016/j.cell.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nanduri S, Carpick BW, Yang Y, Williams BR, Qin J. Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J. 1998;17:5458–5465. doi: 10.1093/emboj/17.18.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 15 *.VanOudenhove J, Anderson E, Krueger S, Cole JL. Analysis of PKR structure by small-angle scattering. J Mol Biol. 2009;387:910–920. doi: 10.1016/j.jmb.2009.02.019. [A study of full-length PKR in its latent form by small angle X-ray scattering (SAXS). The authors uncover that PKR has intrinsically disordered regions that may have special functions upon RNA binding.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–7513. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos A, Grunert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 2000;19:997–1009. doi: 10.1093/emboj/19.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia MA, Gil J, Ventoso I, Guerra S, Domingo E, Rivas C, Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: Virus and cell control. Biochimie. 2007;89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–292. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 21.Bevilacqua PC, Cech TR. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 22.Ucci JW, Kobayashi Y, Choi G, Alexandrescu AT, Cole JL. Mechanism of interaction of the double-stranded RNA (dsRNA) binding domain of protein kinase R with short dsRNA sequences. Biochemistry. 2007;46:55–65. doi: 10.1021/bi061531o. [DOI] [PubMed] [Google Scholar]
- 23.Lemaire PA, Lary J, Cole JL. Mechanism of PKR activation: dimerization and kinase activation in the absence of double-stranded RNA. J Mol Biol. 2005;345:81–90. doi: 10.1016/j.jmb.2004.10.031. [DOI] [PubMed] [Google Scholar]
- 24.Su Q, Wang S, Baltzis D, Qu LK, Wong AH, Koromilas AE. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc Natl Acad Sci U S A. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon CH, Lee ES, Lim DS, Bae YS. PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc Natl Acad Sci U S A. 2009;106:7825–7827. doi: 10.1073/pnas.0812148106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura T, Furuhashi M, Li P, Cao H, Tuncman G, Sonenberg N, Gorgun CZ, Hotamisligil GS. Double-stranded RNA-dependent protein kinase links pathogen sensing with stress and metabolic homeostasis. Cell. 2010;140:338–348. doi: 10.1016/j.cell.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–1945. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5'-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–1458. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 29 **.Dauber B, Martinez-Sobrido L, Schneider J, Hai R, Waibler Z, Kalinke U, Garcia-Sastre A, Wolff T. Influenza B virus ribonucleoprotein is a potent activator of the antiviral kinase PKR. PLoS Pathog. 2009;5:e1000473. doi: 10.1371/journal.ppat.1000473. [This represents the first study to link the presence of a 5'-triphosphate in a viral RNA to activation of PKR.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30 *.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–1213. doi: 10.1261/rna.1007408. [First study to show importance of natural RNA modifications to regulation of PKR by both single- and double-stranded RNA. Interestingly, these modifications also effect TLRs and RIG-I.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemaire PA, Anderson E, Lary J, Cole JL. Mechanism of PKR activation by dsRNA. J Mol Biol. 2008;381:351–360. doi: 10.1016/j.jmb.2008.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 33 **.Heinicke LA, Wong CJ, Lary J, Nallagatla SR, Diegelman-Parente A, Zheng X, Cole JL, Bevilacqua PC. RNA dimerization promotes PKR dimerization and activation. J Mol Biol. 2009;390:319–338. doi: 10.1016/j.jmb.2009.05.005. [This study addresses the long-standing discrepancy of whether TAR activates or inhibits PKR. The authors show that monomeric TAR inhibits PKR, while dimeric TAR activates it. The data support the classical length dependence of dsRNA for activating PKR.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34 *.Cole JL. Analysis of PKR activation using analytical ultracentrifugation. Macromol Biosci. 2010;10:703–713. doi: 10.1002/mabi.201000069. [An excellent overview of the biophysical properties of PKR and its mechanism of activation, with a primary focus on analytical ultracentrifugation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vyas J, Elia A, Clemens MJ. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA. 2003;9:858–870. doi: 10.1261/rna.5330503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36 *.Toroney R, Nallagatla SR, Boyer JA, Cameron CE, Bevilacqua PC. Regulation of PKR by HCV IRES RNA: importance of domain II and NS5A. J Mol Biol. 2010;400:393–412. doi: 10.1016/j.jmb.2010.04.059. [Study demonstrates the importance of domain II from HCV IRES in activating PKR. Domain II is demonstrated to mimc the A-form geometry of dsRNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37 *.Shimoike T, McKenna SA, Lindhout DA, Puglisi JD. Translational insensitivity to potent activation of PKR by HCV IRES RNA. Antiviral Res. 2009;83:228–237. doi: 10.1016/j.antiviral.2009.05.004. [Study demonstrates the importance of domains III-IV from HCV IRES in activating PKR.] [DOI] [PubMed] [Google Scholar]
- 38.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat Struct Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 39.McKenna SA, Lindhout DA, Shimoike T, Aitken CE, Puglisi JD. Viral dsRNA inhibitors prevent self-association and autophosphorylation of PKR. J Mol Biol. 2007;372:103–113. doi: 10.1016/j.jmb.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40 *.Coventry VK, Conn GL. Analysis of adenovirus VA RNAI structure and stability using compensatory base pair modifications. Nucleic Acids Res. 2008;36:1645–1653. doi: 10.1093/nar/gkn020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41 *.Wahid AM, Coventry VK, Conn GL. Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J Biol Chem. 2008;283:17485–17493. doi: 10.1074/jbc.M802300200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42 *.Wahid AM, Coventry VK, Conn GL. The PKR-binding domain of adenovirus VA RNAI exists as a mixture of two functionally non-equivalent structures. Nucleic Acids Res. 2009;37:5830–5837. doi: 10.1093/nar/gkp595. [These three studies by Conn and co-workers establish a cohesive model for VA RNA structure and function. The studies show that this RNA can exist in two separate structures.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis S, Watson JC. In vitro activation of the interferon-induced, double-stranded RNA-dependent protein kinase PKR by RNA from the 3' untranslated regions of human alpha-tropomyosin. Proc Natl Acad Sci U S A. 1996;93:508–513. doi: 10.1073/pnas.93.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nussbaum JM, Gunnery S, Mathews MB. The 3'-untranslated regions of cytoskeletal muscle mRNAs inhibit translation by activating the double-stranded RNA-dependent protein kinase PKR. Nucleic Acids Res. 2002;30:1205–1212. doi: 10.1093/nar/30.5.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Osman F, Jarrous N, Ben-Asouli Y, Kaempfer R. A cis-acting element in the 3'-untranslated region of human TNF-alpha mRNA renders splicing dependent on the activation of protein kinase PKR. Genes Dev. 1999;13:3280–3293. doi: 10.1101/gad.13.24.3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46 *.Wang Y, Samuel CE. Adenosine deaminase ADAR1 increases gene expression at the translational level by decreasing protein kinase PKR-dependent eIF-2alpha phosphorylation. J Mol Biol. 2009;393:777–787. doi: 10.1016/j.jmb.2009.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47 *.Li Z, Wolff KC, Samuel CE. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology. 2010;396:316–322. doi: 10.1016/j.virol.2009.10.026. [These two studies show a negative link between the functions of ADARs and PKR that may be due to competition for RNA substrates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hundley HA, Bass BL. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci. 2010;35:377–383. doi: 10.1016/j.tibs.2010.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R. Human interferon-gamma mRNA autoregulates its translation through a pseudoknot that activates the interferon-inducible protein kinase PKR. Cell. 2002;108:221–232. doi: 10.1016/s0092-8674(02)00616-5. [DOI] [PubMed] [Google Scholar]
- 50 **.Cohen-Chalamish S, Hasson A, Weinberg D, Namer LS, Banai Y, Osman F, Kaempfer R. Dynamic refolding of IFN-gamma mRNA enables it to function as PKR activator and translation template. Nat Chem Biol. 2009;5:896–903. doi: 10.1038/nchembio.234. [This study shows that short helical segments can assemble into a pseudoknot structure that activates PKR. This structure is ~33 bp in length and so fits the expected length dependence for activation.] [DOI] [PubMed] [Google Scholar]
- 51 *.Launer-Felty K, Wong CJ, Wahid AM, Conn GL, Cole JL. Magnesium-dependent interaction of PKR with adenovirus VAI RNA. J Mol Biol. 2010 doi: 10.1016/j.jmb.2010.08.015. [The authors show that binding of Mg2+ leads VAI RNA to bind PKR as a monomer, which helps explain the well-established inhibitory roles of this RNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Y, Mathews MB. Structure, function, and evolution of adenovirus-associated RNA: a phylogenetic approach. J Virol. 1996;70:5083–5099. doi: 10.1128/jvi.70.8.5083-5099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 54.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 55.Puthenveetil S, Whitby L, Ren J, Kelnar K, Krebs JF, Beal PA. Controlling activation of the RNA-dependent protein kinase by siRNAs using site-specific chemical modification. Nucleic Acids Res. 2006;34:4900–4911. doi: 10.1093/nar/gkl464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 57.Armstrong ME, Gantier M, Li L, Chung WY, McCann A, Baugh JA, Donnelly SC. Small interfering RNAs induce macrophage migration inhibitory factor production and proliferation in breast cancer cells via a double-stranded RNA-dependent protein kinase-dependent mechanism. J Immunol. 2008;180:7125–7133. doi: 10.4049/jimmunol.180.11.7125. [DOI] [PubMed] [Google Scholar]
- 58.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, et al. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 59.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 60.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIGI/RNA binding and signaling activities. J Virol. 2009;83:4174–4184. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61 *.Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, Kariko K. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res. 2010;38:5884–5892. doi: 10.1093/nar/gkq347. [Translation of in vitro transcribed mRNA is shown to be enhanced by incorporation of pseudouridine into the transcript. The effect is mediated by diminished activation of PKR.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–532. [PubMed] [Google Scholar]
- 63.Toroney R, Bevilacqua PC. PKR and the ribosome compete for mRNA. Nat Chem Biol. 2009;5:873–874. doi: 10.1038/nchembio.262. [DOI] [PubMed] [Google Scholar]