Abstract
Mitosis requires the concerted activities of multiple microtubule (MT)-based motor proteins. Here we examined the contribution of the chromokinesin, KLP3A, to mitotic spindle morphogenesis and chromosome movements in Drosophila embryos and cultured S2 cells. By immunofluorescence, KLP3A associates with nonfibrous punctae that concentrate in nuclei and display MT-dependent associations with spindles. These punctae concentrate in indistinct domains associated with chromosomes and central spindles and form distinct bands associated with telophase midbodies. The functional disruption of KLP3A by antibodies or dominant negative proteins in embryos, or by RNA interference (RNAi) in S2 cells, does not block mitosis but produces defects in mitotic spindles. Time-lapse confocal observations of mitosis in living embryos reveal that KLP3A inhibition disrupts the organization of interpolar (ip) MTs and produces short spindles. Kinetic analysis suggests that KLP3A contributes to spindle pole separation during the prometaphase-to-metaphase transition (when it antagonizes Ncd) and anaphase B, to normal rates of chromatid motility during anaphase A, and to the proper spacing of daughter nuclei during telophase. We propose that KLP3A acts on MTs associated with chromosome arms and the central spindle to organize ipMT bundles, to drive spindle pole separation and to facilitate chromatid motility.
INTRODUCTION
Mitosis depends on the action of the spindle, a mechano-chemical machine that uses energy released from nucleotide hydrolysis to assemble itself and to coordinate chromosome movements (Compton, 2000; Karsenti and Vernos, 2001; Mitchison and Salmon, 2001; Wittmann et al., 2001; Scholey et al., 2003). Spindle morphogenesis requires the separation of the spindle poles, and correct pole-pole spacing determines the steady state length of the spindle, contributes to chromosome segregation in anaphase, and is important for cytokinesis (Sharp et al., 2000b; Dechant and Glotzer, 2003).
Spindle pole separation is driven by forces generated by dynamic microtubules (MTs), which polymerize and depolymerize to generate pushing and pulling forces and by mitotic motors that can control MT dynamics, regulate mitotic progression, or move spindle MTs relative to adjacent MTs, actin filaments, centrosomes, or chromosomes (Inoué and Salmon, 1995; Sharp et al., 2000b; Karsenti and Vernos, 2001; Scholey and Mogilner, 2002). Current work focuses on the roles of individual MTs and motors in spindle morphogenesis and on how multiple force generators cooperate as ensembles to drive spindle pole motility. These studies suggest that pole-pole spacing and steady state spindle length are controlled by antagonistic or complementary forces generated by dynamic MTs and sets of mitotic motors (Inoué and Sato, 1967; Saunders and Hoyt, 1992; Hoyt and Geiser, 1996; Sharp et al., 1999b; Kapoor and Mitchison, 2001; Karsenti and Vernos, 2001; Scholey et al., 2001; Brust-Mascher and Scholey, 2002; Cytrynbaum et al., 2003).
For example, high-resolution time-lapse microscopy of Drosophila embryos revealed that, during spindle morphogenesis, the spindle progresses through a series of transient steady state structures in which spindle poles are proposed to be maintained at a constant spacing by a balance of inward and outward forces generated by ensembles of mitotic force generators (Sharp et al., 2000a, 2000b; Brust-Mascher and Scholey, 2002; Cytrynbaum et al., 2003). Specifically, it was hypothesized that the bipolar kinesin, KLP61F acting on interpolar (ip) MT bundles, and cortical dynein acting on astral MTs, together generate outward forces on the spindle poles, whereas Ncd in the spindle midzone exerts antagonistic inward-directed forces, acting as a brake that restrains the rate and extent of pole separation. In addition, ipMTs polymerize at the equator and depolymerize at the poles, contributing to the balance of forces acting on the poles. When these outward and inward forces balance one another, the spindle poles are maintained at a constant spacing, and the spindle exists as a steady state structure, using energy from nucleotide hydrolysis to maintain its dynamic but stable morphology. Tipping this balance of forces by up- or downregulating one or more of the force generators causes transitions between steady state structures, which are manifest as the movement of the spindle poles in the direction of the prevailing force.
Although this model is plausible in outline, it is clearly incomplete, because other motors are likely to act on MTs and other structures such as chromosomes to contribute to the balance of forces that positions mitotic spindle poles. One candidate is KLP3A, discovered in Drosophila as a kinesin that localizes to nuclei and meiotic central spindles (Williams et al., 1995). Homozygous klp3a mutants develop into adults but display male and female sterility, and meiotic spindles in klp3a mutant spermatocytes display disorganized central spindles and defects in cytokinesis (Williams et al., 1995, 1997; Giansanti et al., 1998). Central spindles (defined herein as the central region of the spindle) consist of organized arrays of antiparallel MTs formed by the interdigitation of the plus ends of ipMTs and have been proposed to be a site of MT cross-linking and sliding for spindle pole separation (McIntosh et al., 1969; McIntosh and McDonald, 1989; Sharp et al., 1999a, 2000a, 2000b). The absence of well-organized central spindle MTs in klp3a spermatocytes appears to have little effect on pole-pole separation because the average metaphase and telophase spindle lengths obtained from fixed specimens were indistinguishable in wild-type and klp3a mutants (Williams et al., 1995). However, the role of KLP3A in mitosis has not been tested. Moreover, this protein may make subtle contributions to spindle morphogenesis that can only be revealed by real-time analysis.
KLP3A is expressed in mitotic cells, including Drosophila early embryos, cultured S2 cells, and larval neuroblasts, although its functions there are unknown (Williams et al., 1997; Somma et al., 2002). The Drosophila early embryo is an excellent system for studying mitosis in a living system using time-lapse microscopy of large numbers of spindles (Foe et al., 1993; Sullivan and Theurkauf, 1995; Sharp et al., 1999b) and S2 cells are convenient for efficient protein depletion using RNA interference (RNAi; Somma et al., 2002). Therefore, we investigated the localization and function of KLP3A, by exploiting RNAi in S2 cells and the microinjection of KLP3A antibodies and dominant negative proteins in embryos. We find that KLP3A is a chromokinesin, displaying a dynamic distribution in mitotic cells, where it localizes to nuclei, regions on or around chromosomes, central spindles, and telophase midbodies. Moreover, KLP3A is required during mitosis for central spindle organization, spindle pole separation, daughter nuclear spacing, and, surprisingly, to facilitate anaphase chromatid motility.
MATERIALS AND METHODS
Drosophila Stocks and Embryo Collection
Flies were maintained and embryos collected as described (Sharp et al., 1999a). Flies expressing GFP-histone, GFP-tubulin, and GFP-CID were obtained from Drs.William Sullivan (UC Santa Cruz), Allan Spradling (Carnegie Institution of Washington), and Steven Henikoff (Fred Hutchinson Cancer Research), respectively. Claret nondisjunctional (Ncd) null flies (cand) were provided by Dr. Scott Hawley (Stowers Institute for Medical Research).
Recombinant Proteins and Antibody Production
A cDNA encoding full-length KLP3A was given to us by Dr. Michael Goldberg (Cornell University). To produce antibody and dominant negative constructs, two expression plasmids for GST fusions with KLP3A tail and stalk regions were generated. PCR-amplified KLP3A (nts 1002-1212 and 351-1001) encoding the tail and stalk were subcloned into pGEX-JDK (Amersham Pharmacia Biotech, Piscataway, NJ) and corresponding bacterially expressed proteins were purified on glutathione-agarose (Amersham Pharmacia Biotech). Two rabbit polyclonal antibodies against KLP3A were generated by immunization with purified KLP3A tail and affinity-purified on GST-KLP3A tail columns. Preimmune IgG was purified on Affigel protein A columns (Bio-Rad Laboratories, Hercules, CA). All antibodies were acid eluted, neutralized in Tris buffer, dialyzed into PBS buffer, and concentrated.
Immunoprecipitation and Western Blotting
For immunoprecipitation, 0-3 h Drosophila embryonic lysates (300 μl) were precleared with 50 μl rabbit IgG-agarose beads (Sigma). Affinity-purified anti-KLP3A antibodies, 30 μg, were added to the lysates and incubated for 2 h at 4°C, and the immunocomplex was recovered by incubation with protein A agarose (Bio-Rad) for 1 h at 4°C. Mock precipitation was performed with protein A beads only (rather than nonspecific IgG, which was used in the prior preclearing step). S2 cell double-stranded (ds) RNAi-treated or control lysates were prepared with 150 μl of cell lysis buffer (50 mM Tris, pH 7.8, 150 mM NaCl, 1% NP40, protease inhibitors). Samples were run on 7.5% SDS-PAGE gel and corresponding blots were probed with anti-KLP3A or anti-α-tubulin.
S2 Cell Culture and dsRNA
Drosophila S2 cells were cultured at 27°C (Rogers et al., 2002) and dsRNAi-treated (Clemens et al., 2000) except that 1 × 106 cells/ml were treated with 60 μg/plate of dsRNAi. For KLP3A RNAi, we PCR-amplified a fragment of the last 510 base pairs corresponding to KLP3A cDNA (nts 3029-3636) using the primers 5′-TAATACGACTCACTATAGGGGAAAGAATTTCCGCTTGGGCGTG-3′ and 5′-TAATACGACTCACTATAGGGGACCGCCAGAGAAACAGGATGC-3′. PCR products were used as templates for in vitro transcription using the Megascript T7 Kit (Ambion, Austin, TX).
Immunofluorescence Microscopy and MT Depolymerization
Fixation of Drosophila embryos for immunofluorescence was performed as described (Sharp et al., 1999a). Double or triple labeling was performed with rabbit anti-KLP3A (20-200 μg/ml), antiskeletor (mAb1A1 at 1:100, provided by Dr. Kristen Johansen, Iowa State University), FITC-conjugated anti-α-tubulin monoclonal DM1a (at 1:100, Sigma-Aldrich, St. Louis, MO) antibodies and DAPI for DNA. The appropriate secondary antibodies (Jackson ImmunoResearch, West Grove, PA) were used. For MT depolymerization experiments, bleach-dechorionated embryos were preincubated with a 1:1 volume of heptane-PBS-containing colchicine at 5 mM final conc. for 20 min at RT, serially fixed with formaldehyde/methanol, and stained with antibodies. Embryos were mounted and visualized on a Leica TCS NT confocal microscope (Deerfield, IL). Images were acquired by averaging 16-32 scans of a single optical section, and the overlay of pseudocolored micrographs was prepared using Adobe Photoshop (Adobe Systems, San Jose, CA). For S2 cells, immunofluorescence microscopy analysis was performed as described (Rogers et al., 2002) with anti-α-tubulin, anti-KLP3A antibodies, and propidium iodide (DNA). Images are shown as 3D projections generated from stacks of 10-15 sections (0.5 μm thick) using Metamorph (Universal Imaging Corp., West Chester, PA).
Microinjection of Antibody and Recombinant Protein into Embryos
Microinjection of 0-2 h embryos was carried out as described (Sharp et al., 2000a). Briefly, rhodamine-conjugated bovine tubulin (Cytoskeleton, Denver, CO) was injected into GFP-histone, GFP-CID, or Ncd null embryos. After a 5-min recovery period, embryos were injected with antibodies or recombinant proteins. In some cases, GFP-tubulin embryos were directly injected with antibodies or recombinant proteins. Affinity-purified anti-KLP3A tail antibodies or purified recombinant GST-KLP3A stalk proteins were dialyzed into PBS buffer, concentrated, and microinjected at concentrations ranging from 20 to 30 mg/ml to display consistent phenotypes. As controls, embryos were injected with identical concentrations of preimmune IgG or rabbit IgG (Sigma) or GST proteins in the same buffer. Anti-KLP3A or KLP3A stalk proteins were prepared immediately before use and stored for reuse over the next 1-2 weeks at 4°C.
Time-lapse Laser Scanning Confocal Microscopy
Time-lapse images were acquired from one focal plane at 8.3-s intervals using a Leica TCS NT confocal microscope with a 40× 1.3 numerical aperture (NA) objective or at 3-7-s intervals on an Olympus microscope equipped with an Ultraview spinning disk confocal head (Perkin Elmer-Cetus Wallac, Gaithersburg, MD) with a 60× 1.4 NA objective.
Analysis of Interzonal MT Organization; Axial Ratio, and Fluorescence Line Scan
Embryos expressing GFP-CID, a kinetochore marker, were injected with rhodamine tubulin. The axial ratio of a spindle was determined by dividing its width by its length during metaphase (20 s before anaphase A onset when kinetochores start to move poleward), anaphase A (20 s before the end of anaphase A, when all kinetochores reach opposite poles), and anaphase B (20 s after the end of anaphase A). For quantitation of telophase midbody organization, fluorescence line scans (5 pixel wide) were generated along the spindle's long axis using Metamorph. The intensities were normalized to the average intensity of the two poles (defined as 100) in each spindle.
Quantitative Analysis of Spindle Pole Positioning and Nuclear Spacing
Quantitation of spindle pole positioning was carried out as described (Sharp et al., 2000a) using Metamorph. In each plot, spindle pole separation is plotted as a percentage of control metaphase spindle length (100%) to minimize minor variances in spindle length among different embryos. The data shown are averages of 10 spindles from 2 embryos. To measure daughter nuclear spacing, we determined the center of each nucleus in GFP-histone-expressing embryos using the “Centroid” tool and measured the distance between the centroids of daughter nuclei from the time when chromosome decondensation started. For 3D projections, Z series from 0.5-μm-thick confocal sections were obtained and reconstructed using Metamorph.
Kinetochore-to-pole Movement and Congression
GFP-CID-expressing embryos were injected with rhodamine tubulin and time-lapse confocal images were acquired with a 100× 1.35 NA objective. The positions of the poles and kinetochores were logged, and the distance between a kinetochore and the corresponding pole were calculated and plotted as a function of time to obtain the rate of chromatid-to-pole movement. To study congression, the positions of each kinetochore in the four time points just before anaphase A onset were averaged.
Online Supplemental Material
Quicktime videos associated with Figures 2C, 3, 4, and 5C and Supplemental Figure 3 are available. The videos show the effects of KLP3A inhibitors, anti-KLP3A antibody and dominant negative KLP3A protein on the organization of ipMT bundles, the positioning of spindle poles and the spacing of daughter nuclei in GFP-histone, Ncd null, or GFP-CID embryos microinjected with rhodamine tubulin and KLP3A inhibitors.
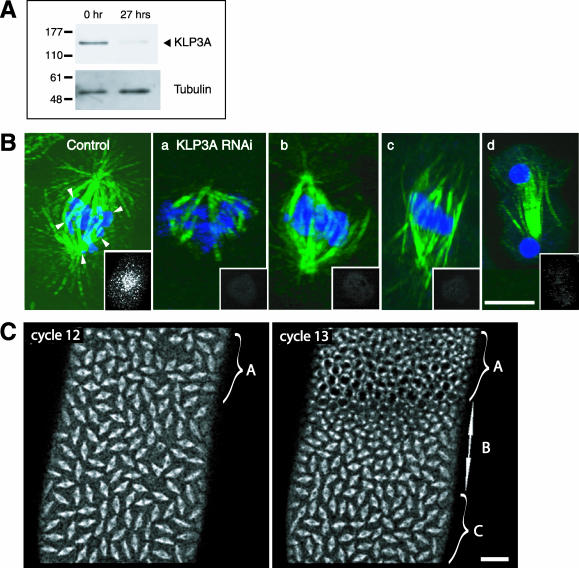
Figure 2.
General effects of KLP3A inhibition on spindle morphology and dynamics in S2 cells and Drosophila embryos during mitosis. (A) Western blot of S2 cell lysates probed with anti-KLP3A antibody showing >90% depletion of KLP3A protein (as confirmed by densitometry) after 27 h of incubation with dsRNAi. α-tubulin was used as a loading control. (B) Examples of S2 cell spindles after RNAi. 3D projections of control and RNAi-treated (a-d) S2 cells stained with anti-α-tubulin, anti-KLP3A antibodies, and propidium iodide and shown as merged images with tubulin in green and DNA in blue. KLP3A staining (insets) show that there was no detectable KLP3A after KLP3A RNAi (a-d), whereas the control cell inset shows KLP3A staining concentrated in the region around the congressing chromosomes (arrowheads indicate the boundary of congressing chromosomes). Note the highly disorganized spindle MTs in a-d. Bar, 10 μm. (C) Gradient of effects of KLP3A inhibition on spindle pole separation in the early embryo. In a GFP-tubulin embryo injected with anti-KLP3A during cell cycle 10, mitosis occurred normally throughout the embryo in cell cycle 11 (unpublished data). However, in cycle 12, the third of the embryo (A) nearest to the injection site displayed short spindles that did not elongate properly, whereas the bottom two thirds of the embryo exhibited normal pole separation. In the following cell cycle (cell cycle 13), spindles with pole separation defects in the previous cycle arrested in prophase (region A), whereas the middle third of the embryo (B) had short spindles, and the bottom third of the embryo (C) had normal spindles. Bar, 20 μm. See Online Supplemental Material, “Movie 1, Gradient effect of antibody inhibition.”
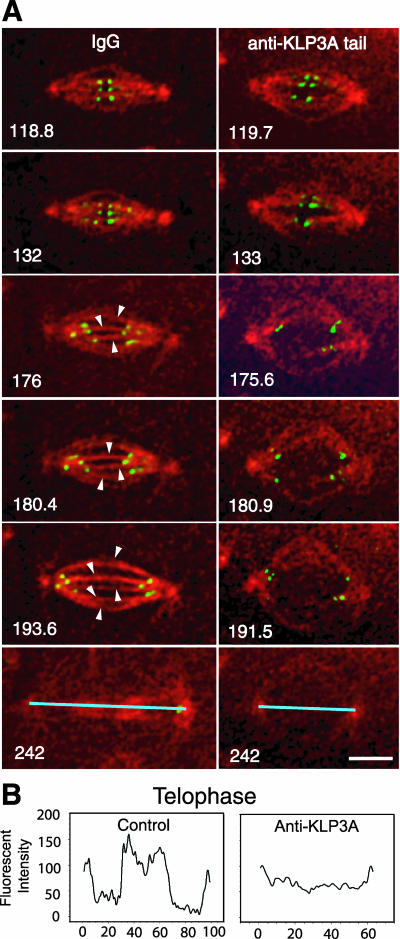
Figure 3.
Disruption of interpolar (ip) MT bundle organization after KLP3A inhibition. (A) High-magnification time-lapse images taken from rabbit IgG (control) or anti-KLP3A-injected GFP-CID embryos coinjected with rhodamine tubulin in cycle 11 (t = 0 at NEB). Kinetochores and tubulin are shown in green and red, respectively. As kMTs disassemble and kinetochores (green dots) move to the poles (176 s), control spindles show robust ipMT bundles (arrowheads). In contrast, KLP3A inhibition results in splayed spindles with very few ipMT bundles connecting two closely spaced poles in anaphase (175.6-191.5 s), which fail to form a tightly bundled central spindle and telophase midbodies (242 s). The absence of tightly bundled central spindles in KLP3A-inhibited embryos was confirmed by 3D projections (our unpublished results). Bar, 5 μm. See Online Supplemental Material, “Movies 2 and 3, IgG or anti-KLP3A-injected GFP-CID embryos.” (B) Linescans of fluorescence intensity in control and KLP3A-inhibited telophase spindles (shown as blue lines at 242 s). The absence of fluorescence in the central region after KLP3A inhibition indicates the absence of a robust telophase midbody. 0.129 μm/pixel.
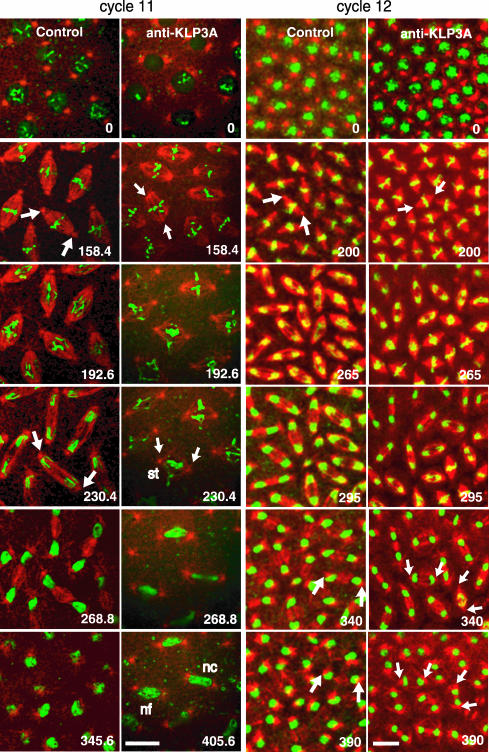
Figure 4.
Inhibition of KLP3A activity using antibody microinjection inhibits spindle pole separation subsequent to NEB. Sequential time-lapse confocal micrographs from control (preimmune IgG) and anti-KLP3A-injected GFP-histone (green) embryos coinjected with fluorescent tubulin (red) at different time points (0 s for NEB) during mitosis in cell cycles 11 and 12. In both control and KLP3A-inhibited embryos, poles separate to opposite sides along the nuclear envelope during interphaseprophase (0 s). However, after NEB, anti-KLP3A-injected embryos assemble short metaphase spindles (compare spindles at 158.4 s in cycle 11 or at 200 s in cycle 12) and less elongated anaphase/telophase spindles (compare spindles at 230.4/268.8 s in cycle 11 or at 265/390 s in cycle 12). The short spindles often have chromosome segregation defects (see Table 1), such as stretched (st) or lagging (lg, see Supplemental Figure 3) chromosomes followed by daughter nuclear fusion (nf) or collapse (nc). Telophase defects are observed not only in cycle 12 but also in cycle 11, where individual spindles are more spaciously distributed. Bar, 10 μm. See Online Supplemental Material, “Movies 4 and 5, IgG or anti-KLP3A-injected GFP-histone embryo.”
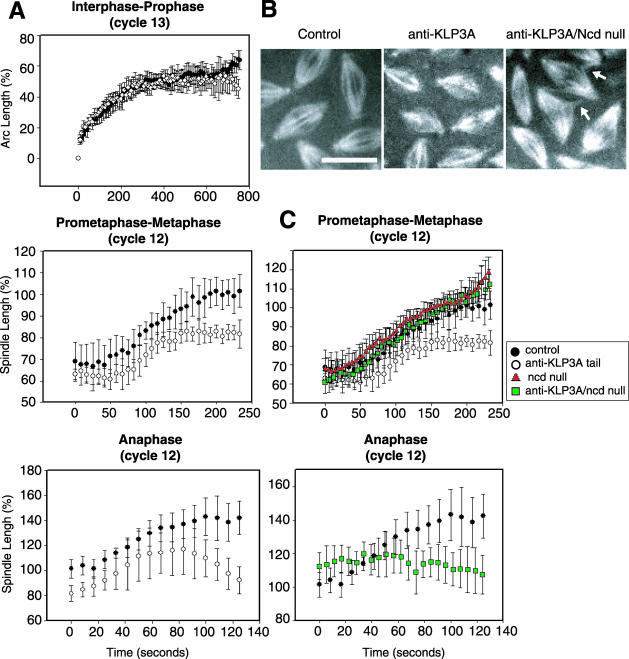
Figure 5.
Real-time analysis showing that KLP3A functions to separate spindle poles during the prometaphase-to-metaphase transition and during anaphase in Drosophila embryos. (A) Plots of spindle pole separation as a function of time during the stages indicated in preimmune IgG-injected (•), or anti-KLP3A-injected (○) embryos. Pole-to-pole distance is shown as (%) spindle length relative to control metaphase spindle length (100%). Anti-KLP3A was injected into wild-type (A) or Ncd null mutant embryos (C). (B) Images of metaphase spindles taken 200 s after NEB in control wild-type embryos (10 μm long), anti-KLP3A-injected embryos (8 μm long), and anti-KLP3A-injected Ncd null embryos (10 μm long). (C) Anti-KLP3A-induced pole separation defects (○) in prometaphase-metaphase are rescued in double inhibited anti-KLP3A-injected Ncd nulls (green squares), although the spindles are somewhat disorganized with MT spurs (B, arrows) between them, similar to those seen in anti-KLP61F-injected Ncd null embryos (Sharp et al., 1999b). The Ncd null is shown as red triangles. In plots, time 0 represents the appearance of two adjacent centrosomes in interphase-prophase (A, top panel), NEB in prometaphase-metaphase (A, center panel; C, top panel), and the separation of sister chromosome masses at the onset of anaphase (A and C, bottom panels). See Online Supplemental Material, “Movie 6, anti-KLP3A-injected Ncd null.” Bar, 10 μm.
RESULTS
Molecular Analysis of KLP3A and Probes for Its Mitotic Roles
Examination of the sequence and domain organization of KLP3A revealed an N-terminal motor domain related to those of XKLP1-subfamily chromokinesins (Vernos and Karsenti, 1995), a neck domain predictive of a plus-end-directed MT motor, a stalk containing two consensus nuclear localization signals (NLSs; 618-635 and 633-650 aa), and a tail containing a zinc finger DNA-binding domain characteristic of the XKLP1 subfamily of chromokinesins (Supplemental Figure 1, A and B; Williams et al., 1995). This predicts that KLP3A may be a plus-end-directed motor that associates with MTs, nuclei, and chromosomes.
To study KLP3A localization and function, we generated two affinity-purified rabbit antibodies against purified recombinant GST-KLP3A tail proteins (Supplemental Figure 1C, left panel), which specifically react with a protein of ∼140 kDa (Supplemental Figure 1D, left panel), consistent with the estimated molecular weight of KLP3A (138 kDa) and immunoprecipitate KLP3A from embryonic extracts (Supplemental Figure 1D, right panel). In addition, recombinant GST-KLP3A stalk proteins (Supplemental Figure 1C, right panel) were purified for use as dominant negative constructs that are predicted to heterodimerize with endogenous KLP3A polypeptides to form nonfunctional products (see Rogers et al., 2000).
KLP3A Localizes to Interphase Nuclei, Chromosomes, Central Spindles, and Telophase Midbodies
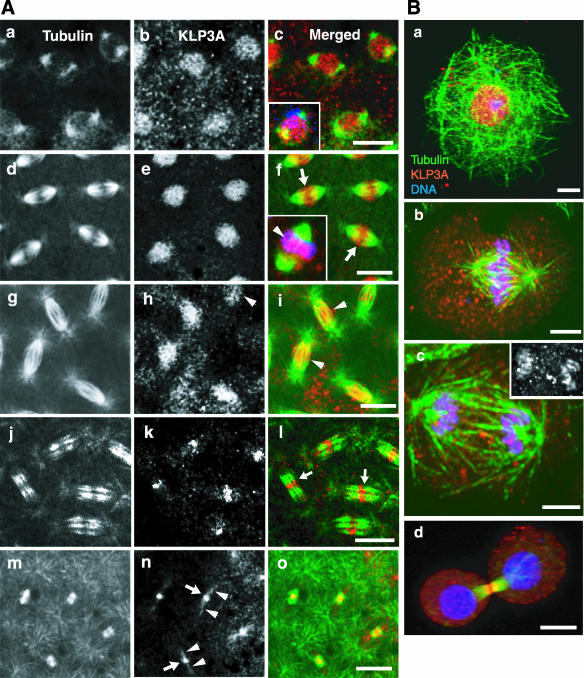
Immunofluorescence microscopy of Drosophila syncytial embryos and cultured S2 cells stained with the aforementioned antibodies revealed that, in both cell-types, KLP3A associates with nonfilamentous punctae that localize to mitotic spindles and interphase nuclei (Figure 1 and Supplemental Figure 2). Anti-KLP3A antibodies from two rabbits displayed identical staining patterns and these were completely abolished by preadsorption of the antibodies with the antigen that was used for their generation, verifying antibody specificity (our unpublished results). In both S2 cells and embryos at interphase and prophase, KLP3A was concentrated in the nucleus (Figure 1, A, a-c, and Ba), similar to its distribution during male meiosis (Williams et al., 1995) and consistent with the NLSs in the stalk domain. During telophase in both embryos and S2 cells, KLP3A was concentrated in the center of the midbody (Figure 1, A, j-o, and Bd). In embryos this midbody staining appeared as a distinct line where the plus ends of MTs overlap (arrows in Figure 1 Al), and later it was constricted to a central dot (Figure 1An, arrows) from which a faint filamentous pattern extended along midbody MTs between daughter nuclei (arrowheads).
Figure 1.
Immunolocalization of KLP3A in Drosophila early embryos and S2 cells. (A) Confocal immunofluorescence images of 0-2-h embryos stained with anti-KLP3A (b, e, h, k, and n) and anti-α-tubulin antibodies (a, d, g, j, and m). (a-c) Before nuclear envelope breakdown (NEB), (d-f) metaphase, (g-i) anaphase, (j-l) early, and (m-o) late telophase. Merged images (c, f, i, l, and o) are shown with KLP3A in red and tubulin in green (with DNA in blue, inset in c and f). Yellow indicates high overlap between tubulin and KLP3A. (f, inset). Metaphase embryonic spindle showing KLP3A on or around chromosomes indicated by overlap between DNA and KLP3A in purple (arrowhead). In embryos the association of KLP3A with chromosomes appears transient and difficult to capture in fixed images. Bar, 10 μm. (B) S2 cells at (a) interphase, (b) metaphase, (c) anaphase A, and (d) telophase stained with anti-KLP3A (red), anti-α-tubulin (green), and propidium iodide (blue). Bar, 5 μm. Note that KLP3A punctae clearly associate with nuclei, (S2 cell) chromosomes (inset in c; KLP3A single label fluorescence displays clear anaphase chromosome localization of KLP3A), central spindles, and midbodies.
During prometaphase, metaphase and anaphase A, we observed clear differences in the localization of KLP3A between embryos and S2 cells. In embryos, the most obvious localization of KLP3A was in the central spindle region where it appears to associate with ipMTs (Figure 1A, d-f, arrows; g-i, arrowheads). In S2 cells, KLP3A appeared to be tightly associated with chromosomes through anaphase A (Figure 1B, b and c), in accordance with the Zn-finger DNA-binding domain in its tail domain, only translocating to the central spindle and midbody at late stages of mitosis.
The punctate staining of KLP3A (Figure 1, A and B) is unlike the obvious filamentous staining of KLP61F (Sharp et al., 1999a), suggesting that KLP3A may associate with non-MT structures in the spindle. Extensive examinations of embryos have not revealed a tight chromosome localization like that seen in S2 cells, although KLP3A does localize to an indistinct region on and around the chromosomes, consistent with a transient association with chromosomes (Figure 1A, inset in f, arrowhead). During these studies we noticed that the staining pattern during prometaphase and metaphase, but not during telophase, is superficially similar to that of the putative spindle matrix component, skeletor (Supplemental Figure 2A; Walker et al., 2000). Consequently, to test if KLP3A, like skeletor, binds spindles in a non-MT-dependent manner, we fixed and stained embryos after depolymerizing MTs with colchicine (confirmed by loss of anti-α-tubulin staining) and found that KLP3A is solubilized along with tubulin, whereas skeletor remains associated with an insoluble spindle remnant (Supplemental Figure 2B). Thus KLP3A requires MTs for its association with mitotic spindles. We did not observe any distinctive KLP3A staining of astral MTs, spindle poles, actin rich caps, or pseudocleavage furrows that surround each spindle (Kellogg et al., 1988), although this motor behaves biochemically as a microtubule and microfilament-associated protein (MMAP) (Sisson et al., 2000). Thus, these localization studies suggest that KLP3A may bind in a MT-dependent manner to a non-MT spindle component, but they do not reveal its identity, and they show that, during mitosis, KLP3A punctae concentrate on or around chromosomes, in the central spindle and in the telophase midbody.
Mitotic Defects in S2 Cells; RNAi-induced KLP3A Depletion
To examine KLP3A function in S2 cells, we generated dsRNA for RNAi. After 27 h of RNAi treatment, KLP3A protein was efficiently depleted, as confirmed by Western blots and immunostaining (Figure 2, A and B, insets in a-d). The corresponding loss of KLP3A function produced short spindles (Supplemental Table 1) and severe defects in spindle MT organization (Figure 2B). By immunofluorescence, these defects include (a) monopolar or asymmetric bipolar spindles containing chromosomes lying toward their periphery, (b) bipolar spindles with disorganized interpolar MTs, (c) disorganized spindles with splayed poles, and (d) disorganized telophase central spindle MTs. Loss of KLP3A by RNAi caused an increase in mitotic index with high prometaphase/metaphase figures (Supplemental Table 1), yet chromosomes successfully congressed (Figure 2B, a-c) and segregated (Figure 2Bd) on these disorganized spindles, and, as reported by Somma et al. (2002), cell proliferation occurred normally. This suggests that the spindle defects caused by loss of KLP3A function could be corrected during a mitotic delay in these S2 cells (Supplemental Table 1).
Survey of Mitotic Defects after KLP3A Inhibition in Embryos
To understand the precise roles of KLP3A during mitosis, real-time, high-resolution analysis of antibody- and dominant negative fragment-microinjected embryos was performed (Figure 2C). In these microinjected embryos we observed a gradient of “quasi” phenotypes, characterized by defects in spindle pole separation and chromosome segregation, presumably reflecting a concentration gradient of injected inhibitors as observed in other studies (Sharp et al., 2000c; Blower and Karpen, 2001) with the most severe phenotypes being proximal to the injection site. Figure 2C shows a typical GFP-tubulin embryo that was microinjected with anti-KLP3A in cycle 10, proceeded normally through mitosis in cycle 11, but displayed mitotic defects in cycle 12. Specifically, the third of the embryo proximal to the injection site (region A, left panel) displayed short bipolar metaphase spindles that did not elongate properly, whereas the distal two thirds displayed normal pole separation. Many of the short spindles in region A were associated with chromosome segregation defects visible during anaphase or telophase (described in Table 1; see Figures 4 and 7 and Supplemental Figure 3). Spindles in region A that were abnormally short during cell cycle 12 subsequently arrested in cycle 13, and spindles in the adjacent region that had appeared normal in cycle 12 became abnormally short in cycle 13 (region B, right panel), whereas the more distal spindles remained normal (region C, right panel). Thus the injected inhibitors appear to cause a temporal as well as a spatial gradient of phenotypes in the embryo. In the sections that follow we focus our discussion on spindles from region A during cell cycle 12.
Table 1.
Survey of mitotic defects after KLP3A inhibition in Drosophila embryosa
| a. Metaphase spindle length defects (% total observed)b
|
||||
|---|---|---|---|---|
| Control IgG | Anti-KLP3A | Control GST | KLP3A stalk | |
| No. of spindles | 277 | 204 | 345 | 222 |
| Normal | 100 | 22.5 | 100 | 37.4 |
| Short | 0 | 77.5 | 0 | 62.6 |
| b. Chromatid motility defects on short versus normal metaphase spindles after KLP3A inhibitionc
|
||||||
|---|---|---|---|---|---|---|
| Anti-KLP3A
|
KLP3A stalk
|
|||||
| Control IgG | Normal | Short | Control GST (%) | Normal | Short | |
| Anaphase A | ||||||
| Normal | 97.8 | 93.5 | 68.4 | 97.4 | 96.4 | 54.7 |
| Stretched chromosomes | 1.8 | 4.3 | 17.7 | 1.4 | 1.2 | 18.0 |
| Lagging chromosomes | 0.4 | 2.2 | 13.9 | 1.2 | 2.4 | 27.3 |
| Anaphase B/Telophase | ||||||
| Normal | 97.5 | 97.8 | 1.9 | 95.1 | 96.4 | 10.8 |
| Daughter nuclear fusion | 1.1 | 2.2 | 23.4 | 2.0 | 1.2 | 35.3 |
| Daughter nuclear collapse | 1.4 | 0 | 74.7 | 1.7 | 2.4 | 52.5 |
Microscopy of GFP-Histone-expressing embryos, coinjected with rhodamine tubulin and KLP3A inhibitors, reveals a gradient of mitotic defects. Data from proximal and distal regions of 4—5 embryos/condition during cell cycles 11 and 12. KLP3A inhibition produced short spindles with chromosome segregation defects, including stretched chromosomes (sister chromatids initially separated poleward but their arms were distributed between two poles), lagging chromosomes (thin bridges of chromatids connecting two chromosome masses), daughter nuclear fusion (stretched or lagging chromosomes decondensed in close proximity so that daughter nuclei fuse), or daughter nuclear collapse (chromosomes separated into two masses but subsequently collapsed to within 5 μm), usually resulting in prophase arrest in the following cell cycle. Nuclear collapse between non—daughter-neighboring nuclei was not significant: 1.2% in control GST injected and 1.4% in KLP3A stalk injected
Spindle length defects: survey of all spindles within the field of view, which corresponded to one third to one half of a typical embryo. Short spindles have pole-to-pole distances <90% of control
Chromatid motility defects are expressed as % of normal or short metaphase spindles
Figure 7.
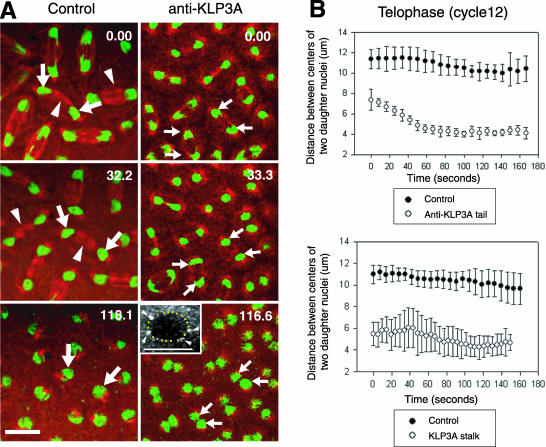
Inhibition of KLP3A activity by microinjection of anti-KLP3A or dominant negative KLP3A stalk protein results in a decrease in the spacing between daughter nuclei during telophase. (A) Time-lapse confocal micrographs from control (preimmune IgG) and anti-KLP3A-injected GFP-histone (green) embryos, coinjected with rhodamine tubulin (red), in telophase. KLP3A inhibition displays a “daughter nuclear collapse” phenotype (compare large and small arrows indicating daughter nuclei). Inset shows 3D projection of anti-KLP3A-injected spindle taken ∼14 min after NEB with dotted circle, indicating the perimeter of fused nuclei that are associated with four centrosomes (arrowheads). Bar, 10 μm. (B) Defects in nuclear spacing in anti-KLP3A (top panel) or KLP3A stalk (bottom panel)-injected embryos. Plots of daughter nuclear spacing during telophase of cell cycle 12 were generated from 20 nuclei in two different embryos, respectively. The onset of telophase (0 s), corresponding to ∼345 s after NEB, is defined by chromosome decondensation and the cessation of control anaphase B spindle elongation.
KLP3A Is Required for the Organization of Interpolar MT Bundles
We hypothesize that mitotic defects associated with KLP3A inhibition result from a primary defect in the organization of ipMT bundles (Figure 3; Supplemental Table 2), based on time-lapse confocal imaging of embryos expressing GFP-CID injected with rhodamine tubulin to distinguish ipMTs from kinetochore MTs (kMTs; Brust-Mascher and Scholey, 2002). After KLP3A inhibition, we found almost no difference in the appearance of robust kMTs, but we observed highly disorganized ipMT bundles very different from the robust ipMT bundles seen in controls (Figure 3A, arrowheads). In the KLP3A-inhibited embryos, the disorganization of ipMT bundles gives rise to short and “splayed” spindles with larger axial ratios than controls (width/length; Supplemental Table 2), which fail to form central spindles and telophase midbodies (Figure 3A, bottom panel, and B).
KLP3A Contributes to the Force-Balance That Separates Spindle Poles during the Prometaphase-to-Metaphase Transition and during Late Anaphase
In KLP3A-inhibited embryos we observed an inhibition of pole-pole separation during specific phases of spindle morphogenesis and elongation, notably during the prometaphase-to-metaphase transition and during anaphase B (Figures 4 and 5 and Supplemental Figure 3).
Sequential time-lapse images of living GFP-histone-expressing embryos labeled with rhodamine tubulin after microinjection of antibody (Figure 4) or the dominant negative protein (Supplemental Figure 3A) reveal that the initial interphase-prophase spindle pole separation was not affected by KLP3A-inhibition, and centrosome separation occurs circumferentially around the nuclear envelope to opposite sides of the nucleus, as in controls (Figure 4; Supplemental Figure 3A, 0 s). However, after NEB, the spindle poles in anti-KLP3A or stalk-injected embryos separate less than in controls, resulting in shorter metaphase spindles (Figure 4; Supplemental Figure 3A). During anaphase B and telophase, spindles in KLP3A-inhibited embryos elongate less extensively than in control embryos (Figure 4; Supplemental Figure 3A), and consequently daughter nuclei lie closer together than usual (discussed later).
To determine when KLP3A acts to drive spindle pole separation, we quantitatively analyzed the kinetics of spindle pole separation after KLP3A inhibition (Figure 5; Supplemental Figure 3B). During interphase-prophase (Figure 5A), the dynamics of spindle pole separation in control and KLP3A-inhibited embryos are identical. However, although the rates of spindle pole separation are similar in controls and KLP3A-inhibited embryos during the prometaphase-metaphase transition (0.03 ± 0.01 μm/s; Figure 5, center panel; Table 2), we consistently observed an ∼20% decrease in the metaphase spindle length, corresponding to a 50% decrease in the extent of pole separation during this period, leading to the formation of short metaphase spindles (Figure 5; Supplemental Figure 3B; Table 2). Although spindles are abnormally short at the onset of anaphase because of the preceding defects in prometaphase-metaphase spindle elongation, spindle pole motility during the first half of anaphase B occurs at a rate similar to the rate in KLP3A-inhibited embryos and in controls (Figure 5A, bottom panel; Table 2), but in late anaphase, spindle poles come closer together, resulting in ∼20-36% decrease in the average length of maximally elongated anaphase spindles compared with controls (Table 2). Generally, the inhibition of spindle pole dynamics was similar in anti-KLP3A- and KLP3A stalk-inhibited spindles, although the anaphase defects produced in the latter case were slightly more severe (Figure 5; Supplemental Figure 3; Table 2).
Table 2.
Observed defects in embryonic spindle pole separation resulting from KLP3A inhibitiona
| Antibody
|
Dominant negative
|
|||
|---|---|---|---|---|
| Control (preimmune IgG) | Anti-KLP3A tail | Control (GST) | KLP3A stalk | |
| Rate of pole separation (μm/sec) | ||||
| Prometaphase-metaphase | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 | 0.03 ± 0.01 |
| Early anaphase | 0.06 ± 0.01 | 0.05 ± 0.02b | 0.07 ± 0.01 | 0.04 ± 0.01b |
| Spindle length (%) | ||||
| Metaphase | 100 | 80.8c | 100 | 78.0c |
| Anaphase | 100 | 81.5c | 100 | 64.1c |
All numbers are averages of measurements on 10 spindles from two embryos (The graphs are shown in Figure 5 and Supplemental Figure 3B for antibody and dominant negative injection, respectively)
The rate of pole separation during early anaphase, corresponding to the first 60—80 s of anaphase (which lasts a total of ∼120 s). During late anaphase, the rate of pole separation becomes negative or 0, indicating that two poles get close together or no longer separate
Percentage spindle length relative to control spindle length (100%). Spindle length was determined from the plateaus in graphs of spindle pole separation during the prometaphase-to-metaphase transition and anaphase
KLP3A Acts Antagonistically to Ncd during the Prometaphase-Metaphase Transition, But Not during Anaphase B or Telophase
We propose that KLP3A acts in concert with dynein and KLP61F to exert an outward force on spindle poles that antagonizes the inward braking force exerted by Ncd (Sharp et al., 1999b, 2000a). To test the hypothesis that KLP3A and Ncd act antagonistically, we microinjected anti-KLP3A into Ncd null mutant embryos (Figure 5, B and C). We observed an almost complete rescue of wild-type spindle pole motility during the prometaphase-metaphase transition (characterized by an elongation from 7 to 10 μm, in control spindles) as displayed clearly in the superimposed plots of control and double-inhibited embryos (Figure 5, B and C, top panel). In the double KLP3A-Ncd inhibited embryos, as in Ncd nulls (Brust-Mascher and Scholey, 2002), spindle poles separate to the same extent as in control embryos, but there is a significant decrease in the duration of the 7- and 10-μm-long isometric spindles that occur during prometaphase and metaphase, respectively (Figure 5C). Interestingly, however, no rescue of spindle pole separation was observed during anaphase B or telophase in KLP3A-Ncd-inhibited embryos (Figure 5C, bottom panel), consistent with our hypothesis that the braking effect of Ncd is turned off during anaphase B (Sharp et al., 2000a, 2002b).
KLP3A Is Required for Efficient Poleward Kinetochore Motility during Anaphase A
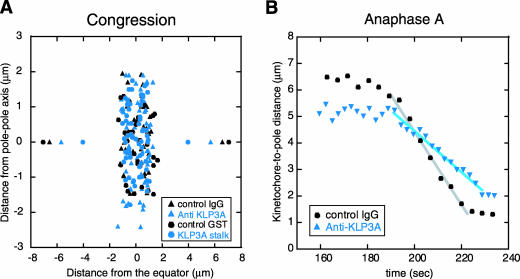
In KLP3A-inhibited short spindles, kMTs identified as rhodamine MT fibers ending in GFP-CID, appear robust (Figure 3). Accordingly, after KLP3A inhibition, final congression of kinetochores to the metaphase spindle equator and the average distance between sister kinetochores are similar to controls (Figure 6A), suggesting that these kinetochores are held under tension through normal bipolar attachment. Surprisingly, the rate of kinetochore-to-pole motility during anaphase A in these spindles decreased ∼30% relative to controls (Figure 6B and Table 3). As kMTs disassemble, KLP3A-inhibited spindles do not display the robust ipMTs seen in control spindles. This suggests that the establishment of proper ipMT organization and metaphase spindle length mediated by KLP3A provide a structural framework that supports normal chromatid-to-pole motility.
Figure 6.
Analysis of kinetochore motility after KLP3A inhibition. (A) Distribution of kinetochore pairs in metaphase spindles from control and KLP3A-inhibited GFP-CID embryos coinjected with rhodamine tubulin. In KLP3A-inhibited spindles, kinetochores congress to the metaphase plate (but spread slightly perpendicular to the long axis of the spindle because of the splayed morphology of such spindles; Figure 3A). Pairs of data points at opposite ends of the X axis indicate the position of spindle poles. (B) Plot of kinetochore-to-pole distance as a function of time (t = 0 at NEB) during anaphase A, showing that poleward kinetochore movement is slowed down after KLP3A inhibition. Each plot is a single representative kinetochore from observations of multiple kinetochores (see Table 3 for averages). Note reduced pole-pole spacing (A) and kinetochore-to-pole spacing (B, 160 s), reflecting short spindles in KLP3A-inhibited embryos.
Table 3.
Rate of kinetochore to pole movement during anaphase Aa
| Antibody inhibition
|
Dominant negative
|
||||
|---|---|---|---|---|---|
| Uninjected | Control IgG | Anti-KLP3A (μm/s) | Control GST | KLP3A stalk | |
| Kinetochore to pole rate | 0.11 ± 0.02 | 0.11 ± 0.02 | 0.07 ± 0.02 | 0.10 ± 0.02 | 0.07 ± 0.01 |
| (3/15/61) | (3/12/45) | (4/13/43) | (3/11/54) | (2/10/42) | |
Parentheses gives number of embryos/spindles/kinetochores
KLP3A Functions to Establish and Maintain the Spacing between Daughter Nuclei during Telophase in Drosophila Embryos
The inhibition of KLP3A function disrupted the proper spacing of daughter nuclei during telophase (Figure 7). In control embryos (Figure 7A, left panels, and B) daughter nuclei remain approximately evenly spaced throughout telophase, but in KLP3A-inhibited embryos, daughter nuclei (spaced only 6-7 μm at telophase onset due to earlier defects in spindle pole separation) further collapse to ∼4 μm (Figure 7A, right panel, and B). In some cases, the collapsed nuclei appear to undergo nuclear fusion (see inset showing fused nuclei with four associated centrosomes), which is often followed by prophase arrest. Ncd null mutants did not display defects in telophase nuclear spacing, and the defects associated with KLP3A-inhibition were not rescued in the KLP3A-Ncd-inhibited embryos, suggesting that Ncd activity is downregulated at anaphase B onset.
The nuclear spacing defect resulting from KLP3A inhibition was strikingly specific to pairs of daughter nuclei (Figure 7) rather than between neighboring nuclei in general, consistent with it resulting from the disruption of the morphology of central spindles (Figure 3A, 242 s; Figure 7A, arrowheads indicate central spindles in control). This is consistent with a role for KLP3A in organizing MTs of the central spindle between daughter nuclei and thus in maintaining telophase nuclear spacing.
DISCUSSION
We investigated the role of KLP3A during mitosis 1) by depleting KLP3A protein using RNAi in S2 cells and examining its loss-of-function “terminal phenotype” by immunofluorescence, and 2) by interfering with KLP3A function using anti-KLP3A antibodies and dominant negative proteins in embryos and examining its role in spindle dynamics by high-resolution, real-time analysis. We find that KLP3A inhibition leads to disorganized spindles in S2 cells and defects in spindle organization, spindle pole separation dynamics, and chromosome motility in embryos and thus we propose that this chromokinesin acts on MTs associated with chromosomes and the central spindle where it contributes to the force-balance that drives spindle pole and nuclear separation, and facilitates chromatid motility (Figure 8).
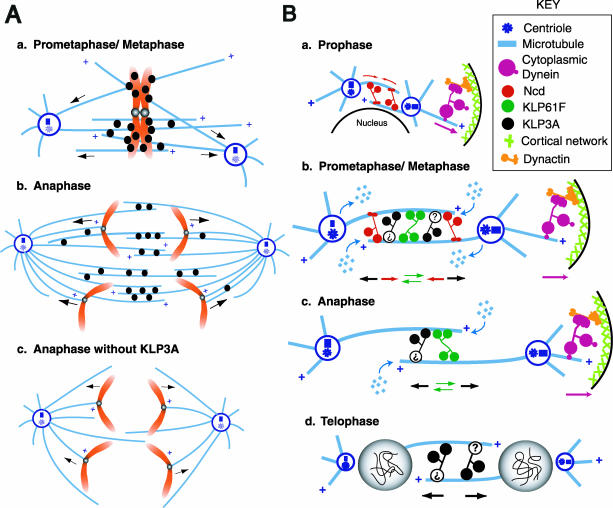
Figure 8.
Model for the functional coordination of KLP3A and other motors in generating the balance of forces that positions spindle poles and daughter nuclei during Drosophila embryonic mitosis. (A) Possible mechanisms for KLP3A action. (a) We hypothesize that KLP3A punctae localized on or around prometaphase/metaphase chromosome arms exert forces that slide MTs with their minus ends outward, thus pushing the spindle poles apart and contributing to spindle elongation during the prometaphase-to-metaphase transition. (b) Subsequently as anaphase A begins, KLP3A translocates to the central spindle where it organizes MTs into ipMT bundles connecting the two poles. Thus the formation of interconnections between the two half spindles facilitates the generation of outward forces acting on the poles, whereas interconnections between ipMT and kMT bundles facilitate chromatid motility. (c) After the inhibition of KLP3A, ipMTs are disorganized, stable interconnections are lost, and the resulting splayed spindles no longer support efficient pole-pole elongation or chromatid-to-pole motility. (B) Model of the pathway for spindle pole positioning determined previously for KLP61F, cytoplasmic dynein, and Ncd (Sharp et al., 2000a), incorporating the data on KLP3A from this study. The KLP3A protein is shown as a hypothetical homodimer possessing MT-binding sites in both the motor domain and the tail region, although its oligomeric state is unknown. (a) During prophase, KLP3A is sequestered in the nucleus like KLP61F and does not participate in initial spindle pole separation. Dynein anchored at the cell cortex pulls astral MTs to separate the poles, whereas Ncd cross-links and slides antiparallel MTs to draw the poles together and to control the rate of spindle pole separation. (b) On NEB, KLP3A (depicted as two motor domains attached to a question mark, because its ultrastructure has not yet been determined) is released from the nucleus and contributes an outward force on the poles during prometaphase/metaphase, thus augmenting the action of KLP61F and dynein. Ncd generates an inward force to counterbalance the outward force due to KLP3A, KLP61F, and dynein. MTs flux poleward with subunit loss at the poles and subunit addition at the plus ends, whereas the poles are maintained at a constant spacing. (c) During anaphase, inhibition of depolymerization at the poles and/or downregulation of Ncd activity allow MT polymerization, and MT sliding by KLP61F, KLP3A, and dynein to exert a force that pushes the poles apart in a temporally coordinated manner. (d) During telophase, KLP3A acts on antiparallel MTs in the midbody region between the newly formed daughter nuclei to establish and maintain the spacing between them, offsetting nuclear collapse or fusion.
Mitotic and Meiotic Functions of KLP3A
Previous genetic studies showed that KLP3A is required for central spindle organization and cytokinesis during male meiosis and is required for pronuclear positioning or specification (Williams et al., 1995, 1997). It was also observed that 10% of embryos derived from klp3a homozygous mothers are viable and displayed mitotic spindle and nuclear spacing defects, but the reported defects were not characterized. The results that we obtained using inhibitor-injected living embryos and RNAi-treated S2 cells are consistent with each other and with the mitotic and nuclear spacing defects observed in the viable subset of klp3a homozygous mutants, suggesting that they are reliable and specific, despite the possible caveats associated with inhibitor microinjection techniques (direct comparisons with mutants have not yet proven feasible because we were unable to obtain viable, healthy klp3a embryos in sufficient quantities for detailed dynamic analysis). Although the KLP3A-negative S2 cells displayed defective spindle morphology by immunofluorescence, the most extensive information on the mitotic functions of KLP3A was obtained by using real-time analysis of living embryos after KLP3A inhibition, which uncovered roles for KLP3A in several aspects of mitosis, and consequently we focus most of the following discussion on embryos.
Localization of KLP3A in Mitotic Cells
Immunofluorescence of KLP3A revealed that it is associated with punctae that are diffuse in the cytoplasm and concentrated on mitotic spindles. This punctate pattern differs from the fibrous, MT-like staining pattern seen with KLP61F, a spindle protein that also exerts outward forces on the poles (Sharp et al., 1999a). Like KLP61F, KLP3A behaves as a MT-binding motor protein, in the sense that it has a canonical kinesin MT-binding site, it cosediments with MTs (Williams et al., 1995; Supplemental Figure 1D), and it requires MTs for spindle localization (Supplemental Figure 2B). However, the nonfilamentous staining suggests that it may also bind [in a MT-sensitive manner] to an unknown non-MT spindle component, plausibly cross-linking it to MTs.
KLP3A contains Zn finger DNA-binding domains characteristic of the XKLP1 subfamily (Vernos and Karsenti, 1995; Williams et al., 1995) and accordingly our immunofluorescence data suggest a chromosomal localization for KLP3A, which is tighter and more persistent in S2 cells than in embryos where we infer a dynamic redistribution of the motor between nuclei, chromosome arms, central spindles, and telophase midbodies (Figure 1). In many respects this pattern resembles that of some other chromokinesins, e.g., XKLP1 (Vernos and Karsenti, 1995) as well as chromosomal passenger proteins, e.g., INCENP/Aurora B/Survivin (Adams et al., 2001a). Thus, KLP3A may exert polar ejection forces on chromosome arms, but we did not observe any defects in chromosome congression and alignment to the metaphase plate after KLP3A inhibition (Figures 2B, 4, and 6A). Although it is possible that this lack of observed defects could be due to functional redundancy with multiple chromokinesins in fly (e.g., Nod, KLP38B, KLP31D/E; Supplemental Figure 1A), we favor the idea that KLP3A's role is to slide MTs relative to chromosomes, thus pushing the poles apart and playing a role in early spindle assembly (Vernos and Karsenti, 1995; Karsenti and Vernos, 2001; see Figure 8Aa). Chromosomal KLP3A could subsequently be carried as a “passenger” to other sites of functional significance, most notably the central spindle and midbody, where it acts on ipMTs to push the poles and daughter nuclei apart (Figures 8, Ab and B). Interestingly mitotic kinases like cdc2 and aurora B have been shown to regulate the redistribution of Xkid and INCENP (Adams et al., 2001b; Ohsugi et al., 2003) and may similarly control the distribution of KLP3A between the central spindle and chromosomes.
Roles of KLP3A in Spindle Pole Separation and Daughter Nuclear Spacing
We observed defects in the structure of ipMT bundles, spindle pole separation, and daughter nuclear spacing after KLP3A inhibition, supporting the hypothesis that KLP3A contributes to the balance of forces that drives spindle pole separation during spindle morphogenesis (Sharp et al., 2000b). There are at least four ways that this could occur (Figure 8). 1) KLP3A could directly cross-link MTs and slide them apart as proposed for KLP61F or it could cross-link MTs into organized arrays to facilitate MT sliding by other motors; 2) KLP3A could slide MTs relative to a non-MT structure such as the enigmatic “spindle matrix” (Walker et al., 2000; Scholey et al., 2001) or an actin network (Giansanti et al., 1998; Sisson et al., 2000); 3) KLP3A may act as a chromokinesin, which pushes chromosome arms plateward and, by action and reaction, pushes the poles apart; or 4) KLP3A could promote the polymerization of ipMTs, thus pushing the poles apart.
It is interesting that the rate of pole separation during the prometaphase-to-metaphase transition is the same in KLP3A-inhibited and control embryos, but the final extent of pole separation is decreased. It is plausible that this reflects the sequential MT sliding activities of the motor proteins contributing to the force-balance, e.g., with KLP61F, and dynein acting early and KLP3A late. It is also possible that KLP3A may control MT polymer mass and thus spindle length by modulating MT dynamics, for example by organizing ipMTs into ordered arrays and thus facilitating MT polymerization at their interdigitating plus ends.
The decrease in the extent of pole separation that accompanies KLP3A inhibition during the prometaphase-to-metaphase transition is suppressed in an Ncd null background. We therefore propose that, during this period, KLP3A generates an outward force on spindle poles that antagonizes the inward force due to Ncd. Previous studies revealed a similar antagonism between KLP61F and Ncd as well as between dynein and Ncd at stages before anaphase B (Sharp et al., 1999b, 2000a). The lack of rescue at anaphase B is consistent with the hypothesis that Ncd is downregulated at this time (Sharp et al., 2000a) or that the KLP3A-inhibited central spindle becomes too disorganized to permit Ncd-induced rescue. An unexplained finding that complicates our simple force-balance model is that metaphase spindles in Ncd nulls are no longer than wild-type spindles, yet the double inhibition of Ncd/KLP3A rescues the spindle length defects that accompany single inhibition of KLP3A. These observations suggest that other spindle length regulatory factors may operate to constrain spindle length in the Ncd nulls.
In KLP3A-inhibited embryos, we observed daughter nuclei collapse and fusion. Nuclear spacing in syncytial blastoderm embryos is regulated by both MTs and the cortical actin network (Zalokar et al., 1975; Zalokar and Erk, 1976; Foe et al., 1993; Sullivan et al., 1993; Sullivan and Theurkauf, 1995; Rothwell et al., 1998). The disruption of cortical actin structures leads to a collision and/or fusion of neighboring nuclei in general. In contrast, the nuclear collapse that follows disrupted central spindles after KLP3A inhibition is specific to daughter nuclei, suggesting that central spindles and midbodies specifically maintain daughter nuclear spacing.
Role of KLP3A in Chromatid to Pole Motility
We were surprised to find that, after KLP3A-inhibition, the velocity of chromatid-to-pole motility on embryonic spindles decreased. We previously observed that the inhibition of the kinetochore motor, dynein, also interfered with chromatid to pole motility, but in that case the most severe phenotype was the detachment of chromosomes from spindle MTs and a complete cessation of chromatid motility (Sharp et al., 2000c). The most severe consequence of KLP3A inhibition was a 30% reduction in the velocity of chromatid-to-pole motility.
Two mechanisms have been proposed to contribute to chromatid-to-pole movement in Drosophila embryos, poleward flux, and dynein transport coupled to MT depolymerization at kinetochores (Sharp et al., 2000c; Brust-Mascher and Scholey, 2002; Maddox et al., 2002). We observed no significant difference in poleward flux during the metaphase/anaphase A isometric state in KLP3A-inhibited embryos (our unpublished data), suggesting that flux-dependent chromatid motility is not altered. However, time-lapse fluorescent imaging revealed that in wild-type embryos, kinetochores moved poleward on kMTs whose trajectories were strikingly coincident with ipMT bundles. In KLP3A-inhibited embryos, these ipMT bundles are not present and kinetochores move more slowly (Figure 8Ac), which leads to the unconventional suggestion that kinetochore-dynein could translocate kinetochores poleward along adjacent ipMT tracks. Alternatively, it is possible that the apparent juxtaposition of kMTs and ipMTs is coincidental and that ipMTs and/or earlier chromosome interactions with non-kMTs serve to maintain pole-pole spacing providing a structural framework for the depolymerizing kMTs that actually move chromatids poleward.
Functional Coordination between KLP3A and Other Mitotic Motors in the Balance of Forces Driving Spindle Pole Separation
Our work suggests that KLP3A contributes to the balance of forces that drives spindle pole separation during spindle assembly and elongation in Drosophila embryos (Figure 8; Sharp et al., 2000a, 2000b; Brust-Mascher and Scholey, 2002; Cytrynbaum et al., 2003). In the absence of KLP3A function, spindle pole separation and nuclear spacing defects are accompanied by the disorganization of ipMT bundles in the central spindle and midbody consistent with the model in Figure 8A. We hypothesize that, before chromosome segregation, KLP3A acts on MTs located on and around chromosomes to slide MTs outward by pushing them relative to chromosome arms and exerts an outward force on spindle poles that contributes to spindle elongation during the prometaphase-to-metaphase transition (Figure 8Aa). After the initiation of chromosome separation (Figure 8Ab), KLP3A is lost from chromosomes and relocalizes to the central spindle and midbody where it mediates ipMT organization, providing a structural framework for the efficient chromatid motility and facilitating the further separation of spindle poles and daughter nuclei that occurs during anaphase B and telophase.
This hypothesis suggests that KLP3A cooperates temporally with other motors, including cortical dynein, the bipolar kinesin, KLP61F, and the C-terminal kinesin, Ncd, to generate the force balance that operates in the pathway of Drosophila embryonic spindle morphogenesis, as shown in Figure 8B. During prophase, KLP3A is sequestered in the nucleus and does not affect spindle morphogenesis. After NEB, KLP3A generates outward MT sliding forces that augment KLP61F and antagonize Ncd. This would allow KLP3A to contribute to spindle elongation specifically during prometaphase-to-metaphase and anaphase B, as observed. Finally, during telophase KLP3A organizes and maintains the central spindle and midbody, which are necessary to maintain daughter nuclear spacing.
Thus, KLP3A is a mitotic chromokinesin that contributes to the spindle assembly force-balance that drives spindle pole separation. To determine its mechanism of action will require biochemical assays that examine its oligomeric state, motor directionality, effect on MT dynamics, and ability to cross-link and move MTs in relation to adjacent MTs, to chromosomes or to other spindle components.
Supplementary Material
Acknowledgments
We thank Drs. Michael Goldberg and Kristen Johansen for generously providing reagents and Dr. Stephen Rogers for advice on S2 cells. We thank Drs. Frank McNally, Ken Kaplan, Lesilee Rose, Bo Liu, Eric Cytrynbaum, Gul Civelekoglu, and members of our laboratory for discussions and comments on the manuscript. This work was supported by Grant GM-55507 from the National Institutes of Health to J.M.S.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03-07-0489. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-07-0489.
Abbreviations used: KLP3A, kinesin-like-protein at 3A; MTs, microtubules; ipMTs, interpolar MTs; GST, glutathione-S-transferase; NEB, nuclear envelope breakdown; kMTs, kinetochore MTs; MMAP, microtubule- and microfilament-associated protein.
Online version of this article contains supplementary figures, tables, and videos for some figures. Online version is available at www.molbiolcell.org.
References
- Adams, R.R., Carmena, M., and Earnshaw, W.C. (2001a). Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11(2), 49-54. [DOI] [PubMed] [Google Scholar]
- Adams, R.R., Maiato, H., Earnshaw, W.C., and Carmena, M. (2001b). Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153(4), 865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower, M.D., and Karpen, G.H. (2001). The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3, 730-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher, I., and Scholey, J.M. (2002). Microtubule flux and sliding in mitotic spindles of early Drosophila embryos. Mol. Biol. Cell 13, 3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, J.C., Worby, C.A., Simonson-Leff, N., Muda, M., Maehama, T., Hemmings, B.A., and Dixon, J.E. (2000). Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA 97, 6499-6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95-114. [DOI] [PubMed] [Google Scholar]
- Cytrynbaum, E.N., Scholey, J.M., and Mogilner, A. (2003). A force balance model of early spindle pole separation in Drosophila embryos. Biophys. J. 84, 757-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechant, R., and Glotzer, M. (2003). Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell 4, 333-344. [DOI] [PubMed] [Google Scholar]
- Foe, V., Odell, G.M., and Edgar B. (1993). Mitosis and Morphogenesis in the Drosophila Embryo. The Development of Drosophila melanogaster, ed. M. Bate and A. Martinez-Arias. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, Vol. 1, 149. [Google Scholar]
- Giansanti, M.G., Bonaccorsi, S., Williams, B., Williams, E.V., Santolamazza, C., Goldberg, M.L., and Gatti, M. (1998). Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev. 12, 396-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., and Geiser, J.R. (1996). Genetic analysis of the mitotic spindle. Annu. Rev. Genet. 30, 7-33. [DOI] [PubMed] [Google Scholar]
- Inoué, S., and Sato, H. (1967). Cell motility by labile association of molecules. The nature of mitotic spindle fibers and their role in chromosome movement. J. Gen. Physiol. 50(Suppl), 259-292. [PMC free article] [PubMed] [Google Scholar]
- Inoué, S., and Salmon, E.D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell. 6, 1619-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., and Mitchison, T.J. (2001). Eg5 is static in bipolar spindles relative to tubulin: evidence for a static spindle matrix. J. Cell Biol. 154, 1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenti, E., and Vernos, I. (2001). The mitotic spindle, a self-made machine. Science 294, 543-547. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., Mitchison, T.J., and Alberts, B.M. (1988). Behavior of microtubules and actin filaments in living Drosophila embryos. Development 103, 675-686. [DOI] [PubMed] [Google Scholar]
- Maddox, P., Desai, A., Oegema, K., Mitchison, T.J., and Salmon, E.D. (2002). Poleward microtubule flux is a major component of spindle dynamics and anaphase A in mitotic Drosophila embryos. Curr. Biol. 12, 1670-1674. [DOI] [PubMed] [Google Scholar]
- McIntosh, K.L., Hepler, P.K., and Van Wie, D.G. (1969). Model for mitosis. Nature 224, 659-663. [Google Scholar]
- McIntosh, J.R., and McDonald, K.L. (1989). The mitotic spindle. Sci. Am. 261, 48-56. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., and Salmon, E.D. (2001). Mitosis: a history of division. Nat. Cell Biol. 3, E17-E21. [DOI] [PubMed] [Google Scholar]
- Ohsugi, M., Tokai-Nishizumi, N., Shiroguchi, K., Toyoshima, Y.Y., Inoue, J., and Yamamoto, T. (2003). Cdc2-mediated phosphorylation of Kid controls its distribution to spindle and chromosomes. EMBO J. 22(9), 2091-2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, G.C., Chui, K.K., Lee, E.W., Wedaman, K.P., Sharp, D.J., Holland, G., Morris, R.L., and Scholey, J.M. (2000). A kinesin-related protein, KRP (180), positions prometaphase spindle poles during early sea urchin embryonic cell division. J. Cell Biol. 150, 499-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers, S.L., Rogers, G.C., Sharp, D.J., and Vale, R.D. (2002). Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J. Cell Biol. 158, 873-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, W.F., Fogarty, P., Field, C.M., and Sullivan, W. (1998). Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organization. Development 125, 1295-1303. [DOI] [PubMed] [Google Scholar]
- Saunders, W.S., and Hoyt, M.A. (1992). Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell 70, 451-458. [DOI] [PubMed] [Google Scholar]
- Scholey, J.M., Rogers, G.C., and Sharp, D.J. (2001). Mitosis, microtubules, and the matrix. J. Cell Biol. 154, 261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholey, J.M., and Mogilner, A. (2002). Mitotic spindle motors. In: Molecular Motors, ed. M. Schliwa, New York: Wiley and Sons, 327-355.
- Scholey, J.M., Brust-Mascher, I., and Mogilner, A. (2003). Cell division. Nature 422, 746-752. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., Brown, H.M., Kwon, M., Rogers, G.C., Holland, G., and Scholey, J.M. (2000a). Functional coordination of three mitotic motors in Drosophila embryos. Mol. Biol. Cell 11, 241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., McDonald, K.L., Brown, H.M., Matthies, H.J., Walczak, C., Vale, R.D., Mitchison, T.J., and Scholey, J.M. (1999a). The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles. J. Cell Biol. 144, 125-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000b). Microtubule motors in mitosis. Nature 407, 41-47. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000c). Cytoplasmic dynein is required for poleward chromosome movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2, 922-930. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., Yu, K.R., Sisson, J.C., Sullivan, W., and Scholey, J.M. (1999b). Antagonistic microtubule-sliding motors position mitotic centrosomes in Drosophila early embryos. Nat. Cell Biol. 1, 51-54. [DOI] [PubMed] [Google Scholar]
- Sisson, J.C., Field, C., Ventura, R., Royou, A., and Sullivan, W. (2000). Lava lamp, a novel peripheral Golgi protein, is required for Drosophila melanogaster cellularization. J. Cell Biol. V151, 905-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somma, M.P., Fasulo, B., Cenci, G., Cundari, E., and Gatti, M. (2002). Molecular dissection of cytokinesis by RNA interference in Drosophila cultured cells. Mol. Biol. Cell 13, 2448-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, W., Fogarty, P., and Theurkauf, W. (1993). Mutations affecting the cytoskeletal organization of syncytial Drosophila embryos. Development 118, 1245-1254. [DOI] [PubMed] [Google Scholar]
- Sullivan, W., and Theurkauf, W.E. (1995). The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr. Opin. Cell Biol. 7, 18-22. [DOI] [PubMed] [Google Scholar]
- Vernos, I., and Karsenti, E. (1995). Chromosomes take the lead in spindle assembly. Trends Cell Biol. V5, 297-301. [DOI] [PubMed] [Google Scholar]
- Walker, D.L., Wang, D., Jin, Y., Rath, U., Wang, Y., Johansen, J., and Johansen, K.M. (2000). Skeletor, a novel chromosomal protein that redistributes during mitosis provides evidence for the formation of a spindle matrix. J. Cell Biol. 151, 1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, B.C., Dernburg, A.F., Puro, J., Nokkala, S., and Goldberg, M.L. (1997). The Drosophila kinesin-like protein KLP3A is required for proper behavior of male and female pronuclei at fertilization. Development 124, 2365-2376. [DOI] [PubMed] [Google Scholar]
- Williams, B.C., Riedy, M.F., Williams, E.V., Gatti, M., and Goldberg, M.L. (1995). The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 129, 709-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann, T., Hyman, A., and Desai, A. (2001). The spindle: a dynamic assembly of microtubules and motors. Nat. Cell Biol. 3, E28-E34. [DOI] [PubMed] [Google Scholar]
- Zalokar, M., Audit, C., and Erk, I. (1975). Developmental defects of female-sterile mutants of Drosophila melanogaster. Dev. Biol. 47, 419-432. [DOI] [PubMed] [Google Scholar]
- Zalokar, M., and Erk, I. (1976). Division and migration of nuclei during early embryogenesis of Drosophila melanogaster. J Microscopie Biologie Cellulaire 25, 97-108. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.