Abstract
Epigenetic therapy for solid tumors could benefit from an in vivo model that defines tumor characteristics of responsiveness and resistance to facilitate patient selection. Here we report that combining the histone deacetylase inhibitor entinostat with the demethylating agent vidaza profoundly affected growth of K-ras/p53 mutant lung adenocarcinomas engrafted orthotopically in immunocompromised nude rats by targeting and ablating pleomorphic cells that occupied up to 75% of the tumor masses. A similar reduction in tumor burden was seen with epigenetic therapy in K-ras or EGFR mutant tumors growing orthotopically. Increased expression of pro-apoptotic genes and the cyclin dependent kinase inhibitor p21 was seen. Hundreds of genes were demethylated highlighted by the re-expression of polycomb-regulated genes coding for transcription factor binding proteins and the p16 gene, a key regulator of the cell cycle. Highly significant gene expression changes were seen in key regulatory pathways involved in cell cycle, DNA damage, apoptosis, and tissue remodeling. These findings demonstrate the promise for epigenetic therapy in cancer management and provide an orthotopic lung cancer model that can assess therapeutic efficacy and reprogramming of the epigenome in tumors harboring different genetic and epigenetic profiles to guide use of these drugs.
Keywords: lung cancer, DNA methylation, polycomb, epigenetic therapy, nude rat
Introduction
Lung cancer is the leading cause of cancer-related death in the U.S. and will soon reach epidemic levels (1). The minimal efficacy of conventional chemotherapy has prompted a renewed focus on targeted therapy based on pathways altered during the pathogenesis of lung cancer. Two targets being exploited are the epidermal growth factor receptor family and vascular endothelial growth factor (2-6). Although some patients have shown dramatic and sustained response to these therapies, overall response and survival advantage for non-small cell lung cancer (NSCLC) patients in Phase III trials have been modest, albeit statistically significant (2-6). Unfortunately, even preselecting patients based on dysfunction within the targeted pathway is unlikely to yield sustained response in most cases due to the molecular heterogeneity of lung tumors (7).
The silencing of genes through promoter hypermethylation is a major and causal epigenetic event that occurs during lung cancer initiation and progression and affects the function of hundreds of genes. Gene silencing involves methylation of cytosines in the gene promoter region, recruitment of transcriptional co-repressors, and modification of histone tails that culminate in the establishment of chromatin modifications that block transcription (8, 9). Cytosine methylation appears dominant in transcriptional repression, and inhibitors of the cytosine DNA-methyltransferases, vidaza (5-azacytidine [5-Aza]), and decitabine (5-deoxy azacytidine [DAC]) can induce in vitro re-expression of genes silenced through promoter hypermethylation (8, 9). Importantly, while inhibitors of histone deacetylation (HDAC) are not very effective in inducing re-expression of genes silenced by promoter hypermethylation, such inhibitors can synergize with demethylating agents to relieve transcriptional repression (10). Thus, the reversal of gene silencing by pharmacological agents may offer an effective strategy for primary and adjuvant cancer therapy.
Rather than targeting single pathway alterations in cancer, epigenetic therapy may circumvent the problem of tumor heterogeneity by inducing the re-expression of multiple tumor suppressor genes essential for abrogating cancer cell survival and proliferation. The initial history of demethylating agents suggested they were too toxic and not efficacious as cancer cytotoxic agents. However, as a result of laboratory findings, these drugs were reassessed and proved to be a potent therapy for myelodyplasia (MDS), a precursor state to acute myelogenous leukemia (AML), when used at doses much lower than the maximum tolerated dose leading to the approval of these drugs by the FDA for treatment of these diseases (11). Clinical trials with DNA demethylating agents combined with HDAC inhibitors are showing promising responses in the treatment of myeloid malignancies (12). The extension of this targeted approach to solid tumors including lung also may hold promise as a therapy. Our work in which combined treatment with DAC and sodium phenylbutyrate reduced the number of developing lung tumors in a murine model by > 50% supports this supposition (13).
The expansion of epigenetic therapy in the clinic makes it critical to define tumor characteristics that will be most responsive or resistant to epigenetic therapy in order to better select patients with the highest potential to respond. This is a challenge in clinical trials where access to tumor tissue pre- and post-therapy is uncommon. We have developed an orthotopic lung cancer model in which xenografts of human lung cancer-derived cell lines are efficiently engrafted throughout the lungs of the Rowett nude rat (14). The purpose of this study was to determine the efficacy of a demethylating agent alone or in combination with an HDAC inhibitor on the growth of tumors engrafted in the lungs of the nude rat and on reprogramming of the epigenome.
Materials and Methods
Tumor cell implantation and treatment
Male Rowett nude rats (Cr:NIH-ru), 8-10 weeks old were obtained from Frederick Cancer Research and Development (Frederick, MD). Calu-6, A549, and H1975 cells obtained from American Type Culture Collection (Manassas, VA) were cultured and instilled via orotracheal intubation as described (14). Rats (n=20/group) were treated with vehicle (saline or DMSO [10 each]), 5-Aza (Sigma, St. Louis, MO [2 mg/kg, dissolved in saline] or entinostat (MS-275, gift from Syndax Pharmaceutical, Waltham, MA [1 mg/kg, dissolved in DMSO]) by intraperitoneal injection.
Tissue collection, estimation of tumor volume and photomicroscopy
Prior to sacrifice, four animals from each treatment group were randomly selected for collection of tumors for molecular assays. Five tumors were collected from each animal with exception of one rat treated with combination therapy that had no tumors (instillation likely missed the trachea). After weighing the remaining lungs, they were inflated with 10% neutral-buffered formalin at a constant hydrostatic pressure of 25 cm for 6 hours. Lung volume (VL) was determined by fluid displacement. Lung lobes were trimmed at 3-4 mm intervals in a direction perpendicular to the axial airways with the first slice placed randomly near the cranial and/or hilar end. Paraffin embedded lungs were sectioned at 5 μm thickness and stained with hematoxylin and eosin. Approximately 30 histology sections were generated per set of lung lobes. For measurement of tumor size, a mylar morphometry grid with points spaced one mm apart was overlaid upon each of the microscope slides. The total area of the lung sections and the area occupied by tumor were estimated by counting points overlying tissue (15). An estimate of total tumor volume was determined by the percentage of tissue points overlying tumor multiplied by the VL.
Gross pathology images from a PowerShot S3IS digital camera (Canon USA, Inc.) were processed with Adobe Photoshop CS3 (v. 10.0.1; Adobe Systems, Inc.). Contrast was adjusted to +50; images were cropped, rotated, and resized. Measurement bars of 4 mm length were added with the line tool of Photoshop. Photomicrographs were made using Olympus equipment and software (BX41 with UPlanFL objectives, DP25 camera; Olympus America, Inc.). Automatic exposure settings were used, contrast was adjusted to +50, images were resized, and image magnification was indicated using measurement bars of known length.
Gene expression and protein analysis
RNA was isolated following TRI-reagent (Sigma, St. Louis, MO) instructions. Total RNA (1 μg) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) according to the manufactures protocol. RT-qPCR was performed with the ABI PRISM 7900HT and inventoried Taqman assays (Applied Biosystems). Experiments were normalized to GAPDH. Results were repeated in three separate experiments. Data were analyzed with respect to a calibrator sample using the 2-ΔΔCt method and reported as relative quantity (RQ). DNMT1 protein levels were quantified by Western blot as described (16).
Gene methylation and expression profiling
Bisulfite modified DNA isolated from vehicle, 5-Aza, or 5-Aza + MS-275 treated Calu-6 tumors were hybridized to the Methylation Promoter27 beadchip (Illumina, San Diego, CA). Calu-6 DNA treated with SSS1 methylase that converts all CpGs to 5-methylcytosine was included on the array to define signal intensity for each probe and bisulfite modified DNA from normal bronchial epithelial cells was also included to identify genes methylated in normal cells. Average signal intensity between the methylated and unmethylated probes was determined, and a β-value from 0-1 (fully methylated) was calculated. B-values ≥ 0.45 were scored as positive for methylation in vehicle-treated tumors and a reduction in β-value of ≥ 20% for a methylated gene was score as demethylation. Total RNA isolated from the tumors interrogated by the Methylation Promoter27 beadchip were hybridized to Agilent 41K expression arrays using a standardized protocol to identify changes in gene expression.
Statistical analyses
The two-sample t-test and analysis of variance were used to compare the two treatment groups and the two groups with the vehicle, respectively. To check appropriateness of the statistical methods, analyses were rerun with transformed data if appropriate or using the Kruskal-Wallis test. When multiple tumors per rat were used, mixed effects models with a random effect for rat were used. Microarray data were background corrected and analyzed using the linear model analysis Bioconductor package limma that generated mRNA levels and measures of significance. The limma output for each tumor was used for analysis with Metacore (GeneGo Inc., St. Joseph, MI) to identify the functional pathways and networks statistically over represented in the set of changed genes. Only genes increasing or decreasing at least 1.5-fold in two or more tumors for each treatment were included in the analysis. The raw data of the microarray analysis have been deposited in the GEO database (GSE24732). Analyses were done with SAS9.2 unless otherwise indicated.
Results
Epigenetic therapy reduces tumor burden and targets poorly differentiated tumor cells
The Calu-6 cell line, established from an adenocarcinoma, was selected for these studies because it contains mutant K-ras and p53 genes, a genetic profile common to NSCLC (17). Our group has also validated through methylation specific PCR (MSP) and bisulfite sequencing that 44 genes, most notably p16, are silenced through promoter methylation in this cell line (18, 19).
A dose of 2.0 mg/kg 5-Aza was used since it was well tolerated in pilot studies (no systemic toxicity) and approximates the human dose of 40 mg/m2 used in a Phase I/II clinical trial for advanced lung cancer (Juergens, unpublished). The dose selected for the HDAC inhibitor MS-275 (1.0 mg/kg) has been used in animal studies and is within the range being tested in human clinical trials (20, 21). Three groups (n = 20/group) vehicle, 5-Aza, and 5-Aza + MS-275, comprised this efficacy study. MS-275 was not tested alone due to the lack of effectiveness of HDAC inhibitors on murine lung tumor growth and re-expression of methylated genes (10, 13). In addition, no objective responses were seen in lung cancer patients treated with the HDAC inhibitor Romidepsin (22). An additional six rats that did not receive tumor cells were included to determine tumor free lung weights for comparison to treatment groups. The dose schedule selected was four consecutive days of treatment with 5-Aza followed with vehicle or MS-275 on day 5 for four consecutive weeks. This dosing schedule was selected since HDAC inhibitors can cause cell cycle arrest and if given during 5-Aza treatment could reduce the maximal demethylation response (20). Treatment was initiated three weeks after instillation of tumor cells, a time point at which small nodules are scattered throughout the lung parenchyma (14). Without intervention, these nodules grow to coalesce and form large masses that efface >75% of the parenchyma at 9 weeks post-instillation.
Tumor burden was quantified seven weeks following instillation of Calu-6 cells as the change in normal lung weight compared to tumor bearing lung weights. The sample sizes are reduced slightly due to exclusion of animals with no evidence of tumors that occurs when instillation through the orotracheal intubation presumably misses the trachea resulting in placement into the esophagus instead. In addition, two rats were terminated early due to infection unrelated to treatment. Rats that remained in the study showed an 18 – 22% increase in body weight over the 4 weeks, irrespective of treatment. Treatment with 5-Aza significantly reduced tumor burden by 31%, while the combination of 5-Aza + MS-275 had a dramatic effect, reducing tumor burden by 60% (Table 1). Smaller tumor burden was manifested as a reduction in lung weight that in turn was highly correlated with estimates of tumor volume (r = 0.95, p < 0.0001 [n = 14 – 19/group]). The dramatic difference in tumor burden and volume can be seen in the photographs of the dorsal aspects of the lungs from a vehicle and 5-Aza + MS-275 treated rat (Fig. 1A [a, b]). This finding was replicated in A549 (K-ras mutant) and H1975 (EGFR mutant) tumors growing orthotopically in which tumor burden was reduced by 40% and 53%, respectively (p < 0.001; n = 20/group) with combination therapy (not shown).
Table 1.
Epigenetic Therapy Dramatically Reduces Lung Tumor Burden
| Treatment Group1 | N | Lung Weight (grams) (Mean ± SD) | Adjustment for Control Weight (± SD) | Comparison to Vehicle (p- value*) |
|---|---|---|---|---|
| Control2 | 6 | 1.76 ± 0.21 | NA | NA |
| Vehicle | 18 | 4.23 ± 1.9 | 2.48 ± 1.67 | Reference |
| 5-AZA | 19 | 3.45 ± 0.97 | 1.70 ± 0.86 | 0.05 |
| 5-AZA + MS275 | 14 | 2.74 ± 0.54 | 0.99 ± 0.47 | ≤ 0.0001 |
Size of treatment groups differ due to engraftment.
Control animals received no tumor cells.
NA, not applicable
Overall p-value = 0.009
Figure 1.
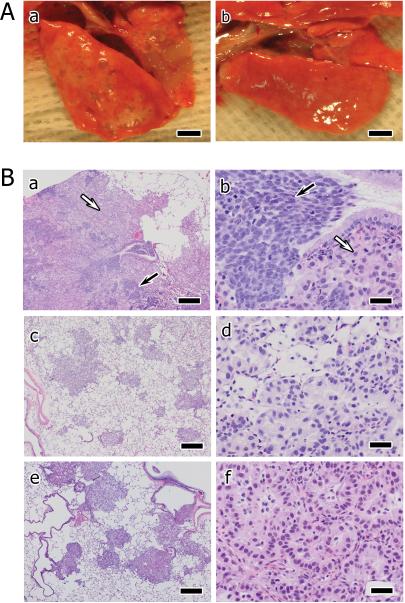
Gross (A) and (B) microscopic pathology is shown for orthotopically xenografted human lung adenocarcinomas growing in the lungs of nude rats. (A): The lungs of an untreated control animal engrafted with Calu-6 cells [a] show multinodular, tan-gray, mucoid masses that bulge from the pleural surfaces. Lungs of an animal treated with 5-Aza + MS-275 [b] showing reduced tumor burden (bars for [A] and [B] = 4 mm). (B): Much of the parenchyma and smaller airways and blood vessels are effaced in lung sections from a vehicle treated animal by [a, b] coalescent masses composed of two distinct cell populations. Cords and nests of darker, basophilic pleomorphic cells (black arrows) are interspersed with more extensive areas of paler, well-differentiated adenocarcinomas (white arrows). The pleomorphic cell population displays moderate anisokaryosis, scant cytoplasm, disorientation, and scattered apoptotic bodies and mitotic figures. The more differentiated cells display typical adenocarcinoma morphology including tubuloacinar arrangement, basally oriented nuclei, and abundant vacuolated to foamy cytoplasm with mucus-secreting features. Less apoptosis and mitotic activity are evident among the more differentiated population of cells. Lung sections from an animal treated with 5-Aza [c, d] show scattered small nodules composed of the well-differentiated adenocarcinoma cells. Alveoli in affected foci are partially to completely filled, but not entirely effaced by cancer cells. The cells have abundant vacuolated cytoplasm. Lung sections from an animal treated with 5-Aza + MS-275 [e, f] show little coalescence of tumor nodules and little to none of the pleomorphic cell population. Cells are aligned along and partially destroy alveolar septa and have somewhat smaller amounts of cytoplasm than cells in [c, d]. Histology sections for [a] through [f] are stained with hematoxylin and eosin (bars for [a], [c], [e] = 400 μm; bars for [b], [d], [f] = 40 μm).
All subsequent histopathology and molecular characterization was focused on the Calu-6 tumors. Histopathology findings in vehicle treated animals revealed large, coalesced masses that occupied up to 50% of the parenchyma within sections of the lung and were often located near bronchioloalveolar duct junctions (Fig. 1B [a, b]). The masses were composed of two morphologically distinct cell populations. One population was composed of well-differentiated mucus secreting adenocarcinoma cells with abundant and finely vacuolated cytoplasm. Tubule lumens and remnants of alveolar airspace were often filled with mucinous material. Occasional nuclear and cytolytic fragments were scattered among these cells along with infiltrates of neutrophils and rare mitotic figures. A second interspersed cell population was comprised of solid cords and nests of pleomorphic cells that occupied up to 75% of the tumor mass (Fig. 1B [c, d]). These cells were disoriented, displayed moderate anisokaryosis, and had little cytoplasm and indistinct cell margins. Mitotic activity in these foci was high (up to 10 mitotic figures per 40X field) along with apoptosis and liquifactive necrosis within the centers of the cell nests. Tumor mass was greatly reduced in lungs treated with 5-Aza alone or in combination with MS-275 corroborating results from assessment of overall tumor burden and volume. Epigenetic therapy largely ablated the pleomorphic cell population (< 5% of the residual tumor) with a portion of the well-differentiated tumor cells remaining (Fig. 1B [c – f]). Mitotic activity was reduced by approximately 50% in the lung tumors from 5-Aza-treated animals and was rare in animals receiving the combination therapy. Only mild inflammation similar to untreated lungs was seen with the epigenetic therapy.
Epigenetic therapy increases expression of pro-apoptotic genes and p21
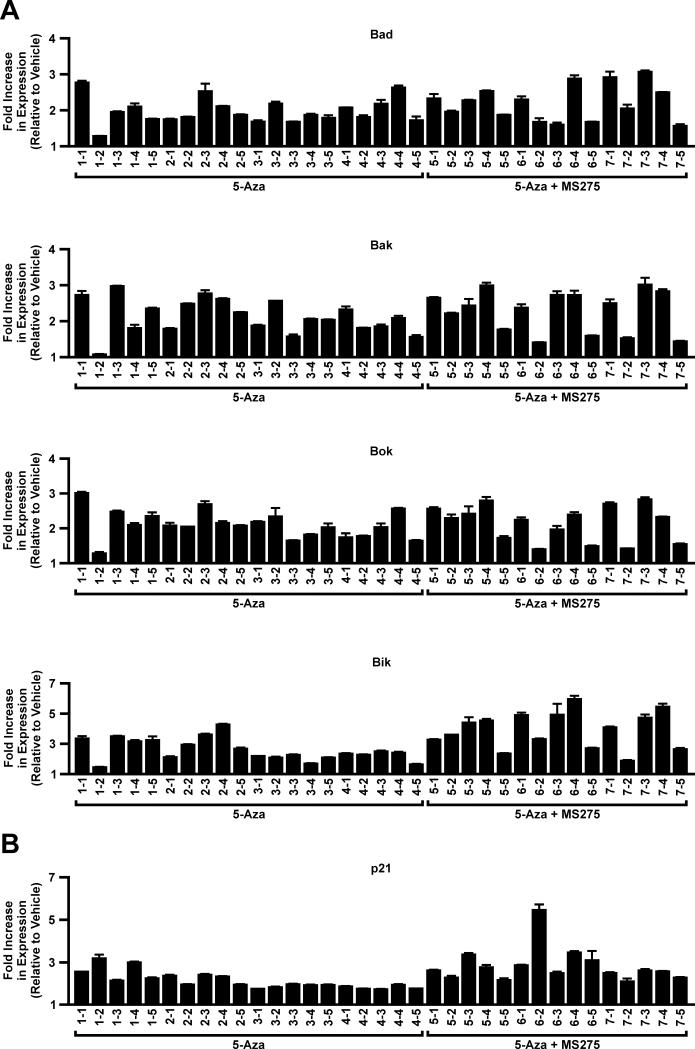
Tumors were collected from lungs of rats treated with vehicle, 5-Aza, or 5-Aza + MS-275 for molecular assays. Expression of the pro-apoptotic genes Bad, Bak, and Bok were increased up to 3-fold in tumors, irrespective of treatment. The magnitude of expression of these genes was similar within a tumor, but varied across tumors within and between lungs from treated animals (Fig. 2A). The largest increase in expression was seen for Bik, a major pro-apoptotic gene in lung epithelial cells (23), and this was greater with combined treatment than 5-Aza alone (3.9 versus 2.6-fold on average, p < 0.001; Fig. 2A). The cyclin-dependent kinase inhibitor p21 (WAF1) functions as a regulator of cell cycle progression and senescence and is induced by HDAC inhibitors that include MS-275 (24). The expression of p21 was increased 1.6 – 3.1-fold and 2.0 – 5.5-fold in tumors from animals treated with 5-Aza or combined treatment, respectively. Overall, the increase in p21 expression was significantly greater in tumors treated with 5-Aza + MS-275 versus 5-Aza (2.8-fold [n = 20] versus 2.0-fold [n = 15] on average, p < 0.01; Figure 2B).
Figure 2.
Epigenetic therapy increases the expression of pro-apoptotic genes and p21. TaqMan PCR was used to quantify the change in expression of the pro-apoptotic genes Bad, Bak, Bok, and Bik (A) and p21 (B) in tumors from animals (animal number-tumor number) treated with 5-Aza or 5-Aza + MS-275. The fold increase in expression is relative to expression seen in vehicle tumors and normalized to GAPDH. Values are the mean ± SEM from triplicate experiments.
Inhibition of cytosine DNA-methyltransferase 1 (DNMT1) and reprogramming of the epigenome
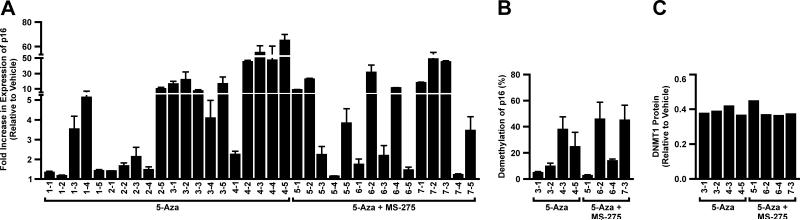
The effect of epigenetic therapy on expression of p16INK4a (p16) whose CpG island is densely methylated leading to complete loss of expression in Calu-6 tumors was used to initially evaluate response to therapy. Considerable heterogeneity within and across animals was seen for re-expression of p16 that ranged from 1.5 – 50-fold and did not differ significantly between treatment groups (Fig. 3A). Bisulfite sequencing of the p16 CpG island in four tumors randomly selected from each treatment group, revealed average demethylation of 5 – 45% across the 27 CpGs interrogated (Fig. 3B) that in turn correlated with increased gene expression (r = 0.72; p = 0.04). Finally, DNMT1 protein levels were reduced by approximately 60%, irrespective of treatment (Fig. 3C).
Figure 3.
Epigenetic therapy induces re-expression and demethylation of the p16 gene and reduces DNMT1 protein levels in lung tumors. (A) TaqMan PCR was used to quantify the change in expression of p16 in tumors from animals treated with 5-Aza or 5-Aza + MS-275. The fold increase in expression is relative to expression seen in vehicle treated tumors and normalized to GAPDH. Values are the mean ± SEM from triplicate experiments. (B) Bisulfite sequencing of 27 CpGs within the p16 promoter of 4 tumors each from animals treated with 5-Aza or 5-Aza + MS-275. Values are the mean percent of demethylation of 10 clones relative to vehicle treated tumors across the CpG island ± SEM. (C) Fold reduction of DNMT1 protein levels relative to vehicle tumors and normalized to β-actin in four tumors each (animal number-tumor number) from animals treated with 5-Aza or 5-Aza + MS-275 is shown.
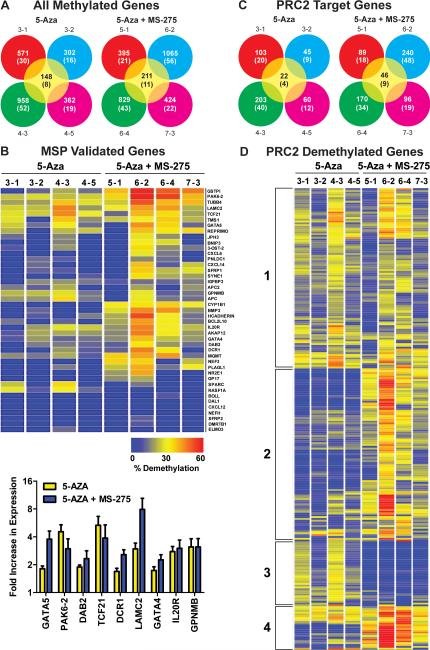
Studies were extended in these eight treated tumors and four vehicle tumors to address the effect of epigenetic therapy on global reprogramming of the epigenome. Bisulfite modified DNA was hybridized to the Methylation27 Beadchip (Illumina, Inc. San Diego, CA) that interrogates 27,600 CpG sites spanning promoter regions of more than 14,000 genes. The number of methylated genes in vehicle-treated tumors varied by < 1% with 1914 genes methylated in all tumors. Demethylation ≥ 20% was seen for 302 – 988 and 395 – 1065 genes in tumors treated with 5-Aza and 5-Aza + MS-275, respectively (Fig. 4A). Differences in the number of demethylated genes were evident across tumors and within animals. The number of genes not methylated across tumors is also shown (Figure S1). A heatmap was generated to display by tumor the extent of demethylation seen in 43 genes methylated on this array (p16 is excluded due to uninformative interrogation by the Beadchip) that had been validated previously using MSP (18, 19). This heatmap shows a similar prevalence for gene demethylation across the two treatment groups, but a slightly greater degree of demethylation with combined therapy (Fig. 4B). TaqMan assays for nine of the demethylated genes corroborated results from the array with increased expression of 2 – 8-fold that correlated well with extent of demethylation (Fig. 4B; r = 0.5 – 0.9; n = 8). The difference in magnitude between re-expression of these genes and p16 is due to the fact that p16 expression was not detected, while low levels of expression was seen for the other genes in vehicle tumors.
Figure 4.
Epigenetic therapy induces reprogramming of the epigenome. (A) Total number of genes showing ≥ 20% demethylation in 4 tumors each (animal number-tumor number) from animals treated with 5-Aza or 5-Aza + MS-275. Values in parentheses reflect the percentage of methylated genes that were demethylated. Center circle depicts the number and percent of genes commonly demethylated across tumors and by treatment. (B) Heatmap showing the extent of demethylation by tumor of 43 genes positive for methylation on the Methylation27 Beadchip and validated by MSP. The fold increase in expression of nine genes showing demethylation in the heatmap by treatment is shown. Values are mean ± SEM from triplicate TaqMan PCR assays. (C) PRC2 target genes showing ≥ 20% demethylation in 4 tumors each from animals treated with 5-Aza or 5-Aza + MS-275. (D) Heatmap showing an unsupervised cluster analysis of PRC2 demethylated genes across tumors and by treatment. Four clusters were identified.
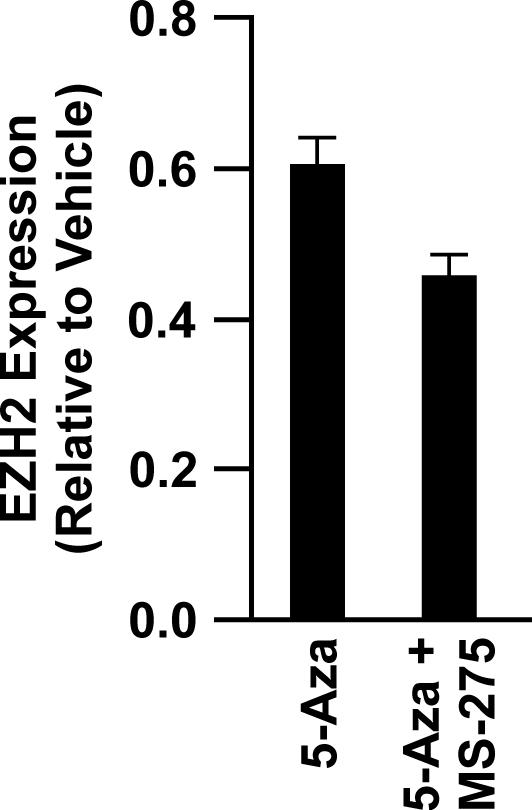
The histone methyltransferase EZH2 is a member of the Polycomb group (PcG) of proteins and catalyzes the trimethylation at Lysine27 of histone H3 (H3K27me3). Methylated K27 along with EZH2, EED, and SUZ12 comprise the PRC2 complex that contributes to the formation of a repressive chromatin state. This PRC2 complex can also recruit the DNMTs (DNMT1, 3a, and 3b) and thus, may play an important role in tumor-specific targeting of de novo methylation at specific gene promoters (25-27). Stem cells rely on PcG proteins to reversibly repress genes encoding transcription factors required for differentiation (28). Several recent studies support the hypothesis of a stem cell signature in cancer in which differentiated cells undergoing transformation reacquire stem cell characteristics through a process of dedifferentiation thereby acquiring the H3K27me3 mark and de novo cytosine methylation (29, 30). Probe sets for 1472 PRC2 target genes are annotated on the Methylation27 Beadchip and 503 (34%) of these genes were methylated in Calu-6 tumors. Thus, PRC2 target genes account for 25% of the genes positive for methylation in this cell line. Moreover, these genes were readily demethylated by both treatments (Figure 4C). Eighty-four of these genes coded for transcription factors. Interestingly, unsupervised cluster analysis of the 258 PRC2 target genes that were demethylated revealed four distinct clusters. The 95 genes found in cluster 2 had a much greater extent of demethylation with the combined therapy compared to treatment with 5-Aza alone (Fig. 4D, Table S1). Thirty-five of these genes code for transcription factor binding proteins (e.g., FOX, HOX gene families) and their extent of demethylation was significantly greater in tumors exposed to combined versus single therapy (29 ± 1% versus 9 ± 1%, respectively; p < 2 × 10-16). In addition, expression of EZH2 was reduced 55 and 40% in tumors exposed to combined versus single therapy, respectively (Fig. 5).
Figure 5.
Epigenetic therapy reduces expression of EZH2. TaqMan PCR was used to quantify the change in expression of EZH2 in tumors from animals treated with 5-Aza (n= 20) or 5-Aza + MS-275 (n = 15) relative to expression in vehicle tumors (n = 4). Values are mean ± SEM.
Epigenetic therapy reprograms major cancer signaling pathways
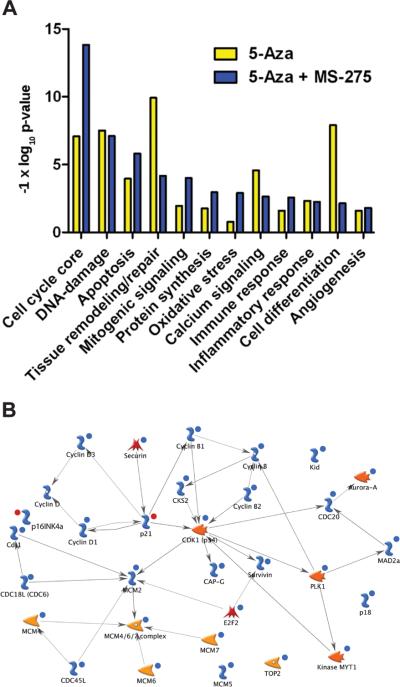
Malignant transformation involves the disruption of many pathways within the cell. Metacore analysis of Agilent expression arrays generated from vehicle and the eight tumors interrogated on the Methylation27 Beadchip was used to identify the major pathways modulated by epigenetic therapy. The most significant changes were seen in expression of genes within the cell cycle, DNA damage, apoptosis, and tissue remodeling (Fig. 6A, Table S2). Other key cell functions altered were inflammatory and immune response, cell differentiation, and angiogenesis. The effect on expression of genes within the cell cycle and apoptotic pathways was greater with the combined therapy. Genes altered in the core cell cycle by single and combined therapy (Fig. 6B, Table S2) included reduced expression of cyclin and cell division kinase (cdk) gene families along with increased expression of p16 and p21 as validated by TaqMan assays. Expression of the minichromosome maintenance (MCM) genes involved in the initiation and elongation phases of DNA replication, Aurora A-kinase that regulates chromosome segregation and cytokinesis and Survivin, a regulator of mitosis and apoptosis were all reduced (Fig. 6B) (31-33).
Figure 6.
Epigenetic therapy affects the expression of genes in key cell signaling pathways. (A) Pathways with the greatest number of genes displaying significant changes in expression in response to 5-Aza or 5-Aza + MS-275 are ranked by -1 × log10 p-value. (B) Metacore depiction of genes within the core cell cycle with significant changes in expression. Red circles indicate increased expression, while blue circles reflect reduced expression relative to tumors from vehicle treated animals. Other symbols:  (transcription factor),
(transcription factor),  (generic binding protein),
(generic binding protein),  (generic phospholipase),
(generic phospholipase),  (generic phosphatase),
(generic phosphatase),  (lipid kinase).
(lipid kinase).
Discussion
These studies demonstrate that epigenetic therapy can profoundly affect the growth of lung adenocarcinomas engrafted in the lungs of the nude rat by reprogramming the epigenome and awakening key regulatory pathways within the cell. The poorly differentiated aggressive cells characterized by high mitotic activity were virtually eliminated within these tumors by epigenetic therapy. Paramount to the effectiveness of this therapy may be the re-expression of polycomb-regulated genes, many of which code for transcription factor binding proteins, and the p16 gene that plays a key role in cell proliferation as a negative regulator of cyclin/CDK complexes (34).
Several studies demonstrate the targeting of PcG proteins for silencing by promoter hypermethylation in malignant tumors and this was also observed in the Calu-6 tumors (25, 29, 30). The effectiveness of epigenetic therapy in demethylation of these genes may relate to the dual role of DNMT1 in cytosine methylation and its interaction with the PRC2 proteins (27). The depletion of DNMT1 and reduced expression of EZH2 seen with 5-Aza should lead to disruption of this repressive chromatin state and this may augment the incorporation of this nucleoside analog during replication to facilitate cytosine demethylation. The heterogeneity of demethylation seen in the response of individual lung tumors to epigenetic therapy is not surprising given the difference in size (< 1 – 3 mm 5 weeks after engraftment) and proportion (25-75%) of pleomorphic tumor cells growing in the lungs of vehicle treated animals (14). Size differences of 4-fold were also seen in tumors collected from treated animals. Thus, the microenvironment of the nude rat lung clearly impacts tumor composition and growth to provide a setting that more closely recapitulates the human lung, and is superior to subcutaneous xenograft models. Remarkably, irrespective of this plasticity, hundreds of genes were still demethylated in each tumor, substantiating the in vivo efficacy of this therapy.
The increased reduction in tumor burden seen with addition of the HDAC inhibitor MS-275 may relate to its recognized anti-proliferative activity and synergism with 5-Aza to facilitate gene re-expression of selected PRC2 target genes (10). HDAC inhibitors strongly activate expression of the CDK inhibitor p21 that regulates cell cycle progression at G1 through enhanced histone deacetylation around its promoter (24). The greater expression of p21 in tumors exposed to 5-Aza + MS-275 versus 5-Aza alone was likely one contributor to the marked effect on genes within the cell cycle. In addition, genes regulating apoptosis as noted by the increased expression of Bik, a pro-apoptotic tumor suppressor gene that is epigenetically regulated in cancer, were also activated to a greater extent with combined therapy (35). The most dramatic effects seen with the addition of MS-275 was the enhanced re-expression of a subset (Cluster 2) of PRC2 target genes that include members of the Hox family recognized for their role in regulating apoptosis, receptor signaling, differentiation, motility, and angiogenesis networks that were increased by this therapy (36).
The extension of this low dose combination epigenetic therapy to solid tumors in the clinic is just beginning with Phase II trials in lung, breast, and colon cancer starting or in progress. The dramatic response seen in this orthotopic model has been replicated in patients participating in a Phase II trial of 5-Aza + MS-275. Complete, partial, or long-term stabilization responses were observed in three patients, respectively with advanced stage NSCLC that had progressed after several other therapies (Juergens, unpublished). These exciting findings highlight the promise for epigenetic therapy in cancer management. However, it will be critical to define tumor characteristics and biomarkers that may be most responsive or resistant to epigenetic therapy in order to better select patients with the highest potential to respond. This validated orthotopic lung cancer model, through its ability to evaluate epigenetic therapy and reprogramming of the epigenome in lung tumors harboring different genetic mutations and epigenetic profiles has already identified key regulatory genes targeted for re-expression and should provide important new insights to guide and further the development of human clinical trials for the treatment of lung cancer.
Supplementary Material
Supplemental Figure Legend. Figure S1. Total number of genes showing no demethylation in 4 tumors each (animal number-tumor number) from animals treated with 5-Aza or 5-Aza + MS-275. Values in parentheses reflect the percentage of methylated genes. Center circle depicts the number and percent of genes commonly remaining methylated across tumors.
Acknowledgements
This study was supported by the Waxman Foundation and by a grant from NIH (R01 CA095568).
Footnotes
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Azim HA, Jr., Ganti AK. Targeted therapy in advanced non-small cell lung cancer (NSCLC): where do we stand? Cancer Treat Rev. 2006;32:630–636. doi: 10.1016/j.ctrv.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 3.Dy GK, Adjei AA. Angiogenesis inhibitors in lung cancer: a promise fulfilled. Clin Lung Cancer. 2006;7(Suppl 4):S145–149. doi: 10.3816/clc.2006.s.006. [DOI] [PubMed] [Google Scholar]
- 4.Sandler A, Herbst R. Combining targeted agents: blocking the epidermal growth factor and vascular endothelial growth factor pathways. Clin Cancer Res. 2006;12:4421s–4425s. doi: 10.1158/1078-0432.CCR-06-0796. [DOI] [PubMed] [Google Scholar]
- 5.Spicer J, Chowdhury S, Harper P. Targeting novel and established therapies for non-small cell lung cancer. Cancer Lett. 2007;250:9–16. doi: 10.1016/j.canlet.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 6.Auberger J, Loeffler-Ragg J, Wurzer W, Hilbe W. Targeted therapies in non-small cell lung cancer: proven concepts and unfulfilled promises. Curr Cancer Drug Targets. 2006;6:271–294. doi: 10.2174/156800906777441780. [DOI] [PubMed] [Google Scholar]
- 7.Bunn PA, Jr., Dziadziuszko R, Varella-Garcia M, et al. Biological markers for non-small cell lung cancer patient selection for epidermal growth factor receptor tyrosine kinase inhibitor therapy. Clin Cancer Res. 2006;12:3652–3656. doi: 10.1158/1078-0432.CCR-06-0261. [DOI] [PubMed] [Google Scholar]
- 8.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 9.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 10.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 11.Yang AS, Doshi KD, Choi SW, et al. DNA methylation changes after 5-aza-2'-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–5503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 12.Gore SD, Baylin S, Sugar E, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–6369. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 13.Belinsky SA, Klinge DM, Stidley CA, et al. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 14.March TH, Marron-Terada PG, Belinsky SA. Refinement of an orthotopic lung cancer model in the nude rat. Vet Pathol. 2001;38:483–490. doi: 10.1354/vp.38-5-483. [DOI] [PubMed] [Google Scholar]
- 15.Bolender RP, Hyde DM, Dehoff RT. Lung morphometry: a new generation of tools and experiments for organ, tissue, cell, and molecular biology. The American journal of physiology. 1993;265:L521–548. doi: 10.1152/ajplung.1993.265.6.L521. [DOI] [PubMed] [Google Scholar]
- 16.Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68:9005–9014. doi: 10.1158/0008-5472.CAN-08-1276. [DOI] [PubMed] [Google Scholar]
- 17.Lehman TA, Bennett WP, Metcalf RA, et al. p53 mutations, ras mutations, and p53-heat shock 70 protein complexes in human lung carcinoma cell lines. Cancer Res. 1991;51:4090–4096. [PubMed] [Google Scholar]
- 18.Tessema M, Willink R, Do K, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 19.Tessema M, Yu YY, Stidley CA, et al. Concomitant promoter methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis. 2009;30:1132–1138. doi: 10.1093/carcin/bgp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracker TU, Sommer A, Fichtner I, Faus H, Haendler B, Hess-Stumpp H. Efficacy of MS-275, a selective inhibitor of class I histone deacetylases, in human colon cancer models. Int J Oncol. 2009;35:909–920. doi: 10.3892/ijo_00000406. [DOI] [PubMed] [Google Scholar]
- 21.Ryan QC, Headlee D, Acharya M, et al. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol. 2005;23:3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 22.Schrump DS, Fischette MR, Nguyen DM, et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res. 2008;14:188–198. doi: 10.1158/1078-0432.CCR-07-0135. [DOI] [PubMed] [Google Scholar]
- 23.Mebratu YA, Dickey BF, Evans C, Tesfaigzi Y. The BH3-only protein Bik/Blk/Nbk inhibits nuclear translocation of activated ERK1/2 to mediate IFNgamma-induced cell death. J Cell Biol. 2008;183:429–439. doi: 10.1083/jcb.200801186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ocker M, Schneider-Stock R. Histone deacetylase inhibitors: signalling towards p21cip1/waf1. Int J Biochem Cell Biol. 2007;39:1367–1374. doi: 10.1016/j.biocel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 25.Schlesinger Y, Straussman R, Keshet I, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 26.Jin B, Yao B, Li JL, et al. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vire E, Brenner C, Deplus R, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 28.Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Widschwendter M, Fiegl H, Egle D, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 30.Ohm JE, McGarvey KM, Yu X, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum DeltaH contains DNA helicase activity. Proc Natl Acad Sci U S A. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2004;96:215–229. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 33.Caldas H, Jiang Y, Holloway MP, et al. Survivin splice variants regulate the balance between proliferation and cell death. Oncogene. 2005;24:1994–2007. doi: 10.1038/sj.onc.1208350. [DOI] [PubMed] [Google Scholar]
- 34.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–112. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 35.Chinnadurai G, Vijayalingam S, Rashmi R. BIK, the founding member of the BH3-only family proteins: mechanisms of cell death and role in cancer and pathogenic processes. Oncogene. 2008;27(Suppl 1):S20–29. doi: 10.1038/onc.2009.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure Legend. Figure S1. Total number of genes showing no demethylation in 4 tumors each (animal number-tumor number) from animals treated with 5-Aza or 5-Aza + MS-275. Values in parentheses reflect the percentage of methylated genes. Center circle depicts the number and percent of genes commonly remaining methylated across tumors.