Abstract
Opioid agonists produce analgesia in humans and other mammals by binding to three distinct types of G protein-coupled receptors; mu (MOR), delta (DOR), and kappa (KOR) opioid receptors. A fourth member of the opioid receptor family is the nociceptin or orphanin FQ receptor (ORL), however the role of the ORL receptor in analgesia is less clear. In the Northern grass frog, Rana pipiens, systemic and central administration of morphine and selective MOR, DOR, and KOR agonists produced dose-dependent antinociceptive effects blocked by the general opioid antagonist, naltrexone. The present study reports on the sequence, expression, and bioinformatics of four opioid receptor cDNAs cloned from Rana pipiens; rpMOR, rpDOR, rpKOR, and rpORL. These were the first opioid receptors cloned from a species of Class Amphibia, are selectively expressed in brain tissue, and show 70–84% identity to their homologous mammalian opioid receptors. Comparisons within species showed that MOR, DOR, and KOR proteins are significantly less divergent in earlier-evolved vertebrates compared to humans and other mammals. Among the four types of opioid receptors, MOR proteins show the least sequence variation among the six vertebrate species. Additionally, phylogenetic analysis supports the hypothesis that the family of opioid receptor proteins are coded by four genes that arose from two gene duplications of a single ancestral opioid receptor gene.
Keywords: Opioid receptors, Analgesia, Amphibian, Cloning, Bioinformatics
In humans and other mammals, it is clear that opioid analgesia begins with the binding of an opioid agonist to one or more types of opioid receptor proteins. There are four members of the opioid receptor family; the classic mu, delta, and kappa opioid receptors (MOR, DOR, KOR) and the opioid receptor-like protein (ORL) that is the target of the neuropeptide, nociceptin (orphanin FQ). The opioid family assignment is based on sequence and functional homology, and conservation of exon structure and gene regulation [12]. The four opioid receptor genes are also mapped to paralogous regions of human chromosomes, i.e. regions with genes that appear to be duplicated on other chromosomes [7].
Much less is known about the types of opioid receptors mediating antinociceptive effects in non-mammalian species. The antinociceptive potency of selective MOR, DOR, and KOR opioid agonists after systemic [25], intraspinal [24], or intracerebroventricular [27] administration in amphibians was highly correlated to that observed in mammals and to the relative potency of opioid analgesics in human clinical studies. However, emerging results from both behavioral and binding studies in Rana pipiens suggested that the selectivity of opioid ligands for the different types of opioid receptors was different in mammalian and non-mammalian species. The MOR, DOR, and KOR type-selective opioid antagonists, β-FNA [28], naltrindole [21], and nor-BNI [31] did not show type-selectivity in blocking the antinociceptive effects of selective opioid agonists in amphibians [26]. In radioligand binding studies, β-FNA, naltrindole, and nor-BNI competed with [3H]-naloxone binding from brain homogenates with equal affinity [19]. Still lacking is the ultimate identification of the types of opioid receptor proteins expressed in Rana pipiens from molecular cloning studies. Thus, the present studies were undertaken to determine what types of opioid receptors might be mediating the opioid antinociception observed in the amphibian, Rana pipiens.
The use of vertebrate animals was approved by the OSU-COM Animal Use Committee (IACUC) in accordance with the Guide from National Institutes of Health. Northern grass frogs, Rana pipiens were obtained (Sullivans, Nashville, TN, USA) and brain tissue, heart, stomach (GI), liver, and muscle tissue (gastrocnemius) removed, flash-frozen in liquid nitrogen and stored at −80 °C until used.
Total RNA was isolated using Trizol and manufacturer’s instructions and purified with Fast Track 2.0 mRNA Isolation Kit (Invitrogen, Carlsbad, CA). cDNA was obtained using SMART RACE cDNA Kit (Clontech, Palo Alto, CA, USA). PCR reactions were done using opioid receptor degenerate oligonucleotides [13] which gave partial sequences elongated by 3′ and 5′ RACE reactions. For each receptor type, at least eight independent PCRs were performed with High-Fidelity 2 Polymerase (Clontech, Palo Alto, CA). TOP10 chemically-competent E. coli (Invitrogen, Carlsbad, CA) were transformed, plated on LB/Amp (100 μg/ml) and incubated (37 °C overnight). Plasmid DNA was purified with QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) and clones sequenced at the OSU Molecular Biology CORE facility. Sequences were identified by comparing to existing vertebrate opioid receptors using BLAST [1].
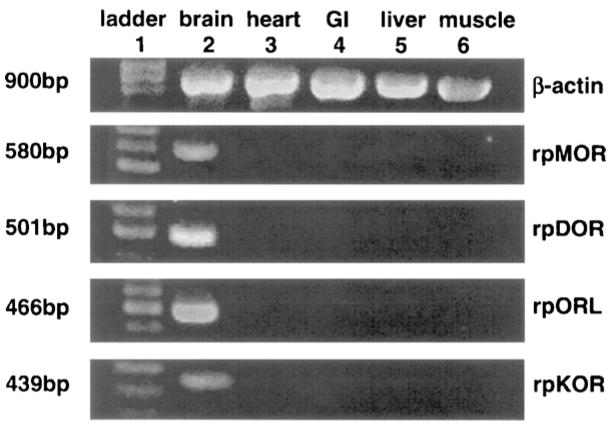
Tissue expression of opioid receptor mRNA in heart, GI, liver, muscle, and brain was assayed in PCR reactions using oligonucleotide primers specific for each type of amphibian opioid receptor. The PCR products were examined by 1.5% agarose gel electrophoresis and ethidium bromide staining for the rpMOR-specific amplicon (580 bp), rpDOR-specific amplicon (501 bp), rpKOR-specific amplicon (439 bp) and the rpORL-specific amplicon (443 bp). As a measure of control for the quality of cDNA used, PCR of a frog β-actin-specific amplicon (900 bp) was also performed.
Bioinformatics focused on comparing protein sequences (percent identity and similarity) by species (MOR vs. DOR vs. KOR in a single species) and across opioid receptor types (MORs vs. DORs vs. KORs). Analyses were done using the predicted amino acid sequence for each frog opioid receptor compared pair-wise against other opioid receptor sequences from species that have all four opioid receptors sequences deposited in Gen-Bank. The software program BLAST-P for two sequences [29] was used to generate comparative data. Differences between species means were assessed by a one-way ANOVA and the post-hoc Newman–Kuels test. Differences between non-mammalian and mammalian mean values were assessed by Student’s t-test. Significant differences were considered at p < 0.05.
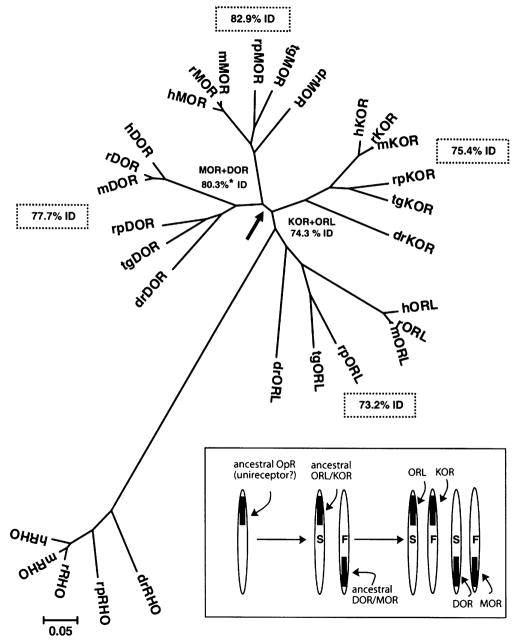
Phylogenetic analysis of vertebrate opioid receptor sequences was made by inputting the predicted amino acid sequences of the cloned R. pipiens opioid receptors and opioid receptor sequences from the other vertebrate species (see legend Fig. 2). The software program MEGA [11] was used to generate neighbor-joining (NJ) dendrograms (trees) of the vertebrate opioid receptor sequence data shown in Fig. 2. The NJ method has a high degree of accuracy [33] and was used previously in studies examining receptor phylogeny [8,9,14]. A set of rhodopsin protein sequences (RHO) were used to provide an outgroup sequences or ‘root’ the tree [6].
Fig. 2.
Phylogenetic analysis of MOR, DOR KOR and ORL sequences in six vertebrates. MEGA software was used to generate a radial phylogenetic tree using the neighbor-joining method, rooted with the available matching sequences of rhodopsin (RHO). Protein sequences from Rana pipiens (rp) were provided by conceptual translation of the cloned cDNA sequences deposited in GenBank. Abbreviations and access codes for MOR, DOR, KOR, and ORL sequences from other vertebrates were: dr: Dano rerio (zebrafish; AAK01143, AAP86771, AAG60607, AAN46747), tg: Taricha granulosa (newt; AAV28689, AAV28690, AAU15126, AAU26067), m: Mus musculus (mouse; P42866, P32300, P33534, P35377), r: Rattus norvegicus (rat; P33535, P33533, P34975, P35370), h: Homo sapiens (human; P35372, P41143, P41145, AAH38433). The arrow shows the bifurcation of MOR + DOR sequences from KOR + ORL. Values on plot are mean percent identity (%ID). Asterisk (*) indicates that these two values were significantly different by t-test. Branch length is equal to the proportional difference among the sequences (scale bar = 0.05 or 5% difference in amino acid sequence). Inset box: The molecular evolution of vertebrate opioid receptors. For simplicity, the genes are referred to by the same acronym as the opioid receptor proteins they encode. ‘S’ denotes slow and ‘F’ fast rate of adaptive evolution. See text for further details.
Four full-length cDNAs homologous to each of the four types of opioid receptors were cloned and sequenced from Rana pipiens brain tissue. Based on the BLAST results (data not shown), the Rana pipiens (rp) cDNA sequences were named rpMOR, rpDOR, rpKOR, and rpORL. The predicted protein length of the frog opioid receptors was 388, 370, 391, and 361 amino acids, respectively. The nucleotide sequences of rpMOR, rpDOR, rpKOR, and rpORL were submitted to GenBank and assigned the access codes AF530571, AF530572, AF530573, and AY434690, respectively.
The tissue expression of amphibian opioid receptors was assayed by PCR reactions using gene-specific primers and tissue cDNA preparations as the template. As shown, the β-actin amplicon (900 bp) was expressed in all tissues assayed (Fig. 1). Each of the four opioid receptor cDNAs were amplified by PCR from frog brain tissue cDNA, but not from heart, GI, liver, or muscle tissue cDNA templates. Each PCR product was consistent with the length of the predicted amplicon (number of bp labeled on the left).
Fig. 1.
Tissue expression of rpMOR, rpDOR, rpKOR, and rpORL in amphibian brain, heart, GI, liver, and muscle tissues (lanes 2–6). Lane 1 is the standardized bp ladder. For control, β-actin expression was also assayed in each of the tissues. Numbers on the left margin are the predicted size for each of the receptor amplicons, based on gene-specific primers used in the PCR reactions.
MOR, DOR, and KOR proteins shared more common amino acids in non-mammalian species than in humans and other mammalian species (Table 1). The mean percent similarity of MOR, DOR, and KOR was significantly more in each of the non-mammalian species compared to a greater divergence in mammals (p < 0.05, one-way ANOVA plus post-hoc Newman–Keuls test). The mean values for both identity and similarity of MOR, DOR, and KOR cloned from non-mammalian species were significantly greater than corresponding means in sequences from mammalian species (p < 0.01, Student’s t-test).
Table 1.
Comparison of vertebrate MOR, DOR, and KOR protein sequences within vertebrate species and by group
| Group/Species | MOR vs. DOR | MOR vs. KOR | DOR vs. KOR | Species mean (S.E.M.)a | Group mean (S.E.M.) |
|---|---|---|---|---|---|
| Percent amino acid identity | |||||
| Non-mammals | |||||
| Danio rerio | 70 | 62 | 65 | 65.7 (2.3) | |
| Rana pipiens | 73 | 65 | 63 | 67.0 (3.1) | |
| Taricha granulose | 70 | 66 | 68 | 68.0 (1.2) | 66.9 (1.2)† |
| Mammals | |||||
| Rattus norvegicus | 66 | 61 | 61 | 62.7 (1.7) | |
| Mus musculus | 61 | 60 | 61 | 60.7 (0.3) | |
| Homo sapiens | 62 | 60 | 59 | 60.3 (0.9) | 61.2 (0.7) |
| Percent amino acid similarity | |||||
| Non-mammals | |||||
| Danio rerio | 82 | 79 | 79 | 80.0 (1.0)* | |
| Rana pipiens | 85 | 81 | 77 | 81.0 (2.3)* | |
| Taricha granulose | 82 | 80 | 82 | 81.3 (0.7)* | 80.8 (0.7)† |
| Mammals | |||||
| Rattus norvegicus | 77 | 75 | 75 | 75.6 (0.7) | |
| Mus musculus | 72 | 75 | 74 | 73.6 (0.9) | |
| Homo sapiens | 73 | 74 | 72 | 73.0 (0.6) | 74.1 (0.5) |
Ver. BLASTP 2.2.14, settings: matrix: Blossum62, gap open: 11, gap extension: 1, x-drop-off: 50, expect: 10.00, wordsize: 3, and filter off.
Standard error of the mean.
Denotes significantly different % similarity than rat, mouse, and human mean values (p < 0.05, one-way ANOVA followed by post-hoc Newman–Kuels test).
Denotes group means (N = 9) different for identity and similarity at p < 0.01, Student’s t-test.
A phylogenetic dendrogram (‘tree’) of all available sets of opioid receptor amino acid sequences from vertebrate species is shown in Fig. 2. There was distinct clustering of each opioid receptor type, confirming the designation of Rana pipiens opioid receptor cDNA to the appropriate opioid receptor protein. The branching pattern of opioid receptor sequences mirrored the accepted evolutionary relationship of the vertebrate species. The sequences of the four groups of opioid receptors were best resolved into a dyad (bifurcation at arrow), with a common node for MOR and DOR and for KOR and ORL. Additionally, ORL sequences were rooted to the RHO outgroup (Fig. 2). A similar phylogenetic tree was produced when the nucleotide sequences were analyzed (data not shown).
The mean sequence identity (%ID) among all vertebrate MOR sequences was 82.9% (shown in dashed boxes in Fig. 2), 77.7% for DOR sequences, 75.4% for KOR sequences, and 73.2% for all vertebrate ORL receptors. When the sequence identity of the combined MOR + DOR group was compared to the combined group of KOR + ORL, the MOR + DOR group had 80.3% mean identity which was significantly greater than that of the KOR + ORL sequences (74.3%). Percent similarity values showed an identical pattern (data not shown).
The cloning and sequencing of rpMOR, rpDOR, and rpKOR cDNA from brain tissue identifies the receptor proteins mediating opioid antinociception following the administration of mu, delta, and kappa opioid agonists in an adjunct pain model using the amphibian, Rana pipiens. Additional behavioral studies are needed to determine the role of each type of opioid receptor in the observed antinociception following administration of selective opioids. With regard to the function of rpORL in amphibians, spinal administration of nociceptin in frogs produced a dose-dependent antinociceptive effect that was blocked by an ORL receptor antagonist but not by the non-selective opioid antagonist, naltrexone (unpublished data).
The basic bioinformatics tool, BLAST, was used to identify cDNA sequences obtained from Rana pipiens. Four frog opioid receptor sequences were identified as homologous to their respective opioid receptor type; as members of the Type A rhodopsin-like superfamily of GPCR, also known as the Rγ-type in the GRAFS classification system [7].
Tissue expression of rpMOR, rpDOR, rpKOR, and rpORL mRNA in the amphibian brain, heart, stomach (GI), liver and muscle showed that the four opioid receptor cDNAs were present in brain, but not in the peripheral tissues. Tissue expression of amphibian opioid receptors in brain is consistent with opioid binding studies in Rana pipiens brain homogenates [18]. Each of the PCR reactions used gene-specific primers and amplified a single, major product from brain tissue cDNA, suggesting that alternative transcripts were not present, or if present, did not contain the sequence targeted by the primers. The selective expression of opioid receptor message in frog brain tissue, with no detectable expression in peripheral tissues, parallels the results found in other non-mammalian species [2,30].
Results from previous behavioral studies showed that selective MOR, DOR, and KOR opioid agonists produced antinociception in Rana pipiens. Whereas the relative antinociceptive potency of MOR, DOR, and KOR agonists was conserved in amphibians and mammals, differences were noted in the action of highly-selective opioid antagonists, β-FNA, NTI and nor-BNI (see above). The hypothesis that opioid receptors from earlier-evolved vertebrates show less opioid type-selectivity than those found in humans and other mammals [26] is supported by the sequence analysis shown in Table 1. MOR, DOR, and KOR proteins expressed in Rana pipiens and other non-mammalian species were more similar to each other than those found in mammalian species.
The finding that there is less divergence of primary sequence among opioid receptor proteins in non-mammalian species suggests that opioids may show less type-selectivity at non-mammalian opioid receptors than at mammalian opioid receptors. This is supported by data on the first non-mammalian opioid receptor, ccMOR, cloned from the white suckerfish, Catostomus commersoni [4]. In this study, ccMOR expressed in HEK cells bound the non-selective opioid antagonist naloxone with high-affinity to HEK cell membranes; however the mu-selective opioid agonist, DAMGO, competed with naloxone with low affinity. Additionally, non-selective opioid ligands bound well to zebrafish delta opioid receptors (ZFOR1, labeled drDOR in Fig. 2) expressed in HEK cells but selective mu, delta, or kappa opioid ligands bound with low affinity [23]. Studies of opioid receptors in a second amphibian species, the newt Taricha granulose, led to the conclusion that the type-selectivity of newt opioid receptors was less stringent than in mammals [3]. In data presented elsewhere, parallel studies of amphibian (rpMOR) and human (hMOR) opioid receptors expressed in CHO cells showed significant differences in the affinity of selective opioid ligands supporting the hypothesis that opioid receptors in earlier-evolved vertebrates are less type-selective (manuscript in preparation).
The phylogenetic analysis of vertebrate opioid receptors confirms the assignment of frog cDNA sequences to the correct opioid receptor type (Fig. 2). Each type of opioid receptor (each set of orthologs) provided a pattern of vertebrate evolution consistent with established fossil evidence and phenotypic characteristics. Overall, the four groups of opioid receptor sequences formed a dyad, with MOR and DOR sequences sharing a common ancestor (node) and KOR and ORL sharing a different common node. A similar, but unrooted tree was generated after all four types of opioid receptors were cloned and sequenced in the newt [3].
There was an apparent rank order in the mean identity of each group of opioid receptor type with MOR proteins more conserved among the six species (see dotted boxes, Fig. 2), followed by DOR, KOR, and the set of ORL proteins being the most divergent. However, only when MOR and DOR sets were collapsed did a significant difference appear compared to KOR + ORL. More sequences from the cloning of additional vertebrate opioid receptors are needed to confirm these differences in group sequence identity related to opioid receptor type.
In harmony with the present data, the molecular evolution of vertebrate opioid receptors by gene duplication is proposed (see boxed inset, Fig. 2). Although not widely known, it is accepted by most evolutionary biologists that two rounds of genome-wide duplication (quadrupling the genes) occurred early in vertebrate evolution, called the 2R hypothesis [20]. The phylogenetic pattern of (AB)(CD), shown in Fig. 2, is expected from the application of the 2R hypothesis to a single ancestral gene [15].
The evidence for gene duplication is based partly on the location of MOR, DOR, KOR and ORL genes on paralogous regions of human chromosomes. Human MOR and DOR genes are mapped to chromosome 6 and 1, respectively, while KOR and ORL genes map to chromosomes 8 and 20 [7]. The specific pairing of DOR/MOR and ORL/KOR is supported by a genomic study whereby the greatest number of gene duplicates were found between paralogous regions on chromosomes 1 and 6, and between paralogous regions on chromosomes 8 and 20 [16]. Thus, the gene loci and whole genome studies support the assignment of ORL/KOR and DOR/MOR as the ancestor genes.
Although many GPCR and other gene families in the human genome appear deficient in an even number of duplicate genes due to gene deletion or mutation into pseudogenes [32], it is possible that the opioid receptor gene family avoided this fate due to a gene dosage effect that provided an immediate selective advantage (increased opioid receptor expression and function), as noted for other gene families [10].
Finally, it is also known that duplicate genes undergo asymmetrical divergence such that one gene is under relaxed constraint, showing an increased rate of adaptive evolution (positive selection), while the other gene maintains ancestral structure and function [10,17,22]. The gene encoding hMOR, and not any of the other opioid receptor types, was one of only nine genes controlling brain size or behavior that showed a significantly increased rate of evolution in the Homo sapiens genome compared to primate and rodent genomes [5]. Thus, MOR is assigned an ‘F’ (fast) and conversely DOR is the ‘S’ (slow) member of the pair (see inset, Fig. 2). Likewise, because ORL is most closely related to the rhodopsin sequences in the dendrogram, it is assigned an ‘S’ (slower rate of adaptive evolution) and KOR is the faster member of the pair. In the first duplication event, the finding that vertebrate ORL maintained most ancestral characteristics (most closely related to RHO) and the significant difference between MOR + DOR mean identity versus KOR + ORL identity (see above), supports the assignment of the ORL/KOR duplicate as the ‘slow’ ancestor gene and MOR/DOR as the ‘fast’ duplicate.
It is of greater interest to note that the target of most clinically used opioids is the mu opioid receptor (MOR) and that, in general, the rank order of analgesic potency is MOR > DOR > KOR (e.g. [24]). It is hypothesized that the rank order of opioid analgesic potency is correlated to the rank order of the mean protein sequence identity of opioid receptor types among vertebrates. This suggests that the greatest degree of adaptive evolution occurred with MOR in the six vertebrates examined. Thus, the molecular evolution of vertebrate opioid receptors may give a striking example of Darwinian positive selection with an evolutionary vector of increased classic opioid analgesia function.
In summary, cloning the four opioid receptors expressed in Rana pipiens brain tissue identified the opioid receptor proteins most likely mediating antinociception in this amphibian species. The degree of opioid receptor sequence divergence within species was correlated with vertebrate evolution. As primary amino acid sequence (structure) is a determinant of the selective binding (function) of opioid receptors, the correlation of sequence divergence with vertebrate evolution shows that opioid receptor proteins exhibit an evolutionary vector of increased type-selectivity. Additionally, the rapid rate of adaptive evolution (positive selection) of vertebrate MOR suggests an evolutionary vector of increased opioid receptor function, at least with regard to analgesia. If confirmed, these hypotheses provide a unique understanding of the pharmacology of vertebrate opioid receptors and the many other families of GPCR proteins encoded by putative duplicate genes.
Acknowledgments
Supported in part by the Oklahoma Center for the Advancement of Science and Technology, Health Research Contract HR02-122R and NIH grant DA12248 to CWS. The authors appreciate the advice and encouragement from Dr. Greg W. Sawyer at OSU-CHS.
References
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Barrallo A, González-Sarmiento R, Porteros A, Gracia-Isídoro M, Rodríguez RE. Cloning. molecular characterization, and distribution of a gene homologous to δ opioid receptor from zebrafish (Danio rerio) Biochem Biophys Res Commun. 1998;245:544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- 3.Bradford CS, Walthers EA, Stanley DJ, Baugh MM, Moore FL. Delta and mu opioid receptors from the brain of a urodele amphibian, the rough-skinned newt, Taricha granulosa: cloning, heterologous expression, and pharmacological characterization. Gen Comp Endocrinol. 2006;146:275–290. doi: 10.1016/j.ygcen.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 4.Darlison MG, Greten FR, Harvey RJ, Kreienkamp H, Stuhmer T, Zwiers H, Lederis K, Richter D. Opioid receptors from a lower vertebrate (Catostomus commersoni): sequence, pharmacology, coupling to a G-protein-gated inward-rectifying potassium channel (GIRK1), and evolution. Proc Natl Acad Sci USA. 1997;94:8214–8219. doi: 10.1073/pnas.94.15.8214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorus S, Vallender EJ, Evans PD, Anderson JR, Gilbert SL, Mahowald M, Wyckoff GJ, Malcom CM, Lahn BT. Accelerated evolution of nervous system genes in the origin of Homo sapiens. Cell. 2004;119:1027–1040. doi: 10.1016/j.cell.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 6.Filipek S, Teller DC, Palczewski K, Stenkamp R. The crystallographic model of rhodopsin and its use in studies of other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 2003;32:375–397. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families: Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 8.Iwama H, Gojobori T. Identification of neurotransmitter receptor genes under significantly relaxed selective constraint by orthologous gene comparisons between humans and rodents. Mol Biol Evol. 2002;19:1891–1901. doi: 10.1093/oxfordjournals.molbev.a004013. [DOI] [PubMed] [Google Scholar]
- 9.Josefsson LG. Evidence for kinship between diverse G-protein coupled receptors. Gene. 2000;239:333–340. doi: 10.1016/s0378-1119(99)00392-3. [DOI] [PubMed] [Google Scholar]
- 10.Kondrashhov FA, Rogozin IB, Wolf YI, Koonin EV. Selection in the evolution of gene duplications. Genome Biol. 2002;3:0008.1–0008.9. doi: 10.1186/gb-2002-3-2-research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S, Tamura K, Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 12.Law PY, Loh HH, Wei LN. Insights into the receptor transcription and signaling: implications in opioid tolerance and dependence. Neuropharm. 2004;47(Suppl 1):300–311. doi: 10.1016/j.neuropharm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Keith DE, Jr, Evans CJ. Multiple opioid receptor-like genes are identified in diverse vertebrate phyla. FEBS Lett. 1996;397:25–29. doi: 10.1016/s0014-5793(96)01126-x. [DOI] [PubMed] [Google Scholar]
- 14.Madsen O, Willemsen D, Ursing B, Arnason U, De Jong W. Molecular evolution of the mammalian alpha 2B adrenergic receptor. Mol Biol Evol. 2002;19:2150–2160. doi: 10.1093/oxfordjournals.molbev.a004040. [DOI] [PubMed] [Google Scholar]
- 15.Makalowski W. Are we polyploids? A brief history of one hypothesis. Genome Res. 2001;11:667–670. doi: 10.1101/gr.188801. [DOI] [PubMed] [Google Scholar]
- 16.McLysaght A, Hokamp K, Wolfe KH. Extensive genomic duplication during early chordate evolution. Nat Genet. 2002;31:200–204. doi: 10.1038/ng884. [DOI] [PubMed] [Google Scholar]
- 17.Merritt TJ, Quattro JM. Evidence for a period of directional selection following gene duplication in a neurally expressed locus of triosephosphate isomerase. Genetics. 2001;159:689–697. doi: 10.1093/genetics/159.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman LC, Sands SS, Wallace DR, Stevens CW. Characterization of mu, kappa, and delta opioid binding in amphibian whole brain tissue homogenates. J Pharmacol Exp Ther. 2002;301:364–370. doi: 10.1124/jpet.301.1.364. [DOI] [PubMed] [Google Scholar]
- 19.Newman LC, Wallace DR, Stevens CW. Selective opioid agonist and antagonist displacement of [3H]-naloxone binding in amphibian brain. Eur J Pharmacol. 2000;397:255–262. doi: 10.1016/s0014-2999(00)00265-x. [DOI] [PubMed] [Google Scholar]
- 20.Ohno S. Evolution by Gene Duplication. George Allen and Unwin; London: 1970. [Google Scholar]
- 21.Portoghese PS, Sultana M, Takemori AE. Naltrindole, a highly selective and potent non-peptide delta opioid receptor antagonist. Eur J Pharm. 1988;146:185–186. doi: 10.1016/0014-2999(88)90502-x. [DOI] [PubMed] [Google Scholar]
- 22.Prince VE, Pickett FB. Splitting pairs: the diverging fates of duplicated genes. Nat Rev Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez RE, Barrallo A, Garcia-Malvar F, McFadyen IJ, Gonzalez-Sarmiento R, Traynor JR. Characterization of ZFOR1, a putative delta-opioid receptor from the teleost zebrafish. Neurosci Lett. 2000;288:207–210. doi: 10.1016/s0304-3940(00)01239-8. [DOI] [PubMed] [Google Scholar]
- 24.Stevens CW. Relative analgesic potency of mu, delta and kappa opioids after spinal administration in amphibians. J Pharmacol Exp Ther. 1996;276:440–448. [PubMed] [Google Scholar]
- 25.Stevens CW, Klopp AJ, Facello JA. Analgesic potency of mu and kappa opioids after systemic administration in amphibians. J Pharmacol Exp Ther. 1994;269:1086–1093. [PubMed] [Google Scholar]
- 26.Stevens CW, Newman LC. Spinal administration of selective opioid antagonists in amphibians: evidence for an opioid unireceptor. Life Sci. 1999;64:PL125–PL130. doi: 10.1016/s0024-3205(99)00013-2. [DOI] [PubMed] [Google Scholar]
- 27.Stevens CW, Rothe KS. Supraspinal administration of opioids with selectivity for μ–, δ–, and κ–opioid receptors produces analgesia in amphibians. Eur J Pharmacol. 1997;331:15–21. doi: 10.1016/s0014-2999(97)01026-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takemori AE, Ho BY, Naeseth JS, Portoghese PS. Norbinaltorphimine, a highly selective kappa-opioid antagonist in analgesic and receptor binding assays. J Pharmacol Exp Ther. 1988;246:255–258. [PubMed] [Google Scholar]
- 29.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 30.Walthers EA, Bradford CS, Moore FL. Cloning, pharmacological characterization and tissue distribution of an ORL1 opioid receptor from an amphibian, the rough-skinned newt, Taricha granulosa. J Mol Endocrinol. 2005;34:247–256. doi: 10.1677/jme.1.01687. [DOI] [PubMed] [Google Scholar]
- 31.Ward SJ, Portoghese PS, Takemori AE. Pharmacological profiles of beta-funaltrexamine (β-FNA) and beta-chlornaltrexamine (β-CNA) on the mouse vas deferens preparation. Eur J Pharmacol. 1982;80:377–384. doi: 10.1016/0014-2999(82)90083-8. [DOI] [PubMed] [Google Scholar]
- 32.Wolfe KH, Li WH. Molecular evolution meets the genomics revolution. Nat Genet. 2003;33:255–265. doi: 10.1038/ng1088. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44:S139–S146. doi: 10.1007/pl00000067. [DOI] [PubMed] [Google Scholar]