Abstract
Major histocompatibility complex (MHC) class I restricted cytotoxic T lymphocytes (CTL) are known to play an important role in the control of Mycobacterium tuberculosis infection so identification of CTL epitopes from M. tuberculosis is of importance for the development of effective peptide-based vaccines. In the present work, bioinformatics technology was employed to predict binding motifs of 9mer peptides derived from M. tuberculosis for the 12 HLA-I supertypes. Subsequently, the predicted peptides were synthesized and assayed for binding to HLA-I molecules in a biochemically based system. The antigenicity of a total of 157 peptides with measured affinity for HLA-I molecules of KD ≤ 500 nm were evaluated using peripheral blood T cells from strongly purified protein derivative reactive healthy donors. Of the 157 peptides, eight peptides (5%) were found to induce T-cell responses. As judged from blocking with HLA class I and II subtype antibodies in the ELISPOT assay culture, none of the eight antigenic peptides induced HLA class I restricted CD8+ T-cell responses. Instead all responses were blocked by pan-HLA class II and anti-HLA-DR antibodies. In addition, CD4+ T-cell depletion before the 10 days of expansion, resulted in total loss of reactivity in the ELISPOT culture for most peptide specificities. FACS analyses with intracellular interferon-γ staining of T cells expanded in the presence of M. tuberculosis peptides confirmed that the responsive cells were indeed CD4+. In conclusion, T-cell immunity against HLA-I binding 9mer M. tuberculosis-derived peptides might in many cases turn out to be mediated by CD4+ T cells and restricted by HLA-II molecules. The use of 9mer peptides recognized by both CD8+ and CD4+ T cells might be of importance for the development of future M. tuberculosis peptide-based vaccines.
Keywords: cytotoxic T lymphocyte epitope, HLA-I, HLA-II, Mycobacterium tuberculosis, vaccine
Introduction
Tuberculosis (TB) is caused by the intracellular pathogen Mycobacterium tuberculosis. Despite the existence of effective chemotherapeutic drugs and the widespread use of the bacillus Calmette–Guérin (BCG) vaccine, TB is still one of the leading causes of morbidity and mortality worldwide, especially in the developing countries. It has been estimated that one-third of the world's population is latently infected with M. tuberculosis, and that about 8 million people develop the disease and 2–3 million die annually (http://www.who.int/tb/publications/global_report/2008/en/index.html). These figures do not include tuberculosis-related deaths in TB–HIV co-infected individuals. Although there is an effective chemotherapeutic treatment, the prolonged period of treatment is associated with non-compliance. The situation is further complicated by the appearance of multidrug-resistant strains.1 Furthermore, the epidemic of HIV infection, which induces progressive immune deficiency, increases the rate for developing TB disease dramatically.2
The current vaccine, BCG, is the most widely used vaccine in the world. To date more than three billion people have received the vaccinations.3 However, BCG only protects efficiently against the early manifestations of TB in children,4 and has limited effect against adult pulmonary TB. The estimates of protection by BCG vaccination have ranged from 0% to 80%.5 Hence, the development of more efficient vaccines capable of offering protection from TB disease is urgently needed.
Cell-mediated immunity is known to be crucial for protection against TB disease.6,7M. tuberculosis resides primarily in the macrophage phagosome,8 a vacuolar compartment associated with MHC II antigen processing and presentation. MHC class II presentation of mycobacterial antigens by macrophages to CD4+ T cells is pivotal for a protective response against the disease.6,7,9–11 In addition, many studies have indicated that MHC class I restricted cytotoxic T lymphocytes (CTL) also play an important role in the control of M. tuberculosis infection.12,12–17 The identification of new CTL epitopes is therefore of importance for the analysis of the involvement of CD8+ T cells in M. tuberculosis infections as well as for vaccine development. The identification of epitopes that have the potential of eliciting a CTL response has been greatly facilitated by the characterization of binding motifs for different MHC-I alleles of the 12 HLA-I supertypes.18 It is estimated that nearly 100% of persons in all ethnic groups surveyed possessed at least one allele within at least one of the 12 supertypes. As a result, just 12 vaccine epitopes representing each of these 12 MHC-I supertypes would lead to almost complete population coverage. To date, however, only CTL epitopes restricted by a limited number of HLA molecules have been identified.19
‘Reverse immunology’ based on immuno-bioinformatics is maturing rapidly and has now reached the stage where genome-, pathogen- and HLA-wide scanning for antigenic epitopes are possible at a scale and speed that makes it possible to exploit the genome information as fast as it can be generated. Immuno-informatic tools have been widely used for the in silico identification of T-cell epitopes from the proteomes of infectious micro-organisms including M. tuberculosis.20–25 We have previously used such approaches successfully to identify T-cell epitopes derived from influenza A virus and vaccinia virus.26–28
In the present study, with the help of immuno-bioinformatics, M. tuberculosis-derived proteins were analysed in silico for CTL cell epitopes within the 12 HLA-I supertypes.18 The 9mer peptides corresponding to predicted epitopes were synthesized and affinity of binding to recombinant HLA class I molecules was measured. One hundred and fifty-seven 9mer peptides, predicted to bind to the 12 HLA class I supertypes, were shown to have high to intermediate binding affinity (KD < 500 nm) for the relevant HLA class I supertypes. These peptides were evaluated in vitro for their ability to stimulate T cells from strongly purified protein derivative (PPD) reactive donors to release interferon-γ (IFN-γ) in an ELISPOT assay. Eight peptides were found to induce IFN-γ release by peripheral T cells from strongly PPD-reactive donors. Strikingly, none of these eight antigenic peptides appear to induce HLA class I restricted responses. Instead all responses could be demonstrated to be HLA class II restricted CD4+ T-cell responses.
Materials and methods
Collection of blood samples
Buffy coats of 500 ml whole blood from individuals in the Danish blood donor corps (age range: 35–65 years; including informed consent) were obtained from The Blood Bank at Rigshospitalet (Copenhagen, Denmark) and used within 24 hr to isolate peripheral blood mononuclear cells (PBMC). The donors were selected, according to serological typing of their HLA-A and HLA-B haplotypes, to maximize coverage of the 12 HLA-I supertypes. High-resolution sequence-based typing of the HLA-A/B/C and HLA-DR/DQ/DP loci was subsequently established (Genome Diagnostics, Utrecht, the Netherlands). Twelve donors, from whom PBMC were responding strongly to PPD in ELISPOT, were included in the present study.
Sampling and use of PBMC were in accordance with the Institutional Review Board, Rigshospitalet, Denmark.
Isolation of PBMC
The PBMC were isolated from buffy coats by density gradient centrifugation using Lymphoprep (Nycomed Pharma AS, Oslo, Norway). The freshly isolated PBMC were cryopreserved for later use at 20 × 106 cells in 1 ml RPMI-1640 containing 20% fetal calf serum and 10% DMSO at −140°.
Bioinformatics search strategy for CTL epitopes derived from TB
The NetCTL prediction method29 was used for predicting 9mer CTL epitopes in 24 M. tuberculosis proteins (Rv0151c, Rv0152c, Rv0159c, Rv0284, Rv0288, Rv0834c, Rv0980c, Rv1037c, Rv1072, Rv1404, Rv1979c, Rv1980c, Rv2557, Rv2711, Rv3144c, Rv3343c, Rv3347c, Rv3350c, Rv3403c, Rv3507, Rv3514, Rv3532, Rv3555c, Rv3873). The predictions were performed for 12 HLA-I supertypes (A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58 and B62). For each protein and each HLA-I supertype, the top-scoring 9mer of the protein was defined as the predicted CTL epitope if it had a NetCTL-score above 1·25. The selection strategy resulted in a total of 206 predicted CTL epitopes.
Peptides
The 9mer peptides were synthesized by standard 9-fluorenylmethyloxycarbonyl chemistry, and purified by reversed-phase high-performance liquid chromatography (at least 80%, usually > 95% purity) and validated by mass spectrometry (Shafer-N, Copenhagen, Denmark). Peptides were distributed at 500 μg/vial and stored lyophilized at −20° until use. Peptides were dissolved just before use.
Biochemical peptide–HLA-I binding assay
The biochemical assay for peptide–MHC-I binding was performed as previously described.30 Briefly, denatured and purified recombinant HLA heavy chains were diluted into a renaturation buffer containing β2-microglobulin and graded concentrations of the test peptide, and were incubated at 18° for 48 hr allowing equilibrium to be reached. We have previously demonstrated that denatured HLA molecules can de novo fold efficiently, however, only in the presence of appropriate peptide.31 The concentration of peptide–HLA complexes generated was measured using a Luminescent Oxygen Channeling Immunoassay and plotted against the concentration of peptide offered. Because the effective concentration of HLA (1–3 nm) used in these assays is below the equilibrium dissociation constant (KD) of most high-affinity peptide–HLA interactions, the peptide concentration leading to half-saturation of the HLA is a reasonable approximation of the affinity of the interaction.
Affinity measurements of peptides to recombinant HLA-DRB1*0101, -DRB1*0301, -DRB1*0302, -DRB1*0401, -DRB3*0301, -DRB5*0101 and DPA1*0103/DPB1*0401 molecules were performed according to previous work.32 Briefly, peptides including reference peptides known to bind the used HLA-II alleles [DR-binding peptide HA 306–318 (sequence: YKYVKQNTLKLAT) and DP-binding peptide, Plasm. Falciparum 239–253 (3D7)33 (sequence: YILLKKILSSRFNQM)] were dissolved and titrated in 25% glycerol, 0·1% pluriol (F68) and 150 mm NaCl. An HLA-II stock solution consisting of bacterially expressed and urea-denatured α- and β-chains, at appropriate concentrations were diluted into refolding buffer: 100 mm Tris/Citrate, 25% glycerol, 0·01% Pluriol F68 containing protease inhibitors (TPCK and Pepstatin both 3·3 μg/ml) at pH 6 (DRB1*0101. DRB5*0101) or pH 7 (remaining HLA-II alleles). The diluted HLA-II stock was subsequently mixed 1 : 1 with peptide titrations and incubated at 18° for 48 hr. Formed HLA-II complexes were detected using a homogeneous proximity assay (Alpha Screen; Perkin Elmer, Waltham, MA, USA); briefly, streptavidin-coated donor beads and L243 (murine monoclonal anti-DR) coupled acceptor beads, both 5 mg/ml, were diluted 500 times into PBS 0·1% Pluriol (F68). Ten microlitres of bead mix was mixed with 10 μl HLA-II/peptide samples in 384 Optiplates (Perkin Elmer). Following 18 hr of incubation at 18° they were read on an Envision Reader (Perkin Elmer) and analysed accordingly.32
Depletion of CD4+ T cells from PBMC
The CD4+ T cells were positively depleted from PBMC according to the manufacturer's instruction using monoclonal anti-CD4-coated Dynabeads from Dynal Biotech ASA (Oslo, Norway). The PBMC were effectively (>98%) depleted of CD4+ T cells as verified by flow cytometry.
IFN-γ ELISPOT assay
The PBMC were thawed, washed and then used for CD4+ or CD8+ T-cell depletion or cultured directly in RPMI-1640 supplemented with 5% heat-inactivated AB serum (Valley Biomedical, Winchester, VA), 2 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. The PBMC (4 × 106 to 6 × 106) or depleted PBMC were cultured in 1 ml culture medium in 24-well plates (Nunc, Roskilde, Denmark) in the presence of individual peptides with a final concentration of 10 μg/ml per well, and incubated for 10 days at 37°, 5% CO2 in humidified air. Recombinant human interleukin-2 (rhIL-2; Proleukin; Chiron, Amsterdam, the Netherlands) 20 U/ml was added on day 1. Cells were harvested on day 10, washed twice in RPMI-1640 and resuspended in complete medium to a final concentration of 1 × 106 to 2 × 106 cells/ml. The IFN-γ ELISPOT assay was performed as described previously26,27 to quantify peptide-specific T cells after in vitro expansion. Briefly, PBMC, 1 × 105 to 2 × 105, were cultured for 20 hr in the presence or absence of indicated peptides with a final concentration of 10 μg/ml in an ELISPOT plate. To block HLA-II-restricted responses, 10 μg/ml anti-pan HLA class II monoclonal antibody IVA12 [American Type Culture Collection (ATCC), Rockville, MD], anti-DP (B7/21; Abcam, Cambridge, MA, USA), anti-DQ (SPV-L3, IgG2a, a kind gift from Dr H. Spits, DNAX, CA) and anti-DR (L243, ATCC) was added, respectively; and to block HLA-I restricted responses anti-HLA-I antibody W6/32 ascites (ATCC) was added at a final dilution of 1 : 40 for 30 min before adding peptides in ELISPOT assays. As positive controls, cells were exposed to 10 μg/ml phytohaemagglutinin (Sigma-Aldrich, Poole, Dorset, UK). The PBMC were also depleted of CD4+ or CD8+ T cells and cultured in the presence or absence of indicated peptides in ELISPOT plates to confirm the dependence of T-cell subsets responsible for peptide-induced responses.
Intracellular IFN-γ staining with FACS analysis
Peripheral blood mononuclear cells restimulated for 10 days with peptide were harvested, washed and incubated with or without the relevant peptide at 1 μm for 4 hr at 37°. Brefeldin A (Sigma-Aldrich) was present for the last 3 hr of incubation. The cells were subsequently stained according to the ‘FastImmune’ protocol (Pharmingen, San Diego, CA, USA) with CD3-allophycocyanin-Cychrome7, CD4-Peridinin chlorophyll protein, CD8-allophycocyanin, CD69-phycoerythrin, and IFN-γ-fluorescein isothiocyanate. The stained cells were analysed on a FACS Aria II.
Statistics
Student's t-test was used to analyse the quantitative differences between the experimental wells and control in ELISPOT assays. A P-value below 0·05 was considered significant.
Results
Prediction of M. tuberculosis CTL epitopes
The complete sequenced genome of M. tuberculosis was deciphered in 199834,35 and revealed the presence of 3985 open reading frames, which are all potential targets for a TB vaccine. The search for CTL epitopes specific for M. tuberculosis were restricted to a subset of 24 M. tuberculosis proteins against which ex vivo reactivity had earlier been found by an IFN-γ ELISPOT assay using pools. Here, a peptide library representing 10% of the M. tuberculosis genome was screened directly for CD8+ T-cell responses ex vivo by IFN-γ ELISPOT in donors with LTBI (positive CD4+ T-cell response to either ESAT-6 or CFP-10; D. M. Lewinsohn, unpublished data). The criteria for including the proteins for CTL epitope prediction were a positive IFN-γ ELISPOT response detected in more than two donors or a positive IFN-γ ELISPOT response detected in two donors, where at least one of the donors had an IFN-γ response of >200 spot-forming cells per 106 PBMC. To identify antigenic M. tuberculosis CTL epitopes, a bioinformatics method (NetCTL) was employed to predict epitopes restricted to at least one of the 12 HLA-I supertypes.18 Based on the predictions, 206 potential CTL epitopes were synthesized.
Biochemical validation of HLA-I binding
To determine whether the synthesized peptides were indeed binders to the relevant HLA-I proteins, they were tested in a biochemical binding assay system (see Materials and Methods). A total of 157 peptides were found to bind to one of the 12 HLA molecules with a measured KD ≤ 500 nm, which is the normally accepted threshold36–38 for being a potential antigenic epitope. The numbers of binding peptides for the individual supertypes are: HLA-A1 (11 peptides), HLA-A2 (15 peptides), HLA-A3 (four peptides), HLA-A24 (14 peptides), HLA-A26 (15 peptides), HLA-B7 (18 peptides), HLA-B8 (seven peptides), HLA-B27 (eight peptides), HLA-B39 (17 peptides), HLA-B44 (20 peptides), HLA-B58 (14 peptides) and HLA-B62 (14 peptides). Consistent with previous classifications, the binding affinity (KD) of the 157 binding peptides can be divided into groups of high-affinity binders (n = 83; KD ≤ 50 nm) and intermediate-affinity binders (n = 74; 50 nm < KD ≤ 500 nm).
Antigenicity of the predicted peptides
The 157 HLA-I binding peptides were tested for their ability to stimulate T cells from a cohort of healthy PPD+ Danish subjects aged 35–65 years. The peptides were evaluated for their ability to stimulate IFN-γ production in an ELISPOT assay by PBMC from those HLA-matched donors who reacted most strongly with PPD. Since many donors' PBMC failed to respond after 2 days of peptide exposure, the sensitivity of the procedure was increased by exposing PBMC for 10 days to peptides before performing the ELISPOT assays. Positive reactivity towards peptides was confirmed at least twice in the same donor as well as in other HLA supertype matched donors. According to this criterion eight peptides (5%) belonging to five different supertypes (A1, A26, B7, B44 and B62) were found to be antigenic. An overview of peptide-reactive donors, their HLA class I type, and their reactivity according to ELISPOT data is shown in Table 1. The number of reactive donors and the actual ELISPOT data are shown in Table 2. Each of the eight antigenic peptides was also tested in 10 donors with low PPD reactivity. Only four of these donors showed reactivity against one or more of the eight antigenic peptides, an observation, which strongly underscores the M. tuberculosis specificity of the responses observed in the present study.
Table 1.
Overview of the donors tested positive for Mycobacterium tuberculosis peptide reactivity
| Sequence-based HLA-I typing | Sequence-based HLA-II typing | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Donor# | Sex | Age | HLA-A | HLA-B | DRB | DQB1 | DPB1 | EPI1 | |||||||
| 6 | M | 41 | *0101 | *0301 | *0801 | *3501 | 1*0101 | 1*0301 | 3*0101 | – | *0201 | *0501 | *0401 | *0101 | 1 |
| 10 | F | 38 | *0301 | *2501 | *0702 | *1801 | 1*0401 | 1*1501 | 4*0103 | 5*0101 | *0302 | *0602 | *0401 | – | 1 |
| 11 | M | 47 | *0201 | *0301 | *0702 | *4501 | 1*0701 | 1*1501 | 4*0103 | 5*0101 | *0303 | *0602 | *0401 | – | 3 |
| 13 | M | 60 | *0201 | – | *3901 | *4402 | 1*0701 | 1*1501 | 4*0103 | 5*0101 | *0303 | *0602 | *0401 | – | 1 |
| 16 | M | 50 | *0201 | *2902 | *5101 | *4501 | 1*0401 | 1*0803 | 4*0103 | – | *0301 | – | *0201 | *0401 | 2 |
| 17 | F | 35 | *0201 | *2902 | *4403 | *1501 | nd 2 | nd | nd | nd | nd | nd | nd | nd | 2 |
| 19 | F | 46 | *0101 | *0201 | *0801 | *4001 | 1*0301 | 1*1302 | 3*0101 | 3*0301 | *0201 | *0604 | *0401 | – | 1 |
| 21 | M | 54 | *0101 | *2402 | *0702 | *3901 | 1*1101 | 1*1501 | 3*0202 | 5*0101 | *0301 | *0602 | *0402 | *0901 | 1 |
| 25 | F | 62 | *2402 | *2601 | *0702 | *3801 | 1*0301 | 1*1501 | 3*0202 | 5*0101 | *0201 | *0602 | *0201 | *0402 | 1 |
| 28 | F | 62 | *0301 | *2601 | *0702 | *1401 | 1*0701 | 1*0803 | 4*0101 | – | *0202 | *0301 | *0201 | *0402 | 2 |
| 32 | M | 55 | *0201 | *3201 | *1501 | *5101 | 1*0401 | 1*1101 | 3*0202 | 4*0103 | *0301 | *0302 | *0401 | – | 4 |
| 38 | F | 47 | *6801 | *2902 | *4402 | *4501 | 1*0701 | 1*1101 | 3*0202 | 4*0101 | *0202 | *0302 | *0201 | *0402 | 1 |
EPI, number of peptide epitopes recognized.
nd, not done.
Table 2.
ELISPOT analysis of interferon-γ-positive peptide-specific donors
| Peptide# | Gene name | Description | Sequence | HLA supertype | HLA allele | KD (nm)1 | Responding donors | −peptide2 | +peptide2 |
|---|---|---|---|---|---|---|---|---|---|
| TB 2 | PPE61 | PPE gene family | LLDGLLAWY | A1 | A*0101 | 13 | 6, 21 and 32 | 3 ± 0 | 152 ± 12* |
| TB 47 | PPE52 | PPE gene family | DVAAMSGYY | A26 | A*2601 | 3 | 25 and 28 | 11 ± 4 | 37 ± 4* |
| TB 60 | Rv1979c | Possible conserved permease | GPRTRGYAI | B7 | B*0702 | 9 | 10, 11, 28, 32 | 2 ± 1 | 41 ± 5* |
| TB 88 | Rv1979c | Possible conserved permease | IVFATAARY | B62 | B*1501 | 161 | 32 | 6 ± 4 | 26 ± 6* |
| TB 92 | Rv0284 | Possible conserved membrane protein | LMLADHPEY | B62 | B*1501 | 313 | 32 | 3 ± 2 | 37 ± 8* |
| TB 124 | PPE61 | PPE gene family | LEEIGILLL | B44 | B*4001 | 11 | 19 | 5 ± 3 | 22 ± 6* |
| TB 129 | PPE52 | PPE gene family | LEVPAMGVL | B44 | B*4001 | 25 | 11, 16, 17, 38 | 4 ± 1 | 27 ± 6* |
| TB 132 | PE_PGRS | PE_PGRS gene family | GEGGVGSIL | B44 | B*4001 | 33 | 11, 13, 16, 17 | 5 ± 2 | 25 ± 7* |
KD, the equilibrium dissociation constant; a measurement of the affinity of peptides binding to the relevant HLA molecules in nm, the lower the value, the stronger the binding.
Spot-forming cell numbers represent an individual donor (in bold) and entries are the means of six individual assay cultures and in the absence or presence of peptide.
Differences are significant at P < 0·01.
Peptide reactivity is blocked by anti-HLA class II antibodies
We have previously demonstrated that variola virus-derived 9mer peptides with high HLA-I binding affinity (KD ≤ 5 nm) are able to induce CD4+ T-cell responses from PBMC of vaccinated donors.39 Likewise, we showed that influenza A virus-derived 9mer peptides with binding affinities for HLA-I allele are capable of stimulating strong CD4+ T-cell responses.28 To ascertain whether, or not, CD4+ T cells are involved in the anti-M. tuberculosis responses documented above, a pan-specific anti-HLA-II blocking antibody IVA12 as well as anti-DP, -DQ and -DR blocking antibodies were added into ELISPOT microcultures (see Materials and methods section). Similarly, cultures were exposed to the pan-specific anti-HLA class I antibody W6/32. As shown in Fig. 1, all of the eight antigenic peptides induced HLA-II-dependent responses as evidenced by being fully inhibited by IVA12 and anti-DR blocking antibodies, but not by the HLA class I blocking antibody W6/32. These data strongly indicate that the eight peptides induce HLA-DR restricted responses. It should be noticed that the presence of IVA12 does not affect HLA class I restricted responses and the presence of anti-DR antibody does not affect HLA-DP restricted responses.28
Figure 1.
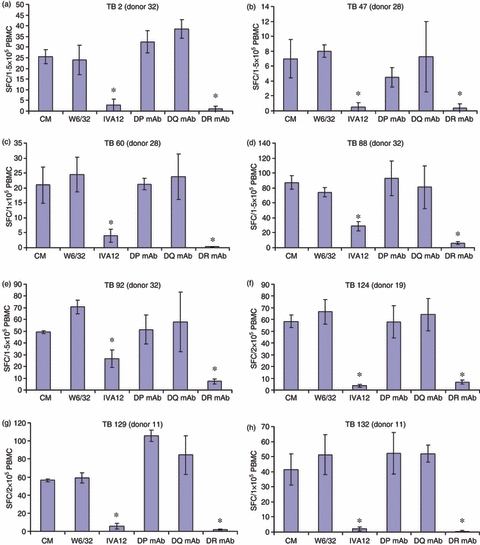
Nine-mer peptides induce HLA-II restricted responses. Peripheral blood mononuclear cells (PBMC) obtained from donors 32 (a, d and e), 28 (b and c), 19 (f) and 11 (g and h), were incubated with the indicated peptides, respectively, for 10 days. Before testing, cells were harvested, washed and stimulated with the indicated peptides in enzyme-linked immunospot (ELISPOT) plates for 20 hr in the absence or presence of anti-HLA class I and class II antibodies (mAb) as shown. Results are expressed as the mean spot-forming cell (SFC) values of four replicate ELISPOT microcultures, each containing 105 PBMC. For each peptide the presence of the anti-HLA class II antibodies IVA12 and anti-DR resulted in significant inhibition of interferon-γ spot formation (*P < 0.01; Student's t-test).
A recently developed assay for peptide binding to recombinant HLA-DR molecules was employed.32 Fourteen recombinant HLA-DR subtypes, representing 33% of all HLA-DR subtypes expressed by the PPD+ donors (Table 2), were assayed for binding of the eight antigenic peptides. However, only three of the eight M. tuberculosis peptides showed binding to HLA-DR subtypes (DRB1*0806, 1*1201, 1*1202), but none of these HLA-DR molecules was expressed by the two donors (no. 19 and 32) who showed reactivity for the three peptides (data not included).
Anti-M. tuberculosis peptide responses are dependent on CD4+ T cells
To obtain direct evidence of the phenotype of M. tuberculosis-peptide-reactive cells, anti-M. tuberculosis reactivity was tested in PBMC depleted of CD4+ T cells before peptide exposure in expansion cultures. As shown in Fig. 2, CD4+ T-cell depletion resulted in a total loss of peptide reactivity in all but one (anti-TB 60 peptide reactivity) of the CD4+ T-cell-depleted PBMC fractions.
Figure 2.
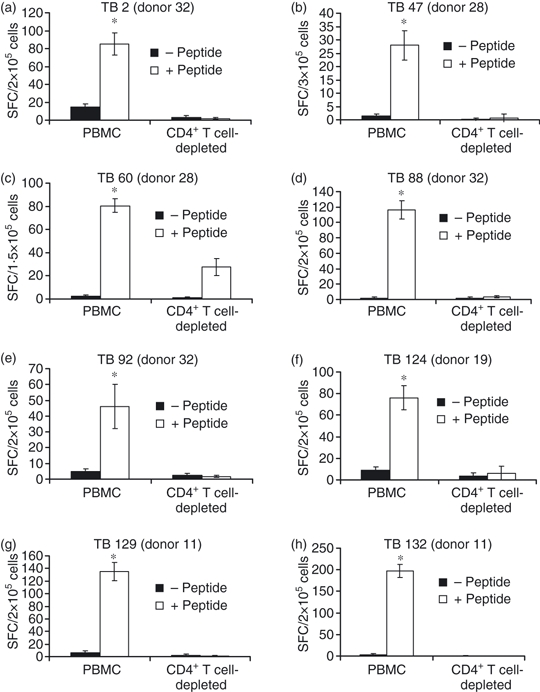
Nine-mer peptide-induced responses are CD4+ T-cell-dependent. Peripheral blood mononuclear cells (PBMC) obtained from Mycobacterium tuberculosis peptide-reactive donors shown in Table 2 were depleted of CD4+ T cells and incubated with the antigenic peptides depicted in Table 2 for 10 days. Before testing, cells were harvested, washed and incubated in enzyme-linked immunospot (ELISPOT) plates for 20 hr in the absence or presence of the indicated peptide. Results are expressed as the mean spot-forming cell values per 105 cells of four replicate ELISPOT microcultures, each containing unfractionated PBMC or CD4+ T-cell-depleted PBMC.
To further validate that the ELISPOT responses were in fact a CD4+ T-cell response and not a mixture of CD4+ and CD8+ T-cell responses, we used a flow cytometry-based intracellular cytokine secretion assay. Two donors were analysed in this assay, Donor 32 stimulated with TB2, TB88 and TB92, and donor 28 stimulated with TB60. After 10 days in vitro restimulation the cells were analysed by intracellular cytokine secretion. For all combinations a low but clear CD4+ T-cell response could be measured, with peptide TB2 and TB92 peptide recognized by donor 32 showing the highest frequency of CD4+-specific T cells (> 1%) (Fig. 3). In all cases no measurable peptide-specific CD8+ T-cell responses could be detected. For the peptide responses in donor 32 this correlates with the finding that the specific ELISPOT response was absent after CD4+ depletion (Fig. 2). The peptide T60 response in donor 28 could only be partially removed by CD4+ depletion (about 30% resides) but only a peptide-specific CD4+ T-cell response and no CD8+ T-cell response could be detected by intracellular cytokine secretion.
Figure 3.
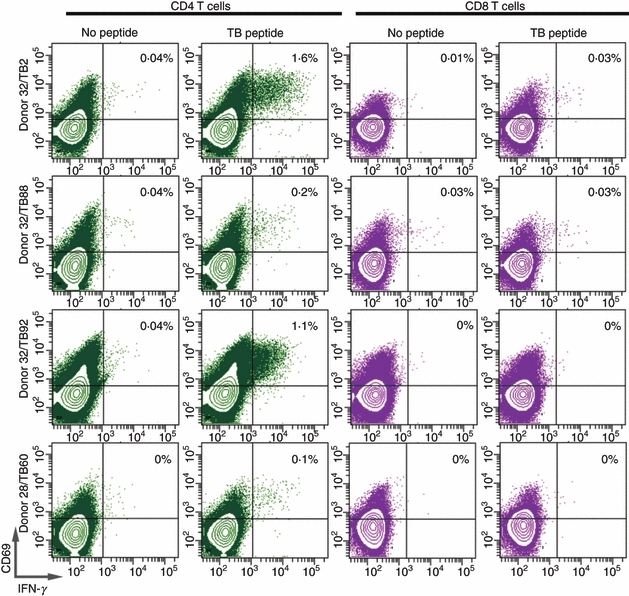
Intracellular interferon-γ (IFN-γ) production by tuberculosis peptide-stimulated peripheral T cells. Peripheral blood mononuclear cells (PBMC) obtained from Mycobacterium tuberculosis peptide-reactive donors shown in Table 2 were incubated with peptide, for 10 days. Cells were harvested, washed and re-stimulated with the indicated peptide for 4 hr in the presence of Brefeldin A. Cells were then stained by anti-CD4, anti-CD8 and IFN-γ monoclonal antibodies, and analysed by flow cytometry. Notice that the majority of IFN-γ producing cells are in the CD8− CD4+ gated lymphocytes.
Discussion
The aim of the present study was to identify CD8+ T-cell epitopes derived from M. tuberculosis using immuno-bioinformatics. We have previously used such an approach to successfully identify T-cell epitopes derived from smallpox virus and influenza A virus.26,27 However, in our previous study 39 and a more recent observation,28 it was shown that HLA-I binding 9mer peptides were able to induce CD4+ T-cell-dependent responses that apparently are restricted by the HLA-II molecules. These observations led us to investigate whether the predicted HLA-I binding peptides derived from M. tuberculosis induced CD4+ T-cell-dependent responses. The key findings of the present study are that eight of a total of 157 peptides selected for HLA class I binding induce T-cell responses in PBMC from one or several PPD+ donors (Table 2). In contrast, only four of 10 donors, with low PPD reactivity in ELISPOT assay, reacted with one or more of the eight antigenic peptides indicating the M. tuberculosis specificity of the responses observed. However, instead of being HLA class I restricted, these responses are apparently restricted by HLA class II molecules because the responses are all blocked by anti-HLA-DR antibody added to the ELISPOT assay culture. In addition, according to results from cell depletion and FACS analyses anti-M. tuberculosis peptide responses are mediated by CD4+ T cells.
The eight epitopes discovered are derived from five different M. tuberculosis proteins. Three of these [Rv1979, Rv3144c (PPE52) and Rv3532 (PPE61)] each express two positive CTL epitopes whereas the remaining two proteins [Rv0284 and Rv3507 (PE_PGRS)] only harbour a single epitope. Interestingly, six of the eight positive epitopes are derived from the PE/PPE gene family of conserved mycobacterial proteins (Table 2). The PE/PPE gene family is interesting from an immunological point of view because they comprise approximately 10% of the M. tuberculosis genome and may be a source of antigenic variation, which the bacterium uses to evade the host immune response.34 These proteins are surface-associated cell wall proteins and may also be accessible to antibodies.40 The B-cell and T-cell immune responses have been reported against both PE and PPE proteins, but their immunological significance remains largely unknown.41–44 Only a few T-cell epitopes have been identified for the PE/PPE gene family. Two have been found in PE-PGRS proteins (Rv1818c and Rv3812) and one in PPE protein (Rv3018c). In the present study we report five new epitopes for the PE/PPE gene family: a single epitope for the Rv3507 (PE_PGRS) and four new epitopes for the PPE proteins [Rv3144c (PPE52) and Rv3532 (PPE61)].
Regarding the phenotype of M. tuberculosis peptide-responding T cells, our data from T-cell depletion of PBMC before ELISPOT and FACS analyses showed that the responding T cells are indeed CD4+ T cells. In our previous studies 26–28 in which we probed for specific T-cell immunity in PBMC against pox and flu virus-derived HLA-I binding peptides, respectively, HLA-I and HLA-II antibody blocking experiments and CD4+ and CD8+ T-cell depletion experiments showed that peptide reactivity was initiated by either CD4+ or CD8+ T cells but never by both subsets in the same ELISPOT culture.
It is generally accepted that HLA class I binding peptides are composed of 8–11 amino acids, whereas HLA class II binding peptides consist of 15–20 amino acids being recognized by CD8+ and CD4+ T cells, respectively.45–47 In contrast, we have previously reported that HLA-I binding peptides nine amino acids long derived from vaccinia virus39 and influenza A virus28 induce CD4+ T-cell responses in PBMC. Here, we have extended these observations by showing that in silico predicted HLA-I binding 9mer peptides derived from M. tuberculosis proteins induce T-cell-dependent responses that appear to be HLA-II restricted because they are totally blocked by a pan HLA-II antibody as well as by an anti-HLA-DR antibody. As in our previous study with vaccinia virus-derived peptides,39 there was a trend of correlation between HLA class II restricted antigenicity and a measured high peptide HLA-I binding affinity, so six of eight antigenic M. tuberculosis peptides bind HLA-I with a KD < 50 nm. However, in accordance with our recent observation on flu epitopes,28 we found that two of the M. tuberculosis peptides with intermediate binding affinities to HLA class I were also capable of stimulating a strong HLA-II restricted T-cell responses.
As the eight antigenic 9mer epitopes appear to be restricted by HLA-II DR molecules (Fig. 1), we tried to predict the binding of all the 157 9mers used in this study to all DR alleles present among the donors using the publicly available MHC-II predictor NetMHCIIpan48 (http://www.cbs.dtu.dk/services). Forty-eight peptides including the two antigenic peptides LEEIGILLL and IVFATAARY were predicted to be either strong binders (SB, predicted KD < 50nm) or weak binders (WB, predicted KD < 500 nm), respectively, to one or more DR alleles present among the donors, (see Supplementary material Tables S1 and S2). However, the two donors (no. 19 and 32) who reacted with these two peptides did not express the predicted HLA-DR alleles.
We have recently developed a technology for assaying the binding of peptides to recombinant HLA-DR molecules.32 However, only three of the eight antigenic M. tuberculosis peptides showed binding to three of the 14 tested HLA-DR molecules, but none of these three HLA-DR molecules were expressed by the two peptide-reactive donors. These negative data might reflect the fact that the number of assayed HLA-DR molecules only represent one-third of the HLA-DR subtypes expressed by the TB peptide immune donors. In addition, the 10-day peptide exposure period might favour low-affinity interactions that might be missed in our biochemical assay. However, so far we have no definitive proof that the eight antigenic 9mer TB peptides discovered in the present study do bind to HLA-DR.
It is well established that CD4+ T cells are instrumental in the control of M. tuberculosis infections.6,7,9–11 For this reason, MHC-II restricted epitopes identified in the present study as capable of stimulating CD4+ T-cell responses may be of importance for the development of effective peptide-based vaccines against TB. In addition, it has been shown that CD4+ T cells are required for priming as well as secondary expansion of CD8+ memory T cells.49–53 It is therefore of critical importance to incorporate both HLA-I restricted and HLA-II restricted epitopes in peptide-based vaccines to obtain optimal vaccination, which may require the participation of both CD4+ and CD8+ T cells to generate a strong and long-lasting immunity against TB. Nine-mer peptides, such as those discovered in the present work, which bind to both HLA-I and HLA-II molecules, may potentially activate both the T helper and CTL arms of the immune system. Our failure to demonstrate CD8-reactive TB peptides in the present study might reflect the fact that many of the our BCG-vaccinated PPD+ donors were not really TB infected. Hence, in contrast to CD4+ T-cell responses, CD8+ T-cell responses are quite specific for TB and would therefore be absent in BCG-vaccinated but non-infected individuals.54
Our present and previous data28,39 suggest that certain HLA-I binding peptides might stimulate CD4+ T-cell immune responses most probably restricted by HLA-II molecules. Hence, ELISPOT-based analyses of reactivity against 9mer class I binding peptides should always include either anti-CD4/CD8 blocking or CD4+/CD8+ T-cell subset depletion experiments or perforin- or granzyme B-based ELISPOT analyses, although CD4+ T cells might occasionally express perforin/granzyme activity.55 Alternatively, proliferation assays and flow cytometry analyses in which PBMC are stained for surface markers specific for T cells should be included to obtain the true phenotype of the antigen-specific T cells.
In conclusion, we have identified eight novel antigenic 9mer M. tuberculosis-derived peptides that activate CD4+ T cells and appear to be restricted by HLA-DR molecules. These results may have important implications for a new design of epitope-based TB diagnostics and vaccines which incorporate both HLA-I and HLA-II restricted epitopes in the same peptide entity.
Acknowledgments
We are grateful to Ms Maja Udsen and Ms Trine Devantier for excellent technical assistance. This work was supported by National Institute of Allergy and Infectious Disease contracts HHSN266200400083C, HHSN266200400025C, EU 6FP 503231 and National Institutes of Health contract HHSN266200400081C (DML).
Disclosures
The authors have no financial disclosures.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1. Predicted binding of peptides from this study to DR alleles present in the donors from this study using NETMHCIIPAN48 (http://www.cbs.dtud.k/services).
Table S2. Predicted binding of peptides from this study (rows) to DR alleles present in the donors from this study (columns).
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- 1.LoBue P. Extensively drug-resistant tuberculosis. Curr Opin Infect Dis. 2009;22:167–73. doi: 10.1097/qco.0b013e3283229fab. [DOI] [PubMed] [Google Scholar]
- 2.Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–21. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 3.Dietrich J, Lundberg CV, Andersen P. TB vaccine strategies – what is needed to solve a complex problem? Tuberculosis (Edinb) 2006;86:163–8. doi: 10.1016/j.tube.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Colditz GA, Berkey CS, Mosteller F, Brewer TF, Wilson ME, Burdick E, Fineberg HV. The efficacy of bacillus Calmette–Guérin vaccination of newborns and infants in the prevention of tuberculosis: meta-analyses of the published literature. Pediatrics. 1995;96:29–35. [PubMed] [Google Scholar]
- 5.Fine PE. Variation in protection by BCG: implications of and for heterologous immunity. Lancet. 1995;346:1339–45. doi: 10.1016/s0140-6736(95)92348-9. [DOI] [PubMed] [Google Scholar]
- 6.Flynn JL, Chan J. Immunology of tuberculosis. Annu Rev Immunol. 2001;19:93–129. doi: 10.1146/annurev.immunol.19.1.93. [DOI] [PubMed] [Google Scholar]
- 7.Scanga CA, Mohan VP, Yu K, Joseph H, Tanaka K, Chan J, Flynn JL. Depletion of CD4+ T cells causes reactivation of murine persistent tuberculosis despite continued expression of interferon gamma and nitric oxide synthase 2. J Exp Med. 2000;192:347–58. doi: 10.1084/jem.192.3.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moreira AL, Wang J, Tsenova-Berkova L, Hellmann W, Freedman VH, Kaplan G. Sequestration of Mycobacterium tuberculosis in tight vacuoles in vivo in lung macrophages of mice infected by the respiratory route. Infect Immun. 1997;65:305–8. doi: 10.1128/iai.65.1.305-308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caruso AM, Serbina N, Klein E, Triebold K, Bloom BR, Flynn JL. Mice deficient in CD4 T cells have only transiently diminished levels of IFN-gamma, yet succumb to tuberculosis. J Immunol. 1999;162:5407–16. [PubMed] [Google Scholar]
- 10.Muller I, Cobbold SP, Waldmann H, Kaufmann SH. Impaired resistance to Mycobacterium tuberculosis infection after selective in vivo depletion of L3T4+ and Lyt-2+ T cells. Infect Immun. 1987;55:2037–41. doi: 10.1128/iai.55.9.2037-2041.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raupach B, Kaufmann SH. Immune responses to intracellular bacteria. Curr Opin Immunol. 2001;13:417–28. doi: 10.1016/s0952-7915(00)00236-3. [DOI] [PubMed] [Google Scholar]
- 12.Cho S, Mehra V, Thoma-Uszynski S, et al. Antimicrobial activity of MHC class I-restricted CD8+ T cells in human tuberculosis. Proc Natl Acad Sci USA. 2000;97:12210–5. doi: 10.1073/pnas.210391497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein MR, McAdam KP. CD8+ T lymphocytes against Mycobacterium tuberculosis. Arch Immunol Ther Exp (Warsz) 1999;47:313–20. [PubMed] [Google Scholar]
- 14.Serbina NV, Liu CC, Scanga CA, Flynn JL. CD8+ CTL from lungs of Mycobacterium tuberculosis-infected mice express perforin in vivo and lyse infected macrophages. J Immunol. 2000;165:353–63. doi: 10.4049/jimmunol.165.1.353. [DOI] [PubMed] [Google Scholar]
- 15.Skinner MA, Yuan S, Prestidge R, Chuk D, Watson JD, Tan PL. Immunization with heat-killed Mycobacterium vaccae stimulates CD8+ cytotoxic T cells specific for macrophages infected with Mycobacterium tuberculosis. Infect Immun. 1997;65:4525–30. doi: 10.1128/iai.65.11.4525-4530.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith SM, Malin AS, Lukey PT, Atkinson SE, Content J, Huygen K, Dockrell HM. Characterization of human Mycobacterium bovis bacille Calmette–Guérin-reactive CD8+ T cells. Infect Immun. 1999;67:5223–30. doi: 10.1128/iai.67.10.5223-5230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenger S. Cytolytic T cells in the immune response to Mycobacterium tuberculosis. Scand J Infect Dis. 2001;33:483–7. doi: 10.1080/00365540110026584. [DOI] [PubMed] [Google Scholar]
- 18.Lund O, Nielsen M, Kesmir C, et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics. 2004;55:797–810. doi: 10.1007/s00251-004-0647-4. [DOI] [PubMed] [Google Scholar]
- 19.Frieder M, Lewinsohn DM. T-cell epitope mapping in Mycobacterium tuberculosis using PepMixes created by Micro-Scale. Methods Mol Biol. 2009;524:369–82. doi: 10.1007/978-1-59745-450-6_27. [DOI] [PubMed] [Google Scholar]
- 20.De Groot AS, Bosma A, Chinai N, Frost J, Jesdale BM, Gonzalez MA, Martin W, Saint-Aubin C. From genome to vaccine: in silico predictions, ex vivo verification. Vaccine. 2001;19:4385–95. doi: 10.1016/s0264-410x(01)00145-1. [DOI] [PubMed] [Google Scholar]
- 21.De Groot AS, Rappuoli R. Genome-derived vaccines. Expert Rev Vaccines. 2004;3:59–76. doi: 10.1586/14760584.3.1.59. [DOI] [PubMed] [Google Scholar]
- 22.Dockrell HM, Brahmbhatt S, Robertson BD, et al. A postgenomic approach to identification of Mycobacterium leprae-specific peptides as T-cell reagents. Infect Immun. 2000;68:5846–55. doi: 10.1128/iai.68.10.5846-5855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammond AS, Klein MR, Corrah T, Fox A, Jaye A, McAdam KP, Brookes RH. Mycobacterium tuberculosis genome-wide screen exposes multiple CD8 T cell epitopes. Clin Exp Immunol. 2005;140:109–16. doi: 10.1111/j.1365-2249.2005.02751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin W, Sbai H, De Groot AS. Bioinformatics tools for identifying class I-restricted epitopes. Methods. 2003;29:289–98. doi: 10.1016/s1046-2023(02)00351-1. [DOI] [PubMed] [Google Scholar]
- 25.McMurry JA, Kimball S, Lee JH, et al. Epitope-driven TB vaccine development: a streamlined approach using immuno-informatics, ELISpot assays, and HLA transgenic mice. Curr Mol Med. 2007;7:351–68. doi: 10.2174/156652407780831584. [DOI] [PubMed] [Google Scholar]
- 26.Tang ST, Wang M, Lamberth K, Harndahl M, Dziegiel MH, Claesson MH, Buus S, Lund O. MHC-I-restricted epitopes conserved among variola and other related orthopoxviruses are recognized by T cells 30 years after vaccination. Arch Virol. 2008;153:1833–44. doi: 10.1007/s00705-008-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang M, Lamberth K, Harndahl M, et al. CTL epitopes for influenza A including the H5N1 bird flu; genome-, pathogen-, and HLA-wide screening. Vaccine. 2007;25:2823–31. doi: 10.1016/j.vaccine.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 28.Wang M, Larsen MV, Nielsen M, et al. HLA class I binding 9mer peptides from influenza A virus induce CD4 T cell responses. PLoS ONE. 2010;5:e10533. doi: 10.1371/journal.pone.0010533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen MV, Lundegaard C, Lamberth K, Buus S, Brunak S, Lund O, Nielsen M. An integrative approach to CTL epitope prediction: a combined algorithm integrating MHC class I binding, TAP transport efficiency, and proteasomal cleavage predictions. Eur J Immunol. 2005;35:2295–303. doi: 10.1002/eji.200425811. [DOI] [PubMed] [Google Scholar]
- 30.Harndahl M, Justesen S, Lamberth K, Roder G, Nielsen M, Buus S. Peptide binding to HLA class I molecules: homogenous, high-throughput screening, and affinity assays. J Biomol Screen. 2009;14:173–80. doi: 10.1177/1087057108329453. [DOI] [PubMed] [Google Scholar]
- 31.Ostergaard PL, Nissen MH, Hansen NJ, et al. Efficient assembly of recombinant major histocompatibility complex class I molecules with preformed disulfide bonds. Eur J Immunol. 2001;31:2986–96. doi: 10.1002/1521-4141(2001010)31:10<2986::aid-immu2986>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 32.Justesen S, Harndahl M, Lamberth K, Nielsen LL, Buus S. Functional recombinant MHC class II molecules and high-throughput peptide-binding assays. Immunome Res. 2009;5:2. doi: 10.1186/1745-7580-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doolan DL, Southwood S, Freilich DA, et al. Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data. Proc Natl Acad Sci USA. 2003;100:9952–7. doi: 10.1073/pnas.1633254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 35.Domenech P, Barry CE, III, Cole ST. Mycobacterium tuberculosis in the post-genomic age. Curr Opin Microbiol. 2001;4:28–34. doi: 10.1016/s1369-5274(00)00160-0. [DOI] [PubMed] [Google Scholar]
- 36.Kast WM, Brandt RM, Sidney J, Drijfhout JW, Kubo RT, Grey HM, Melief CJ, Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol. 1994;152:3904–12. [PubMed] [Google Scholar]
- 37.Sette A, Vitiello A, Reherman B, et al. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–92. [PubMed] [Google Scholar]
- 38.Yewdell JW, Bennink JR. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]
- 39.Wang M, Tang ST, Lund O, Dziegiel MH, Buus S, Claesson MH. High-affinity human leucocyte antigen class I binding variola-derived peptides induce CD4+ T cell responses more than 30 years post-vaccinia virus vaccination. Clin Exp Immunol. 2009;155:441–6. doi: 10.1111/j.1365-2249.2008.03856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, Cole ST. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 41.Skeiky YA, Ovendale PJ, Jen S, et al. T cell expression cloning of a Mycobacterium tuberculosis gene encoding a protective antigen associated with the early control of infection. J Immunol. 2000;165:7140–9. doi: 10.4049/jimmunol.165.12.7140. [DOI] [PubMed] [Google Scholar]
- 42.Okkels LM, Brock I, Follmann F, et al. PPE protein (Rv3873) from DNA segment RD1 of Mycobacterium tuberculosis: strong recognition of both specific T-cell epitopes and epitopes conserved within the PPE family. Infect Immun. 2003;71:6116–23. doi: 10.1128/IAI.71.11.6116-6123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan N, Alam K, Nair S, Valluri VL, Murthy KJ, Mukhopadhyay S. Association of strong immune responses to PPE protein Rv1168c with active tuberculosis. Clin Vaccine Immunol. 2008;15:974–80. doi: 10.1128/CVI.00485-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dillon DC, Alderson MR, Day CH, et al. Molecular characterization and human T-cell responses to a member of a novel Mycobacterium tuberculosis mtb39 gene family. Infect Immun. 1999;67:2941–50. doi: 10.1128/iai.67.6.2941-2950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chicz RM, Urban RG, Lane WS, Gorga JC, Stern LJ, Vignali DA, Strominger JL. Predominant naturally processed peptides bound to HLA-DR1 are derived from MHC-related molecules and are heterogeneous in size. Nature. 1992;358:764–8. doi: 10.1038/358764a0. [DOI] [PubMed] [Google Scholar]
- 46.Rudensky AY, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA., Jr Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991;353:622–7. doi: 10.1038/353622a0. [DOI] [PubMed] [Google Scholar]
- 47.York IA, Goldberg AL, Mo XY, Rock KL. Proteolysis and class I major histocompatibility complex antigen presentation. Immunol Rev. 1999;172:49–66. doi: 10.1111/j.1600-065x.1999.tb01355.x. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen M, Lundegaard C, Blicher T, Peters B, Sette A, Justesen S, Buus S, Lund O. Quantitative predictions of peptide binding to any HLA-DR molecule of known sequence: NetMHCIIpan. PLoS Comput Biol. 2008;4:e1000107. doi: 10.1371/journal.pcbi.1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janssen EM, Droin NM, Lemmens EE, et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 50.Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–6. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 51.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–9. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 52.Sun JC, Williams MA, Bevan MJ. CD4+ T cells are required for the maintenance, not programming, of memory CD8+ T cells after acute infection. Nat Immunol. 2004;5:927–33. doi: 10.1038/ni1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun JC, Bevan MJ. Defective CD8 T cell memory following acute infection without CD4 T cell help. Science. 2003;300:339–42. doi: 10.1126/science.1083317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lewinsohn DA, Winata E, Swarbrick GM, et al. Immunodominant tuberculosis CD8 antigens preferentially restricted by HLA-B. PLoS Pathog. 2007;3:1240–9. doi: 10.1371/journal.ppat.0030127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lewinsohn DM, Bement TT, Xu J, Lynch DH, Grabstein KH, Reed SG, Alderson MR. Human purified protein derivative-specific CD4+ T cells use both CD95-dependent and CD95-independent cytolytic mechanisms. J Immunol. 1998;160:2374–9. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


