Abstract
Actin is implicated in membrane fusion, but the precise mechanisms remain unclear. We showed earlier that membrane organelles catalyze the de novo assembly of F-actin that then facilitates the fusion between latex bead phagosomes and a mixture of early and late endocytic organelles. Here, we correlated the polymerization and organization of F-actin with phagosome and endocytic organelle fusion processes in vitro by using biochemistry and light and electron microscopy. When membrane organelles and cytosol were incubated at 37°C with ATP, cytosolic actin polymerized rapidly and became organized into bundles and networks adjacent to membrane organelles. By 30-min incubation, a gel-like state was formed with little further polymerization of actin thereafter. Also during this time, the bulk of in vitro fusion events occurred between phagosomes/endocytic organelles. The fusion between latex bead phagosomes and late endocytic organelles, or between late endocytic organelles themselves was facilitated by actin, but we failed to detect any effect of perturbing F-actin polymerization on early endosome fusion. Consistent with this, late endosomes, like phagosomes, could nucleate F-actin, whereas early endosomes could not. We propose that actin assembled by phagosomes or late endocytic organelles can provide tracks for fusion-partner organelles to move vectorially toward them, via membrane-bound myosins, to facilitate fusion.
INTRODUCTION
Actin is essential for many cellular processes and dynamic polymerization/depolymerization of actin filaments is a critical property of all eukaryotic cells (Mitchison and Cramer, 1996; Carlier, 1998; Machesky and Insall, 1999; Small et al., 1999; Amann and Pollard, 2000). Much of the F-actin assembled in cells is intimately associated with membranes, especially the plasma membrane (Tilney, 1976; Hoglund et al., 1980; Lindberg et al., 1981; Carraway and Carraway, 1989; Dickinson and Purich, 2002). The first links between F-actin and membranes were reported by Tilney and Cardell (1970) who demonstrated the role of plasma membranes in actin nucleation, and Orci et al. (1972) who showed that cytochalasin can either stimulate or inhibit exocytic fusion, depending on the conditions. In addition to exocytosis (Bernstein et al., 1998; Lang et al., 2000; Bader et al., 2002), actin is now known to be involved in many other trafficking events, including phagocytosis (Swanson et al., 1999; Desjardin and Griffiths, 2003) and transcytosis (Durrbach et al., 2000). In the endocytic pathway, actin and myosins are essential for two different transport steps: clathrin-dependent internalization from the plasma membrane and transport to lysosomes (Durrbach et al., 1996; Riezman et al., 1996; Buss et al., 2001). More recently, actin has also been shown to be directly involved in homotypic fusion between yeast vacuoles (see DISCUSSION).
Our interest in actin emerged from our analysis of an in vitro phagosome-endocytic organelle fusion assay that used latex bead phagosomes (LBP) and early endosomes (EE) and late endocytic organelles (LEO), comprising late endosomes and lysosomes, as fusion partners (LBP-EE/LEO fusion assay) (Jahraus et al., 1998). A number of actin-interfering reagents, such as cytochalasin D and latrunculin A (LatA), inhibited fusion (Jahraus et al., 2001). However, unexpectedly, two actin-binding proteins could stimulate LBP fusion with EE/LEO in the latter study. The first was a gelsolin fragment (G1-3) that can sever actin and bind to free barbed-ends of actin. The second positive effector of fusion, thymosin β4 (Tβ4) was especially difficult to rationalize because an excess of this protein should decrease actin polymerization (Carlier and Pantaloni, 1994; Safer and Nachmias, 1994). This seemed at odds with the ability of cytoD and LatA to inhibit LBP-EE/LEO fusion. Results obtained in the present study provides a possible rationale for this apparent anomaly in the context of the hypothetical models we introduce (Figure 10).
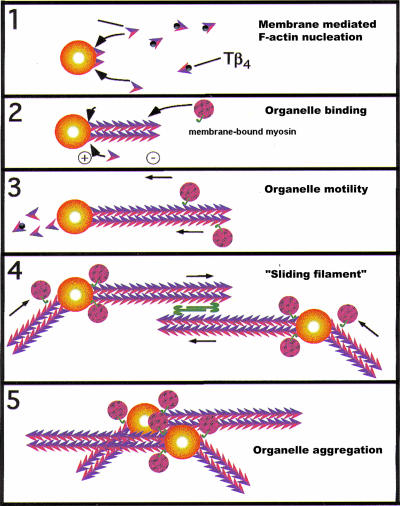
Figure 10.
Hypothetical models to explain how action filaments nucleated from the surface of a membrane organelle could “attract” other myosin-bound organelles toward it. Because the barbed ends of actin are on, or close to the membrane surface, the polarity of the filaments is such that most myosins bound to the organelles would carry their cargo (small spheres) toward the nucleating organelle in the presence of ATP. In a second model, both fusion partners nucleate actin and the double-headed (bipolar) myosin II is proposed to cross-link and slide the filament bundles in opposite directions. This might also facilitate membrane organelle aggregation leading to docking and fusion.
More recently, we have shown that the membranes of LBP can nucleate actin de novo in the absence of cytosol in vitro. This process is an inherent property of the phagosomal membrane and dependent on LBP-bound ezrin and/or moesin (Defacque et al., 2000a), on the phosphinositides P1(4) P(phosphatydilinositol[4]phosphate) and P1(4,5)P2′(phosphatidylinositol[4,5]bisphosphate), (Defacque et al., 2002), and can also be stimulated by gelsolin G1-3 (Defacque et al., 2000b). We recently presented evidence that a large network of lipids and proteins are involved in the regulation of this process, both in latex bead and mycobacterial phagosomes (Anes et al., 2003).
In parallel, we have also analyzed the polymerization of actin in macrophage cytosolic extracts, with and without membranes organelles. Although pure G-actin-ATP can nucleate spontaneously and elongate at concentrations above ≈1 μM, macrophage cytosol (without membranes), containing 5 μM actin, failed to polymerize. However, in the presence of LBP or postnuclear supernatant (PNS) membranes, a significant polymerization of actin was detected (Jahraus et al., 2001).
Here, our main goal was to combine the study of fusion in vitro between phagosomes and different endocytic organelles, and between endocytic organelles themselves, with a detailed description of the polymerization and organization of actin and its association with the membrane organelles. We show that all our earlier results demonstrating effects of F-actin–interfering reagents on the LBP-EE/LEO fusion can now be attributed to late fusion processes: the fusion of a fixed (late) stage of phagosomes with LEO and that between LEO-LEO was similarly facilitated by membrane-assembled actin. Surprisingly, there was no effect of a large spectrum of actin effectors on EE-EE fusion, either in an electron microscopy (EM) assay or in a biochemical fusion assay. A possible explanation for this result was our finding that whereas late endosomes could nucleate actin similarly to LBP, we could not detect any assembly of actin by EE under the same conditions.
MATERIALS AND METHODS
Reagents and Cells
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO), Roche Diagnostics (Mannheim, Germany), and Merck (Darmstadt, Germany). For culture of J774A.1 mouse macrophages and preparation of macrophage cytosol, see Blocker et al. (1996).
PNS and LBP Isolation
PNS and LBP isolation was made according to Jahraus et al., (1998). Throughout the present study only phagosomes isolated from cells that had been pulsed with latex beads for 1 h followed by a 1-h chase were used.
Quantification of F-Actin
For quantitation of F-actin, we used a modification of the assay developed by Cano et al., (1992). Phagosomes and/or PNS were mixed on ice with J774 macrophage cytosol (final concentration 4 mg/ml and an ATP-regenerating system or an ATP-depleting system was added; Jahraus et al., 2001). The mixture was adjusted to contain 60 mM KOAc, 1.5 mM MgOAc, 1 mM dithiothreitol and 12.5 mM HEPES, pH 7.4, and the volumes were balanced to 80 μl with homogenization buffer (HB; 250 mM sucrose, 3 mM imidazole, pH 7.4) and incubated at 37°C for the indicated time periods. The 250-μl ice-cold immunoprecipitation buffer (135 mM KCl, 10 mM NaCl, 2 mM MgCl2, 2 mM EGTA, 10 mM HEPES, pH 7.1), containing 0.4 μM TRITC-phalloidin and 0.1% Triton was added to each tube. After 1 h incubation at room temperature 300 μl of the mixture was loaded on top of a 700 μl 10% sucrose cushion and centrifuged at 80K for 40 min in a TLA 100.2 rotor to separate the F-actin/phagosome mixture in the pellet from the unbound TRITC phalloidin. After an overnight extraction in 1 ml methanol, tetramethylrhodamine B isothiocyanate (TRITC) was quantified in the fluorimeter at 545-nm excitation/575-nm emission by using a detector gain of 850 mV.
Confocal Microscopy Visualization of the Actin Cytoskeleton under In Vitro Fusion Condition
For this, we modified the phagosome-endosome fusion assay described by Jahraus et al. (1998). As an endocytic marker, Oregon Green-labeled horseradish peroxidase (HRP) was added to J774 macrophages for 40 min at 37°C. Phagosomes were prepared using 1 μm of blue fluorescent carboxylatemodified latex beads (Molecular Probes, Eugene, OR) covalently coupled with avidin.
To visualize actin, cytosol extracts were preincubated for 20 min at 37°C with 2 μM muscle G-actin labeled with 5-carboxytetrametyl rhodamine (Molecular Probes), as described in Kellogg et al. (1988). An ATP-regenerating system or an ATP-depleting system was added (Jahraus et al., 2001), and the volumes were balanced to 20 μl with HB (see above). Avidin-conjugated beads were used as a negative control. The incubation chambers consisted of glass microscope slides, and two pieces of double-sided Scotch tape, onto which a 15-mm circular glass coverslip (Menzel, Braunschweig, Germany) was sealed, forming a 7-μl chamber, which was 100 μm in depth and coated with 0.5% fish skin gelatin (Sigma-Aldrich) in H2O and air-dried before use. The samples in the chamber was sealed and incubated up to 80 min at 37°C. Confocal images were acquired on an LSM 510 confocal microscope (Carl Zeiss, Jena, Germany) with a 40× oil Plan-Neofluar lens (numerical aperture 1.30; Carl Zeiss). Optical section were taken 50 μm above the microscope slide by using a pinhole size of 78 μm, giving an optical section of 1 μm to avoid artifacts from surface-induced actin polymerization. Quantification of the size distribution of clusters of phagosomes was performed by taking 10 pictures randomly from two microscope chambers incubated in parallel with identical microscope settings. Images were analyzed using NIH Image software.
In Vitro Actin Nucleation
For the in vitro actin nucleation assay, the endocytic organelles of J774 macrophages were labeled with bodipy-avidin (Molecular Probes) as described by Emans and Verkman (1996). Briefly, cells in suspension were incubated at 37°C with bodipy-avidin (1 mg/ml) for 5-min pulse (EE) or 40-min pulse followed by 40 min of chase (LEO). Cells were washed three times with phosphate-buffered saline, and a PNS was made (Jahraus et al., 1998). Endocytic organelles were then isolated from this PNS according to Diaz et al., (1988). PNS was dilute with HB (1:3) and centrifuged at 37,000 × g for 1 min at 4°C. The supernatant was again centrifuged at 50,000 × g for 5 min at 4°C. The resultant pellet was then suspended in HB and quickly frozen in liquid nitrogen. The actin nucleation assay was performed as described in Defacque et al. (2000a) by using 2 μl of endosomes instead of phagosomes in the assay. The percentage of actin nucleating endosomes was calculated from counting ∼100 endosomes per sample in at least 10 different microscopy fields.
In Vitro Fusion between Endocytic Organelles by EM
For the EM-fusion assay the endocytic organelles of J774 macrophages were labeled with two different sizes of bovine serum albumin (BSA)-gold. One population of macrophages was fed with 5-nm BSA-gold, whereas another population was fed with 10- or 12-nm gold-bovine serum albumin for 60 min at 37°C; both gold preparations were used at an OD600 value of 5. The cells were then washed four times 10 min with phosphate-buffered saline, 0.5% BSA on ice and PNS was made (Jahraus et al., 1998). Labeled organelles were used immediately after preparation.
PNSs (50 μl) (with endocytic organelles containing 5- and 12-nm BSA-gold sizes) were mixed on ice with macrophage cytosol at a final concentration of 4 mg/ml and an ATP-regenerating system or an ATP-depleting system (see above); the volumes were increased to 182 μl with HB. Twenty microliters of this mixture was pipetted onto Parafilm per measurement. A Formvar- and carbon-coated 300-mesh copper grid, which had been coated before use with 0.5% fish skin gelatin, was put on this drop and incubated in a humid chamber at 37°C up to 80 min. Then, each sample was fixed by carefully injecting 80 μl of fixative (2% paraformaldehyde, 10 mM imidazole, 1 mM EGTA, 20 mM KCl, 0.1 mM MgSO4, pH 7) into the drop and left for 10 min (room temperature). The grids were washed briefly on two drops of the above-described buffer (without paraformaldehyde) before in some cases immunogold labeling was performed using an anti-actin antibody and protein A gold (15 nm). The grids were stained using a mixture of 2 parts 3% aqueous uranyl acetate and 8 parts 2% methylcellulose for 10 min on ice before drying in loops (Griffiths, 1993). Moving the grid systematically in the EM and sampling 50 organelles for the presence or absence of mixed gold size populations scored endocytic organelle fusion. The “percentage of fusion” (number of organelles containing two different gold sizes) represents the average of three different grids for each condition. A small signal for fusion at the zero time point (on ice) was subtracted from each time point at 37°C. This fusion signal typically corresponded to the values (usually 2–3%) obtained under low ATP conditions.
In Vitro Endosome-Endosome Fusion Assay
The biochemical assay is a modification of the one described by Horiuchi et al. (1997). Briefly, two populations of J774 macrophages were pulsed for 5 min at 37°C with either biotinylated transferrin or sheep α-human transferrin antibody to predominantly fill up the early endocytic pathway. PNS was then prepared as described in Jahraus et al. (1998), followed by centrifugation for 30 min at 13,000 rpm in an Eppendorf Microfuge at 4°C to separate the membrane organelles from the cytosolic proteins. The membranes were then resuspended in HB and centrifuged once more. The basal fusion reaction was carried out by coincubating the two populations of PNS for 30 min at 37°C in the presence of 4 mg/ml J774 cytosol, an ATP-regenerating system, and unlabeled transferrin. The mixture was adjusted to contain 60 mM KOAc, 1.5 mM MgOAc, 1 mM dithiothreitol, and 12.5 mM HEPES, pH 7.4, and the volumes were balanced to 20 μl with HB. The fusion was monitored and quantified using an Origen analyzer (IGEN Inc., Gaithersburg, MD) (Horiuchi et al., 1997) and expressed as the percentage of the fusion signal obtained relative to the signal seen with 1% Triton X-100 (without quencher; nonbiotinylated transferrin).
Cryo-Field Emission Scanning Electron Microscope (FESEM)
For cryo-FESEM, the grids prepared as described above for EM, were quickly (5–10 s) washed on a drop of water and plunged in liquid-N2–cooled ethane. The frozen grids were then cryotransferred to a Baf 300 freeze etching device (Bal-Tec, Balzers, Principality of Liechtenstein) and partially freeze-dried at a temperature of –95°C for 30 min. Then, the samples were rotary coated with 1.5 nm of tungsten at an angle of 45° by electron beam evaporation under high vacuum (2 × 10-7 mbar). Afterward, the EM-grid was cryotransferred to the precooled cryo-stage (Gatan type 626; Gatan, Pleasanton, CA) and inserted into the FESEM (Hitachi S-5200; Hitachi, Tokyo, Japan). The FESEM was operated at an accelerating voltage of 30 kV and a specimen temperature of –110°C. The images were recorded digitally with a resolution of 1280 × 960 pixel.
RESULTS
F-Actin Assembly during In Vitro Fusion
We have previously shown that the actin cytoskeleton is involved in in vitro fusion events between purified phagosomes and endocytic organelles within a crude postnuclear supernatant (PNS membranes) (Jahraus et al., 2001). That assay used LBP, PNS membranes, and macrophage cytosol at the minimum protein concentration (4 mg/ml), which gives the optimal fusion signal with 1 mM ATP. To study the kinetics of F-actin assembly during in vitro fusion in more detail we first set up a “spin-down” assay to quantify F-actin under in vitro fusion condition. For this, we modified an assay developed by Cano et al. (1992).
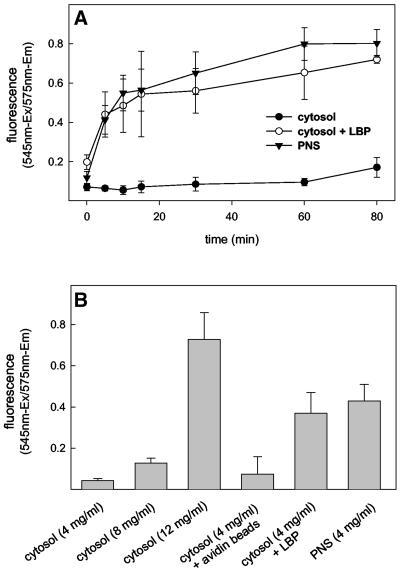
When macrophage cytosol at a concentration of 4 mg/ml was mixed with an ATP-regenerating system (1 mM ATP; Jahraus et al., 2001), but in the absence of membranes, we observed no significant growth of F-actin for up to 60 min incubation at 37°C, with a slight increase seen between 60 and 80 min (Figure 1, A and B). This agrees with rheological measurements of the same system, which failed to detect any change in viscosity/viscoelasticity >60 min, with a small increase apparent between 60 and 80 min (Jahraus et al., 2001).
Figure 1.
F-Actin assembly during in vitro fusion conditions. (A) Kinetics of F-actin assembly. Macrophage cytosol alone (•) or with phagosomes (○) or PNS membranes (▾) were mixed and incubated in the presence of an ATP-regenerating system at 37°C. At the indicated time points, reaction was stopped and F-actin was quantified. (B) Summary of the total actin estimated under the given conditions after 80 min at 37°C. In A, each value represents the mean and range of two parallel tubes from one typical experiment. In B, each value represents the mean and SD of three parallel tubes from one typical experiment
In contrast, when membranes in the form of LBP or PNS membranes were added to the incubation mixture, a detectable increase in F-actin was observed already after 5 min and continued to rise until 15–30 min when a quasi-plateau was reached. Between 30 and 80 min, no further net polymerization of actin was seen in some experiments, with a slight increase seen between 60 and 80 min in others (Figure 1A). Control avidin-coated latex beads had no significant effect on F-actin assembly (Figure 1B). This again demonstrates that phagosomal membranes, as well as a crude membrane fraction, can catalyze F-actin assembly from cytosol in vitro. The prominent increase in viscosity/viscoelasticity seen earlier by rheometry at the 30-min time point (Jahraus et al., 2001), thus corresponds to the time when net F-actin polymerization reached steady state.
When we used higher concentrations of cytosol (especially 12 mg/ml), with an ATP-generating system we found that these extracts could polymerize actin significantly even in the absence of membranes (Figure 1B). We consider this cytosolic nucleation process to be distinct from the membrane-dependent one (Jahraus et al., 2001, and references therein) and will not be considered further in this study.
Organelle Clustering Is Dependent on F-Actin Network Formation and ATP
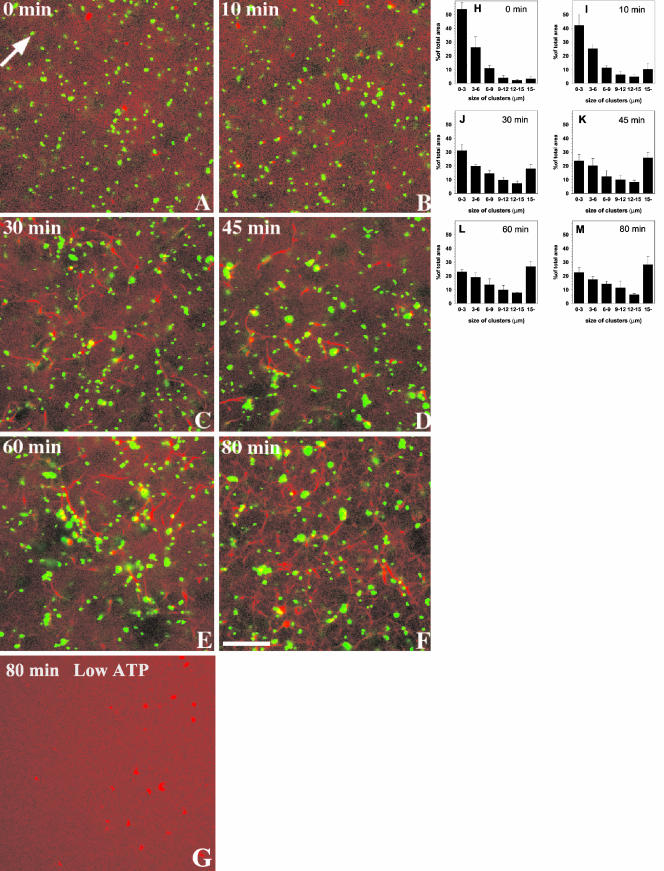
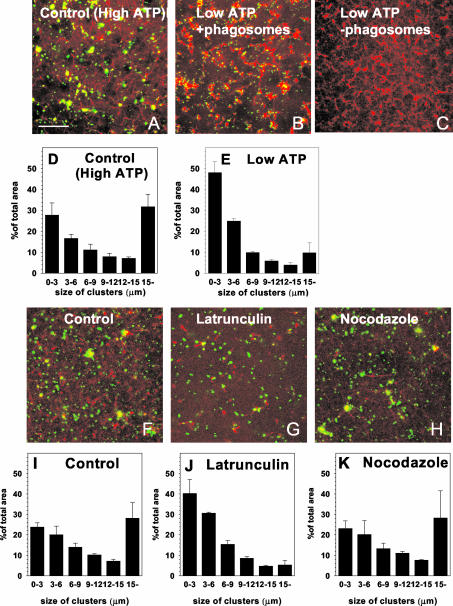
We next investigated the in vitro organization of the membrane organelles and F-actin as it developed in our LBP-EE/LEO assay. A confocal microscope-based assay using rhodamine actin enabled us to simultaneously visualize the actin cytoskeleton that assembles from J774 mouse macrophage cytosol with selectively labeled phagosomes and/or endosomes/lysosomes. When fluorescent-bead LBP were added to the cytosol, a small number of these organelles were associated with distinct puncta of labeled actin after 5- to 10-min incubation (Figure 2B). With increasing incubation times more bundles and networks of actin emerged, concomitant with more clustering of the phagosomes (Figure 2, C and D). Phagosomes aligned along the cables, especially at the sites where the bundles crossed each other (Figure 2, C–F). In agreement with Jahraus et al. (2001), there was no observable effect when cytosol was incubated at 37°C for 80 min (with ATP) without membranes (Figure 2G). A quantitation of the phagosome aggregation (Figures 2, H–M) showed that the process leveled off between 30- and 45-min incubation. Under identical conditions, control avidin-coated beads did not aggregate and no visible actin networks were formed (our unpublished data).
Figure 2.
Clustering of phagosomes along actin in macrophage cytosol in vitro. Phagosomes (green, false color) were mixed with freshly thawed macrophage cytosol mixed with rhodamine-G-actin (red) and incubated in the presence of an ATP-regenerating system at 37°C in a sealed chamber. The confocal microscopy images show a representative field after the indicated incubation times. The arrow in A indicates an individual phagosome. Structures larger than this are presumed to be clustered. Note the first signs of actin associated with LBP after 10-min incubation (arrow in B). (G) Macrophage cytosol mixed with rhodamine-G-actin (red) and incubated in the presence of an ATP-regenerating system at 37°C for 80 min in the absence of phagosomes. (H–M) Quantification of the data in A–F. The size of the clusters is indicated and the values are given as the percentage of the total area of green phagosomes. Bar, 10 μm.
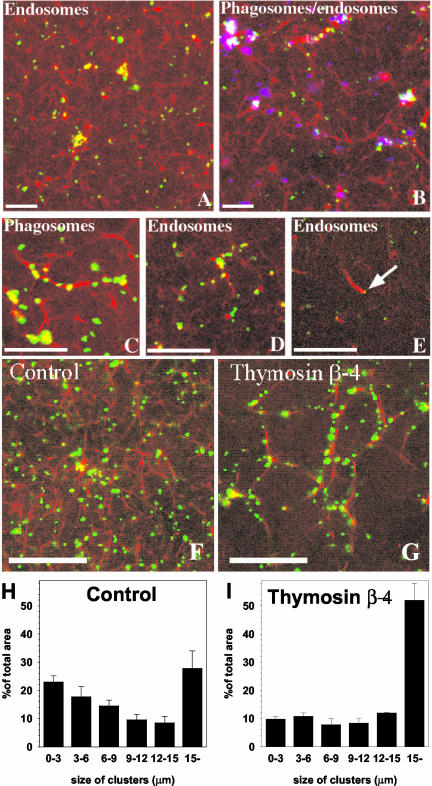
Oregon Green-HRP-labeled EE/LEO (40-min internalization) clustered to the same extent as was observed for LBP. This clustering also correlated with the visible formation of an extensive actin network in the solution (Figure 3A), that was even more prominent with LBP- and PNS-containing labeled endocytic organelles (Figure 3B). Both LBP and endosomes aligned along this actin scaffold and formed large clusters at the junction points where many cables crossed (Figure 3, A–D). Occasionally, we saw actin comet/tail formation on endocytic organelles, in agreement with Merrifield et al. (1999), Rozelle et al. (2000), and Taunton et al. (2000) (Figure 3E). Although comets has also been reported on LBP phagosomes in vivo (Zhang et al., 2002; Southwick et al., 2003), we never observed these on LBP phagosomes in our in vitro system.
Figure 3.
Clustering of endocytic organelles and phagosomes along actin in macrophage cytosol and the role of Tβ4. PNS from cells pulsed for 40 min with Oregon Green-HRP (green) were mixed with macrophage cytosol containing rhodamine-G-actin plus ATP, and incubated at 37°C for 80 min (A). (B) Same experiment is shown with added phagosomes (shown in blue). (C and D) Higher magnifications of phagosomes (green) (C) and endocytic organelles (green) (D), aligning along actin bundles in the presence of ATP after 80-min incubation at 37°C. (E) Endocytic organelle (indicated by arrow) on the tip of an actin tail (comet). (G) Effects of 20 μM Tâ4 relative to untreated control (F). A quantification of the Tâ4 effect is shown in H and I. Bars, 10 μm.
Thus, from the time the incubation is initiated until the 30-min time point when actin polymerization ceases and the viscoelasticity increases exponentially (Jahraus et al., 2001), there was a demonstrable emergence of an organization in the actin cytoskeleton, whose assembly is under the direct control of the cellular membrane organelles.
Tβ4 Induces More Organelle Clustering on a Smaller Actin Platform
The G-actin–buffering protein Tβ4 led to a paradoxical increase in the fusion of LBP with EE/LE (Jahraus et al., 2001). We therefore tested the effects of this actin-sequestering protein on the development of actin and on the clustering of the membrane organelles in the confocal microscopy assay. As shown in Figure 3G, the addition of 20 μM Tβ4 to the assay had a dramatic effect on the system. There were far fewer actin cables relative to the untreated control (Figure 3F), as expected from the known effects of this protein. These bundles grew longer than usual but the overall surface of actin was clearly much less than in the control, and there seemed to be far more clustering of the LBP on the more limited actin platform (cf. Figure 3, E and G; quantified in Figure 3, H and I). This observation provides a rationale for the ability of this protein to increase the extent of fusion (see DISCUSSION).
Under ATP-Depletion Conditions, Actin Polymerizes Efficiently but Organelles Do Not Cluster
Under low ATP (ischemic) conditions, cytosolic actin polymerizes much more than with high ATP, even without membranes (Jahraus et al., 2001, and references therein); there seems to be a deregulation of a system that is normally switched off at physiological levels of ATP (unless actively switched on by signaling effectors; Anes et al., 2003). We therefore analyzed the development of cytosolic actin in the confocal assay under ATP-depleting conditions (which contains 4–20 μM ATP; Jahraus et al., 2001). The actin structures formed under this condition showed an similar pattern with and without membranes (Figure 4, B and C). However, in contrast to the situation with membranes under high ATP conditions (Figure 4A), these formed with ATP-depletion were difficult to resolve clearly. Actin “bundles” were evident that were much shorter and less organized into suprabundle arrays (Figure 4B) than with high ATP (Figure 4A). Importantly, the organelles did not significantly cluster when ATP was depleted (Figure 4, B and E).
Figure 4.
Quantification of ATP- and actin-dependent phagosome clustering in macrophage cytosol and the role of microtubules. Phagosomes (green) were mixed with macrophage cytosol containing rhodamine-G-actin (red) and incubated for 80 min at 37°C with an ATP-regenerating system (A) or an ATP-depleting system (B). (C) Effect of an ATP-depleting system on F-actin in the absence of membranes. The clustering of phagosomes is quantified at high ATP levels (D) and low ATP levels (E). (F–H) Series of experiments with the same conditions as in A; control (F), 10 μM LatA (G), 10 μM nocodazole (H). The corresponding quantitative analyses are shown in I–K. All quantifications represent the mean values and SD of three separate experiments. Bars, 10 μm.
Phagosome Clustering Needs F-Actin but Not Microtubules
To test whether organelle clustering with 1 mM ATP was dependent on the formation of F-actin, we inhibited F-actin assembly by using 10 μM latrunculin A. As expected, this treatment significantly reduced the formation of actin cables, although small bundles, often connected to phagosomes could still be seen (cf. Figure 4, F and G). In addition, we observed a marked decrease in phagosome clustering (Figure 4, I and J). Similar effects was observed by adding high amounts of recombinant human cofilin (20 μM), an actin binding protein that is known to depolymerize F-actin (our unpublished data).
We next asked whether this clustering process was also influenced by microtubules. Labeling attempts with rhodamine-tubulin failed to reveal microtubules in the assay (in agreement with our EM observations; our unpublished data). Moreover, the addition of 10 μM nocodazole, which depolymerizes microtubules, had no effect on either the formation of actin networks or the clustering of phagosomes (Figure 4, H and K). Together, these data indicate that the in vitro process of clustering of endocytic organelles and phagosomes is both actin- and ATP-dependent but independent of microtubules.
Electron Microscopy of Fusion Assays
Methyl Cellulose Embedding. We next investigated the growth of the actin, and its association with membrane organelles by EM under conditions in which we could also monitor fusion. We first used an on-grid adsorption technique developed by Tokuyasu (1978) for preserving thawed cryosections that combines positive (or more strictly a positive-negative) contrast with embedding in methyl cellulose (Tokuyasu, 1978; Griffiths, 1993). We focused here mostly on the fusion between endocytic organelles because the (1 μm) LBP are relatively large are easily broken by specimen preparation. For this, two sets of cells were allowed to internalize two different sizes of gold. We varied the time of internalization to selectively label EE (5-min internalization), or two kinetically different populations of late endocytic organelles, operationally defined as late endosomes (LE; 10-min pulse, 20-min chase) or the total LEO (30-min pulse, 30-min chase). After gentle homogenization, the two sets of PNSs were mixed and incubated with cytosol and an ATP-generating system or an ATP-depletion system for up to 80 min underneath an EM grid before embedding in methyl cellulose/uranyl acetate. The presence of the two sizes of gold in the same vesicle was our indicator of fusion.
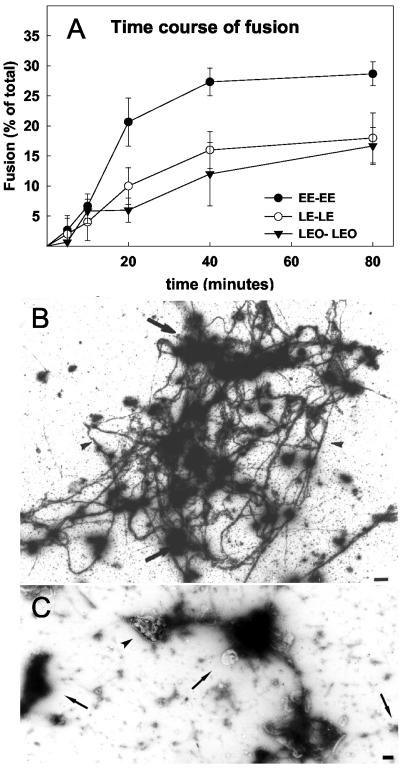
As shown in Figure 5A, the kinetics of fusion between EE-EE, LE-LE, and LEO-LEO fusion with ATP was initially fairly similar to each other; however, the fraction of fusion-competent organelles was always significantly higher for EE-EE than for the other fusion events. In the experiment shown, the bulk of fusion occurred by 40 min; in many other experiments, it was found to be completed by 30–35 min. Thus, the majority of fusion occurred within a time window in which actin polymerized (Figure 1A) and the organelles clustered (Figure 2). It should be noted that the increase in actin polymerization preceded the increase in fusion (cf. Figure 5A with 1A).
Figure 5.
Kinetics of endocytic organelle fusion by on-grid EM and overview of structure. (A) Kinetics of this fusion at 37°C was determined by gold mixing by using EM. Mean and SD are shown from three separate experiments. (B) Low-magnification overview of the prominent actin cables (arrowheads) connected to many organelles (arrows), after 80-min incubation at 37°C with ATP. (C) Example of a fusion assay incubated in the presence of an ATP-depleting system for 80 min; gold-filled late endocytic organelles (arrows), actin (arrowheads). Bars, 1 μm.
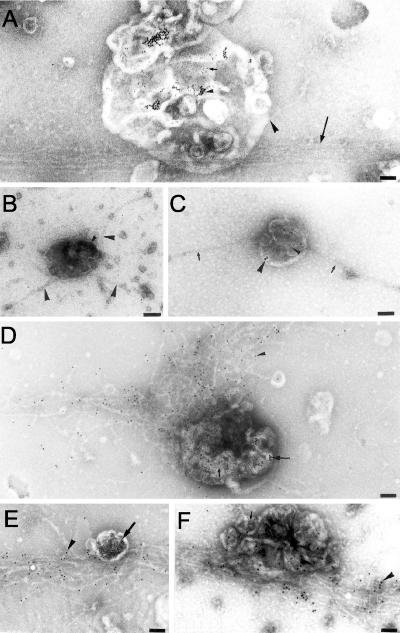
Figures 5B and 6 show examples of images from these experiments. In many of these experiments, the grids were also labeled with anti-actin antibody followed by a third size of gold, conjugated to protein A before drying. At the early, 5- to 10-min incubation times, a small number of filaments could be found connected end-on to endocytic organelles, especially late endocytic ones (Figure 6, A–D). A meshwork of actin could sometimes also be seen adjacent to early endosomes (our unpublished data). At these early times however we had the impression that this approach was not optimally preserving the actin and actin–membrane interactions; for example, the amount of bound actin was variable from experiment to experiment. As the incubation proceeded, we saw increasing amounts of structured actin on the grid, both bundles and networks, and the gold-filled organelles became intimately associated with these actin structures. In addition to what we interpret as actin assembly, all classes of endocytic organelles could also be found apparently bound side-on to actin bundles (Figure 6, A, D, E, and F). After 60- to 80-min incubation, the grids were covered with a huge network of actin, and most of the organelles in the PNS seemed to be associated with these structures (Figure 5B).
Figure 6.
On-grid EM fusion assay. (A) Principle of the EM fusion assay; 5-nm gold particles (small arrow) and 10-nm gold particles (small arrowhead) (derived from different sets of cells) are seen in the same (late) endocytic vesicle after 30-min pulse and 30-min chase in both fusion partners. The large arrowhead indicates a single actin filament oriented end-on to the organelle membrane, whereas the large arrow shows a bundle of actin to which the vesicle seems to be attached. (B–D) Examples of filaments putatively nucleated by late endocytic organelles. In B, three filaments (large arrowheads) are seen in a vesicle having only 5-nm gold particles (after 5-min incubation. (C) Two filaments (arrows) are seen in vesicle that has both small and large internalized gold particles (arrowheads) indicative of a fusion event after 10-min incubation. (D) More extensive filaments after 20-min incubation. This late vesicle contains two sizes of internalized gold (arrow) indicative of fusion, whereas the actin has been labeled with anti-actin and protein A gold (arrowhead). (E and F) Two examples of gold-filled late endocytic organelles (arrows) binding to F-actin bundles; the actin is labeled with anti-actin (arrowheads). Bars, 100 nm.
In the absence of ATP, the organization of actin was different and very difficult to interpret (Figure 5C). Actin bundles were evident, but they were less well structured. The membrane organelles were often grouped together to some extent (and indeed consistently fused at a low rate: 3–6%), but with no obvious organization of the actin in their vicinity
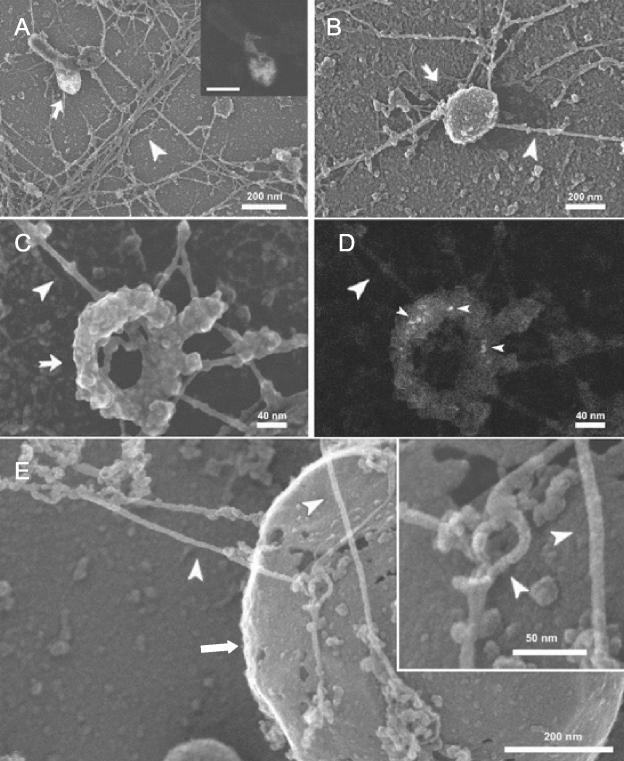
Cryo-FESEM. Cryo-FESEM provided a less perturbing, alternative approach to visualize the structures of actin in association with membranes in the fusion assay. For this, the EM grids incubated on the fusion assay where vitrified, as before, and then freeze-dried. Subsequently, the grids where shadowed with tantalum/tungsten and examined by high-resolution FESEM (Figure 7). Using this method, we could successfully examine not only endocytic organelles in the fusion assay but also phagosomes. Compared with the methyl cellulose approach, scanning electron microscopy (SEM) allowed us to visualize many more end-on connections of actin filaments emanating from the membrane organelles, even at early time points. As seen in Figure 7, the same general features as seen by methyl cellulose embedding are also apparent in the SEM images. Bundles and networks of actin were evident upon which membrane organelles clustered (Figure 7, A and B). The internalized gold particles within endocytic organelles are not evident in the standard imaging mode, they can be seen using the back-scatter electron detector (Hermann et al., 1996) (Figures 7A, inset; and D). Examples of filaments impinging end-on at the membrane surface are shown in Figures 7, A–C. Figure 7E shows details of the connection of actin filaments with LBP at higher resolution. These images provide the first hints that the machinery at the presumed sites of nucleation is quite elaborate, as expected.
Figure 7.
Cryo SEM of fusion assay. (A–D) Assay with endocytic organelles; A and B show an overview at low magnification; edocytic organelles is indicated by arrows; actin filaments are indicated by large arrowheads. The inset in A shows the back-scatter imaging mode to reveal the internalized gold. C shows details of actin filaments (arrowhead) impinging on an endocytic organelle (arrow); the corresponding bach-scatter image is seen in D; gold is indicated by small arrowheads. (E) Details of actin filaments (arrowheads) connected to a LBP (arrow) that is enlarged in the inset.
EM Fusion: Comparison of Fusion between Early and Late Endocytic Organelles with Respect to Actin
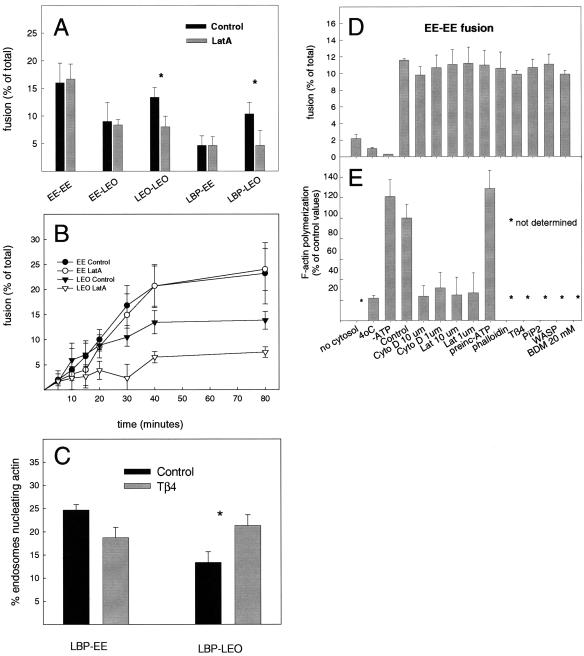
To investigate the role of actin in organelle fusion in more detail, we tested latrunculin A with respect to the different fusion reactions within the phagocytic and endocytic pathways. For this, EM assays were set up to monitor all possible fusion combinations individually (LBP-EE, LBP-LEO, EEEE, EE-LEO, and LEO-LEO), both in the presence and absence of LatA. To be able to include phagosomes in this analysis, the fusion reactions were allowed to proceed in bulk solution followed by chemical fixation and embedding in epoxy resin and sectioning, which better preserves LBP integrity than the methyl cellulose method (Jahraus et al., 1998). As shown in Figure 8A, 10 μM LatA had no effect on the fusion of LBP-EE, nor that between EE and EE. The fusion that occurs between EE and LEO (with macrophage organelles but not with those from baby hamster kidney cells; Jahraus et al., 1998) was also not affected by the drug. In contrast, LatA significantly inhibited the fusion occurring between LEO and LEO, and between LBP and LEO (Figure 8A). These data argue that F-actin facilitates only those fusion events in which both fusion partners are kinetically late stations.
Figure 8.
Quantification of fusion by EM and the biochemical EE fusion assay. (A) Comparison of the fusion between the different partner combinations indicated in the presence and absence of 10 μM LatA after 80-min incubation at 37°C by using Epon sections. (B) Kinetics of fusion of EE-EE and LEO-LEO with and without LatA (10 μM) by using the on-grid EM fusion assay. (C) Effect of 20 μM Tβ4 on the extent of fusion between LBP and either EE or LEO by EM by using Epon sections. Stars in A and C indicates significant differences p < 0.01 by using the Student's t test. (D) Effects of treatments that affect the F-actin on homotypic fusion between early endosomes in vitro using the biochemical assay. (E) Quantification of F-actin during some of the fusion reactions shown in Figure 9D. Each value in D and E represents the mean and SD of three parallel measurements from one typical experiment.
We extended this observation by making a detailed kinetic analysis of the fusion between EE and EE and that between LEO and LEO over an 80-min period, both with and without LatA. For this, we again used the on-grid approach. As shown in Figure 8B, there was again no significant effect of the drug on the whole course of the EE-EE fusion. In the case of LEO-LEO fusion after 10-min incubation with LatA, the extent of fusion was significantly reduced at all time points relative to the untreated controls (Figure 8B). As before, the total extent of fusion was always higher between EE-EE than between LEOLEO (Figure 8B).
Tβ4 Stimulates LBP-LEO but Not LBP-EE Fusion
In our earlier study, we showed that Tβ4 can stimulate the fusion of phagosomes with a mixture of early and late endosomes together (Jahraus et al., 2001). The data in Figure 3, F–I, also argue that this protein leads to more crowding of all endocytic organelles on a smaller actin platform. Because actin does not seem to be involved in EE-EE fusion, we hypothesized that Tβ4 would selectively stimulate fusion events in the late endocytic pathway. In agreement with this notion, the addition of 20 μM Tβ4 could significantly stimulate the fusion of phagosomes with LEO, whereas it did not stimulate LBP-EE fusion; rather, it inhibited the process slightly (Figure 8C).
Biochemical EE Fusion Assay; No Effect of Actin
The apparent lack of effects of actin-depolymerizing drug on EE-EE fusion was unexpected because actin filaments could be seen by EM in apparent intimate association with these organelles. Moreover, the EE also bind to phalloidin-stabilized F-actin in vitro (Kjeken, unpublished observations by using the approach of Al-Haddad et al., 2001). These organelles have significant levels of ezrin (Defacque et al., 2000a) and myosin Va (Kjeken, unpublished observations). We therefore investigated this process in more detail using a sensitive biochemical fusion assay we developed based on an earlier approach by Horiuchi et al. (1997).
The results (Figure 8D) show clearly that there was little or no effect on EE-EE fusion of a wide range of effectors or treatments that are expected to influence the actin cytoskeleton. First, the addition of LatA, or cytochalasin D, which significantly reduced actin polymerization (Figure 8E), had no effect. Second, fusion was not affected by either preincubating cytosol under low ATP condition before fusion to increase the amount of F-actin present in the assay (Figure 8E; see Jahraus et al., 2001), or when we stabilized F-actin by using phalloidin (Figure 8D). We also tested a number of actin binding proteins, or signaling molecules known to regulate membrane or cytosolic actin polymerization. However, the addition of Tβ4, PI(4,5)P2,(phosphatidylinositol[4,5]bisphosphate) or the Wasp-WA domain (which activates the ARP2/3 complex and significantly increased actin polymerization in cytosol; our unpublished data), all failed to influence EE-EE fusion. Together, these data strongly argue that the process of homotypic early endosome fusion as it occurs in vitro is not influenced by F-actin.
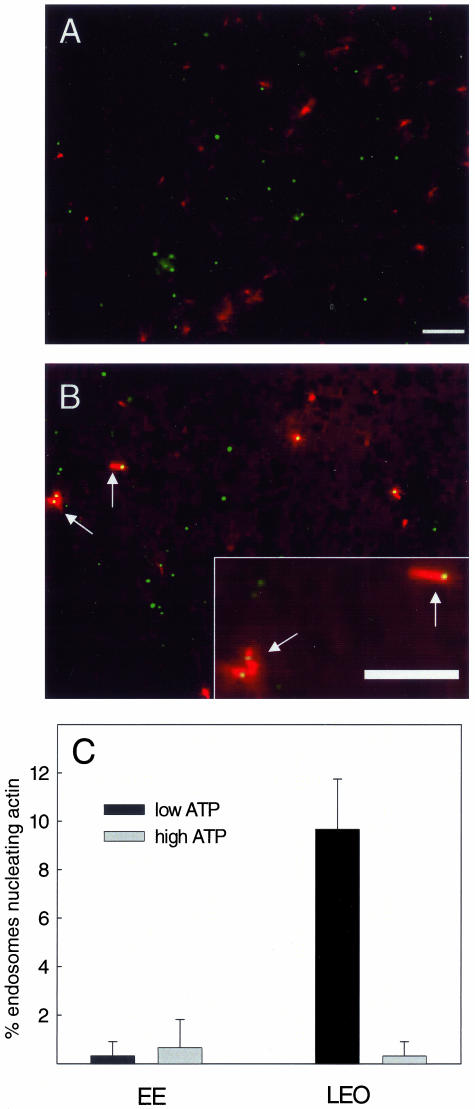
Late, but Not Early Endosomes Can Nucleate Actin In Vitro
A possible explanation for this striking difference between early and late fusion processes might be due to differences in the ability of these organelles to nucleate actin from their membranes. We therefore applied the in vitro assay developed for phagosome actin assembly (Defacque et al., 2000a) to labeled EE and LEO in a PNS mixture. For this, we added bodipy-avidin to J774 cells for either 5 min, to fill EE or a 30-min pulse followed by 30-min chase to load LEO before preparing PNS from both sets of cells. The PNSs were then incubated with rhodamine-G actin, buffered with Tβ4 and the standard (low) level of ATP (0.2 μM); green fluorescent endosomes were evaluated for their labeling with red actin. As shown in Figure 9B, a small but significant fraction (10%) of LEO could nucleate actin similarly to phagosomes. This percentage of positive organelles was at the lower end of the range we routinely obtain with the LBP. The ability of the latter to assemble actin is strongly inhibited by high levels of ATP (5 mM Anes et al., 2003); the same effect was seen with the LEO (Figure 9C).
Figure 9.
Nucleation of actin in the cytosol-free LM assay. Bodipyavidin–labeled EE (A) or LEO (B) were incubated with rhodamine actin and Tβ4 at low (0.2 mM) ATP. Actin is seen associated with a subset of LEO but not EE. Bar, 10 μm. (C) Quantitation of the percentage of EE and LEO that nucleate actin at low and high (5 mM) ATP.
In contrast, we were unable to see any evidence for actin assembly on EE under our standard assay conditions (Figures 9, A and C). This provides further evidence that early and late endocytic organelles are fundamentally different in the manner in which they interact with actin.
DISCUSSION
The interpretation of the data in this study may clarify a number of issued raised by earlier studies on the elusive role of actin in membrane fusion, but as always, they also raise many new questions. In our view, the key process to address to start understanding this role is the process of de novo nucleation and polymerization of actin on membrane surfaces. Actin is universally polymerized on membranes via insertion of monomers at the membrane surface (Tilney, 1975, 1976; Tilney and Mooseker, 1976; Lindberg et al., 1981), the machinery that allows this insertion/polymerization must therefore be a membrane-bound machinery. In the LBP system, we have identified ezrin and other components as being part of this machinery (see INTRODUCTION). It should be noted that although the ARP2/3 complex, Cdc-42, and N-Wasp are present on LBP phagosomes (Desjardins and Griffiths, 2003; our unpublished data), they are unlikely to be involved in the process of actin assembly by phagosomes (Defacque et al., 2002). We observed no clear effect on membrane-catalyzed actin assembly, either using the spin-down assay or in the confocal aggregation assay when we used cytosol depleted for ARP2/3, mixed with LBP whose bound ARP2/3 had been removed by salt-stripping (our unpublished data).
In all known membrane-catalyzed assembly of actin filaments, the standard topology of actin is thus to have the plus-ends at the membrane and the minus-ends growing away from it. Because the majority of myosins move unidirectionally along actin toward the membrane-associated barbed-ends (Hasson and Cheney, 2001), this topology of actin provides a potential universal mechanism for myosin-bound cargo to move vectorially toward the nucleating organelle (Figure 10). When the cargo is a late endosome or a lysosome, it allows these organelles to be attracted, for example toward a phagosome. We suggest that this process can facilitate some fusion events. More evidence in support of this model is given below. If the myosin II present in the cytosol is active in our system (Al-Haddad et al., 2001), it could in addition help to draw the organelles together via filament sliding (Figure 10, steps 4 and 5). In this scheme both fusion partners nucleate actin.
Our data here lead us to the following scenario. A fraction of LBP and late endocytic organelles can nucleate actin (Defacque et al., 2000a; this study). In the presence of cytosol, this polymerizes relatively fast at 37°C, predominantly between 5 and 20 min (Figure 1A). As seen by confocal microscopy and EM, this actin organizes into prominent networks and bundles. Most of the organelles in the PNS associated with these actin structures and the LBP, early, and late endosomes, increasingly aggregated at the actin filament junctions, consistent with the models in Figure 10. Over this time, there was a significant increase in the extent of both aggregation and fusion of all classes of endocytic organelles and phagosomes that we tested. Intuitively, therefore, one would have expected that the membrane-induced polymerization of actin would facilitate all these fusion events. Although our data indeed support this model for phagosomes and late endocytic organelles, to our surprise, they argue strongly against it in the case of the early endosomes.
Our data showed compellingly that the EE behaved differently to phagosomes and LEO with respect to actin, in a number of ways. First, LatA had no effect on EE-EE fusion in all our assays, whereas the fusion of LEO-LEO and phagosomes-LEO was significantly inhibited. Second, the addition of Tβ4, previously shown to stimulate fusion of LBP with a mixture of EE and LEO could be shown here to stimulate LBP-LEO fusion but had little effect on LBP-EE fusion. Third, the availability of a sensitive chemiluminescence assay for detecting EE-EE fusion gave us the opportunity to test a wide range of effectors expected to affect actin (including known effectors of LBP actin); however, none of these had an effect on EE-EE fusion. Finally, under in vitro conditions where LBP and LEO could nucleate the cytosol-independent assembly of actin, the EE failed to do so. Care should be cautioned before concluding that this class of organelle is totally unable to assemble F-actin. Some actin is found associated with EE, both by EM and from biochemical analysis (Harder et al., 1997). It is possible that the machinery for membrane assembly of actin is present on EE but is differently regulated to that on the phagosomes and LEO. As discussed below, phagosomes having pathogenic mycobacteria are also unable to nucleate actin in vitro, or in cells, but can do so when stimulated by some effectors. Further more, a significant fraction of EE can form actin comets in macrophages stimulated with lanthanum and zinc (Southwick et al., 2003). Further studies are obviously needed to investigate the possible interactions of actin with the EE membranes.
The model proposed in Figure 10 for LBP and LEO is supported by a number of our experiments. First, as mentioned, Tâ4 could stimulate the fusion of LBP with LEO. We were initially surprised that a protein whose main role in cells (and in our in vitro actin assembly assay) is to suppress nonspecific nucleation of actin could stimulate a fusion event. The confocal microscopy data provided a rationale for this result. With a fourfold molar excess of Tâ4 relative to total actin in the system, there was less total surface of actin, as expected, but the membrane organelles were now much more heavily clustered on their surface. Apparently, the reduction in available tracks facilitated more clustering of the organelles on the actin platform. This makes it easier to envisage why the late organelles fuse at a higher rate, but another rationale is needed to explain why the fusion of early organelles, which also aggregated more with Tâ4, was not affected. Conceivably, this is related to the inability of the EE to nucleate actin (Figure 10).
Further evidence supporting the view that an increase in membrane-bound assembly of actin can lead to more fusion of some organelles comes from our recent analysis of LBP and mycobacterial phagosomes. The phagosomes enclosing pathogenic mycobacteria are unable to nucleate actin in vitro or in macrophages, but this process can be switched on by the addition of proinflammatory lipids in vitro and in macrophages. Importantly, this treatment also led to a significant stimulation of the fusion of these phagosomes with LEO (leading to more pathogen killing), in agreement with the notion that an increase in membrane-assembled actin facilitates LEO-phagosome fusion (Anes et al., 2003).
Our models (Figure 10) predict that one or more suitable myosins must be present on the fusion partner for the proposed mechanism to operate. In our system, some support for a role of myosins comes from the finding that the lowspecificity myosin ATPase inhibitor BDM (2,3-butanedione monoxime) inhibits phagosome aggregation (Kjeken, unpublished data). One obvious candidate that we tested was myosin V, which by immuno-EM is found on EE, LEO, and phagosomes (with the highest density on EE; unpublished data). This myosin has been shown to be involved in the unidirectional movement of melanosomes, as well as the movement of several other organelles (Reck-Peterson et al., 2000; Wu et al., 2000). On LBP, it is responsible for binding the membrane to F-actin (Al-Haddad et al., 2001). However, removal of this myosin from the system (using macrophages from dilute, myosin V-negative mice) revealed that the microtubule-dependent phagosome movement to the cell center was increased (Al-Haddad et al., 2001). This argues that myosin V may be involved in delaying fusion between late endocytic/phagocytic organelles. It will now be necessary to test the remaining number of other myosins, including myosin I, II, IX, and X that are known to be localized on phagosomes (Allen and Aderem, 1995; Cox et al., 2002; Diakonova et al., 2002; Olazabal et al., 2002). The myosin I class has been shown to be functionally active on endocytic organelles (DePina and Langford, 1999; Neuhaus and Soldati, 2000; Cordonnier et al., 2001).
The models we propose in Figure 10 should only be considered as a first attempt to describe mechanisms by which actin nucleated by membrane organelles can positively influence fusion. Steps 1–3 in Figure 10 are also consistent with data on the role of actin in exocytic fusion in animal cells (Lang et al., 2000), in plant cells (Foissner et al., 1996), and in targeting vesicles to the new buds of budding yeast (Pruyne and Bretscher, 2000; Yin et al., 2000). The actin polymerized transiently by the plasma membrane can allow exocytic vesicles to move toward their fusion target. A recent study of actin in live Dictyostelium cells has provided intriguing evidence that during exocytic fusion of “postlysosomes” a short burst of actin assembly at the site of exocytosis precedes the fusion step (Lee and Knecht, 2002), in agreement with our model. Wang et al. (2002) have recently shown in elegant studies that homotypic membrane fusion between yeast vacuoles is preceded by a tight circumferential ring at the fusion contact sites where SNARE molecules and other fusion machinery accumulate. Intriguingly, actin is also enriched at these vertices and a signaling cascade involving actin, Cdc 42, and the ARP2/3 complex was shown to be somehow necessary for at least two steps in fusion; also here, the precise role of actin is still open (Eitzen et al., 2002; Wang et al., 2003). In these studies, no role for actin in docking was found. However, neither cytosol nor exogenous actin is essential for vacuole fusion in vitro, whereas our models (Figure 10) are dependent on cytosolic actin. The simplest interpretation of our data and those of the Wickner group together is that actin may be required to facilitate three (or more) distinct stages leading to fusion, at least in some fusion processes. Our model clearly does not rule out additional positive mechanisms (e.g., an actin “cage” that squeezes membrane organelles together), nor is it at odds with the long-standing notion that, at high densities, the filaments can provide a barrier to fusion (Burgoyne and Cheek, 1987; Trifaro et al., 1992).
Acknowledgments
We thank Dr. H. Faulstich (MPI, Ladenburg, Germany) for rhodamine-G-actin and many suggestions, Dr. Jens Pfannstiel for the human recombinant cofilin, and Dr. G. Gabbiani (University of Geneva, Geneva, Switzerland) for the anti-actin antibody. The early stages of this work were supported by a network grant from Human Frontier Science Program Organization. R.K. was supported by a fellowship from the Norwegian Cancer Society.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–05–0334. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-05-0334.
Abbreviations used: BSA, bovine serum albumin; EE, early endosome(s); FESEM, field emission scanning electron microscope; HB, homogenization buffer; HRP, horseradish peroxidase; LatA, latrunculin A; LE, late endosome(s); LEO, late endocytic organelle(s); LBP, latex bead phagosome(s); PNS, membranes endocytic organelles within a crude postnuclear supernatant PMEE, 35 mM PIPES-KOH, 5 mM MgSO4, 1 mM EGTA, 0.5 mM EDTA, pH 7.4; Tβ4, thymosin β4.
References
- Anes, E., Kuhnel, M.P., Bos, E., Pereira, J.M., Habermann, A., and Griffiths, G. (2003). Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat. Cell Biol. 5, 793-802. [DOI] [PubMed] [Google Scholar]
- Al-Haddad, A., et al. (2001). Myosin Va bound to phagosomes binds to F-actin and delays microtubule-dependent motility. Mol. Biol. Cell 12, 2742-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, L.H., and Aderem, A. (1995). A role for MARCKS, the alpha isozyme of protein kinase C and myosin I in zymosan phagocytosis by macrophages. J. Exp. Med. 182, 829-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann, K.J., and Pollard, T.D. (2000). Cellular regulation of actin network assembly. Curr. Biol. 10, R728-730. [DOI] [PubMed] [Google Scholar]
- Bader, M.F., Holz, R.W., Kumakura, K., and Vitale, N. (2002). Exocytosis: the chromaffin cell as a model system. Ann. N.Y. Acad. Sci. 971, 178-183. [DOI] [PubMed] [Google Scholar]
- Bernstein, B.W., DeWit, M., and Bamburg, J.R. (1998). Actin disassembles reversibly during electrically induced recycling of synaptic vesicles in cultured neurons. Brain Res. Mol. Brain Res. 53, 236-251. [DOI] [PubMed] [Google Scholar]
- Blocker, A., Severin, F.F., Habermann, A., Hyman, A.A., Griffiths, G., and Burkhardt, J.K. (1996). Microtubule-associated protein-dependent binding of phagosomes to microtubules. J. Biol. Chem. 271, 3803-3811. [DOI] [PubMed] [Google Scholar]
- Burgoyne, R.D., and Cheek, T.R. (1987). Reorganisation of peripheral actin filaments as a prelude to exocytosis. Biosci. Rep. 7, 281-288. [DOI] [PubMed] [Google Scholar]
- Buss, F., Arden, S.D., Lindsay, M., Luzio, J.P., and Kendrick-Jones, J. (2001). Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. EMBO J. 20, 3676-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano, M.L., Cassimeris, L., Joyce, M., and Zigmond, S.H. (1992). Characterization of tetramethylrhodaminyl-phalloidin binding to cellular F-actin. Cell Motil. Cytoskeleton 21, 147-158. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F. (1998). Control of actin dynamics. Curr. Opin. Cell Biol. 10, 45-51. [DOI] [PubMed] [Google Scholar]
- Carlier, M.F., and Pantaloni, D. (1994). Actin assembly in response to extracellular signals: role of capping proteins, thymosin beta 4 and profilin. Semin. Cell Biol. 5, 183-191. [DOI] [PubMed] [Google Scholar]
- Carraway, K.L., and Carraway, C.A. (1989). Membrane-cytoskeleton interactions in animal cells. Biochim. Biophys. Acta 988, 147-171. [DOI] [PubMed] [Google Scholar]
- Cordonnier, M.N., Dauzonne, D., Louvard, D., and Coudrier, E. (2001). Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol. Biol. Cell 12, 4013-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, D., Berg, J.S., Cammer, M., Chinegwundoh, J.O., Dale, B.M., Cheney, R.E., and Greenberg, S. (2002). Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat. Cell Biol. 4, 469-477. [DOI] [PubMed] [Google Scholar]
- Defacque, H., Bos, E., Garvalov, B., Barret, C., Roy, C., Mangeat, P., Shin, H.W., Rybin, V., and Griffiths, G. (2002). Phosphoinositides regulate membrane-dependent actin assembly by latex bead phagosomes. Mol. Biol. Cell 13, 1190-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defacque, H., Egeberg, M., Antzberger, A., Ansorge, W., Way, M., and Griffiths, G. (2000b). Actin assembly induced by polylysine beads or purified phagosomes: Quantitation by a new flow cytometry assay. Cytometry 41, 46-54. [PubMed] [Google Scholar]
- Defacque, H., et al. (2000a). Involvement of ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J. 19, 199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePina, A.S., and Langford, G.M. (1999). Vesicle transport: the role of actin filaments and myosin motors. Microsc. Res. Tech. 47, 93-106. [DOI] [PubMed] [Google Scholar]
- Desjardins, M., and Griffiths, G. (2003). Phagocytosis: latex leads the way. Curr. Opin. Cell Biol. 15, 498-503. [DOI] [PubMed] [Google Scholar]
- Diakonova, M., Bokoch, G., and Swanson, J.A. (2002). Dynamics of cytoskeletal proteins during Fcgamma receptor-mediated phagocytosis in macrophages. Mol. Biol. Cell 13, 402-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz, R., Mayorga, L., and Stahl, P. (1988). In vitro fusion of endosomes following receptor-mediated endocytosis. J. Biol. Chem. 263, 6093-6100. [PubMed] [Google Scholar]
- Dickinson, R.B., and Purich, D.L. (2002). Clamped filament elongation model for actin-based motors. Biophys. J. 82, 605-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrbach, A., Louvard, D., and Coudrier, E. (1996). Actin filaments facilitate two steps of endocytosis. J. Cell Sci. 109, 457-465. [DOI] [PubMed] [Google Scholar]
- Durrbach, A., Raposo, G., Tenza, D., Louvard, D., and Coudrier, E. (2000). Truncated brush border myosin I affects membrane traffic in polarized epithelial cells. Traffic 1, 411-424. [DOI] [PubMed] [Google Scholar]
- Eitzen, G., Wang, L., Thorngren, N., and Wickner, W. (2002). Remodeling of organelle-bound actin is required for yeast vacuole fusion. J. Cell Biol. 158, 669-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emans, N., and Verkman, A.S. (1996). Real-time fluorescence measurement of cell-free endosome fusion: regulation by second messengers. Biophys. J. 71, 487-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner, I., Lichtscheidl, I.K., and Wasteneys, G.O. (1996). Actin-based vesicle dynamics and exocytosis during wound wall formation in characean internodal cells. Cell Motil. Cytoskeleton 35, 35-48. [DOI] [PubMed] [Google Scholar]
- Griffiths, G. (1993). Fine Structure Immunocytochemistry. Springer, Berlin.
- Harder, T., Kellner, R., Parton, R.G., and Gruenberg, J. (1997). Specific release of membrane-bound annexin II and cortical cytoskeletal elements by sequestration of membrane cholesterol. Mol. Biol. Cell 8, 533-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasson, T., and Cheney, R.E. (2001). Mechanisms of motor protein reversal. Curr. Opin. Cell Biol. 13, 29-35. [DOI] [PubMed] [Google Scholar]
- Hermann, R., Walther, P., and Muller, M. (1996). Immunogold labeling in scanning electron microscopy. Histochem. Cell Biol. 106, 31-39. [DOI] [PubMed] [Google Scholar]
- Horiuchi, H., et al. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149-1159. [DOI] [PubMed] [Google Scholar]
- Hoglund, A.S., Karlsson, R., Arro, E., Fredriksson, B.A., and Lindberg, U. (1980). Visualization of the peripheral weave of microfilaments in glia cells. J. Muscle Res. Cell Motil. 1, 127-146. [DOI] [PubMed] [Google Scholar]
- Jahraus, A., Egeberg, M., Hinner, B., Habermann, A., Sackmann, E., Pralle, A., Faulstich, H., Rybin, V., Defacque, H., and Griffiths, G. (2001). ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol. Biol. Cell 12, 155-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahraus, A., Tjelle, T.E., Berg, T., Habermann, A., Storrie, B., Ullrich, O., and Griffiths, G. (1998). In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J. Biol. Chem. 273, 30379-30390. [DOI] [PubMed] [Google Scholar]
- Kellogg, D.R., Mitchison, T.J., and Alberts, B.M. (1988). Behaviour of microtubules and actin filaments in living Drosophila embryos. Development 103, 675-686. [DOI] [PubMed] [Google Scholar]
- Lang, T., Wacker, I., Wunderlich, I., Rohrbach, A., Giese, G., Soldati, T., and Almers, W. (2000). Role of actin cortex in the subplasmalemmal transport of secretory granules in PC-12 cells. Biophys. J. 78, 2863-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, E., and Knecht, D.A. (2002). Visualization of actin dynamics during macropinocytosis and exocytosis. Traffic 3, 186-192. [DOI] [PubMed] [Google Scholar]
- Lindberg, U., Hoglund, A.S., and Karlsson, R. (1981). On the ultrastructural organization of the microfilament system and the possible role of profilactin. Biochimie 63, 307-323. [DOI] [PubMed] [Google Scholar]
- Machesky, L.M., and Insall, R.H. (1999). Signaling to actin dynamics. J. Cell Biol. 146, 267-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield, C.J., Moss, S.E., Ballestrem, C., Imhof, B.A., Giese, G., Wunderlich, I., and Almers, W. (1999). Endocytic vesicles move at the tips of actin tails in cultured mast cells. Nat. Cell Biol. 1, 72-74. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J., and Cramer, L.P. (1996). Actin-based cell motility and cell locomotion. Cell 84, 371-379. [DOI] [PubMed] [Google Scholar]
- Neuhaus, E.M., and Soldati, T. (2000). A myosin I is involved in membrane recycling from early endosomes. J. Cell Biol. 150, 1013-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olazabal, I.M., Caron, E., May, R.C., Schilling, K., Knecht, D.A., and Machesky, L.M. (2002). Rho-kinase and myosin-II control phagocytic cup formation during CR, but not FcgammaR, phagocytosis. Curr. Biol. 12, 1413-1418. [DOI] [PubMed] [Google Scholar]
- Orci, L., Gabbay, K.H., and Malaisse, W.J. (1972). Pancreatic beta-cell web: its possible role in insulin secretion. Science 175, 1128-1130. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000). Polarization of cell growth in yeast. J. Cell Sci. 113, 571-585. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson, S.L., Provance, D.W., Jr., Mooseker, M.S., and Mercer, J.A. (2000). Class V myosins. Biochim. Biophys. Acta 1496, 36-51. [DOI] [PubMed] [Google Scholar]
- Riezman, H., Munn, A., Geli, M.I., and Hicke, L. (1996). Actin-, myosin- and ubiquitin-dependent endocytosis. Experientia 52, 1033-1041. [DOI] [PubMed] [Google Scholar]
- Rozelle, A.L., Machesky, L.M., Yamamoto, M., Driessens, M.H., Insall, R.H., Roth, M.G., Luby-Phelps, K., Marriott, G., Hall, A., and Yin, H.L. (2000). Phosphatidylinositol 4, 5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr. Biol. 10, 311-320. [DOI] [PubMed] [Google Scholar]
- Safer, D., and Nachmias, V.T. (1994). Beta thymosins as actin binding peptides. Bioessays 16, 590. [DOI] [PubMed] [Google Scholar]
- Small, J.V., Rottner, K., and Kaverina, I. (1999). Functional design in the actin cytoskeleton. Curr. Opin. Cell Biol. 11, 54-60. [DOI] [PubMed] [Google Scholar]
- Southwick, F.S., Li, W., Zhang, F., Zeile, W.L., and Purich, D.L. (2003). Actin-based endosome and phagosome rocketing in macrophages: activation by the secretagogue antagonists lanthanum and zinc. Cell Motil. Cytoskeleton 54, 41-55. [DOI] [PubMed] [Google Scholar]
- Swanson, J.A., Johnson, M.T., Beningo, K., Post, P., Mooseker, M., and Araki, N. (1999). A contractile activity that closes phagosomes in macrophages. J. Cell Sci. 112, 307-316. [DOI] [PubMed] [Google Scholar]
- Taunton, J., Rowning, B.A., Coughlin, M.L., Wu, M., Moon, R.T., Mitchison, T.J., and Larabell, C.A. (2000). Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148, 519-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L.G. (1975). The role of actin in nonmuscle cell motility. Soc. Gen. Physiol. Ser. 30, 339-388. [PubMed] [Google Scholar]
- Tilney, L.G. (1976). Actin: its association with membranes and the regulation of its polymerization. In: International Cell Biology 1976–1977, eds. B.R. Brinkley and K.R. Porter, New York: Rockefeller University Press, 388-402.
- Tilney, L.G., and Cardell, Jr., R.R. (1970). Factors controlling the reassembly of the microvillous border of the small intestine of the salamander. J. Cell Biol. 47, 408-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney, L.G., and Mooseker, M.S. (1976). Actin filament-membrane attachment: are membrane particles involved? J. Cell Biol. 71, 402-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu, K.T. (1978). A study of positive staining of ultrathin frozen sections. J. Ultrastruct. Res. 63, 287-307. [DOI] [PubMed] [Google Scholar]
- Trifaro, J.M., Rodriguez del Castillo, A., and Vitale, M.L. (1992). Dynamic changes in chromaffin cell cytoskeleton as prelude to exocytosis. Mol. Neurobiol. 6, 339-358. [DOI] [PubMed] [Google Scholar]
- Wang, L., Merz, A.J., Collins, K.M., and Wickner, W. (2003). Hierarchy of protein assembly at the vertex ring domain for yeast vacuole docking and fusion. J. Cell Biol. 160, 365-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L., Seeley, E.S., Wickner, W., and Merz, A.J. (2002). Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell 108, 357-369. [DOI] [PubMed] [Google Scholar]
- Wu, X., Jung, G., and Hammer, J. A., 3rd. (2000). Functions of unconventional myosins. Curr. Opin. Cell Biol. 12, 42-51. [DOI] [PubMed] [Google Scholar]
- Yin, H., Pruyne, D., Huffaker, T.C., and Bretscher, A. (2000). Myosin V orientates the mitotic spindle in yeast. Nature 406, 1013-1015. [DOI] [PubMed] [Google Scholar]
- Zhang, F., Southwick, F.S., and Purich, D.L. (2002). Actin-based phagosome motility. Cell Motil. Cytoskeleton 53, 81-88. [DOI] [PubMed] [Google Scholar]