Abstract
The membrane-bound Toll-like receptors (TLRs) trigger innate immune responses following recognition of a wide variety of pathogen-derived compounds. Despite the wide range of ligands recognized by TLRs, the receptors share a common structural framework in their extracellular, ligand-binding domains. These domains all adopt horseshoe-shaped structures built from leucine-rich repeat motifs. Typically, upon ligand binding, two extracellular domains form an “m”-shaped dimer sandwiching the ligand molecule bringing the transmembrane and cytoplasmic domains in close proximity and triggering a downstream signalling cascade. Although the ligand-induced dimerization of these receptors has many common features, the nature of the interactions of the TLR extracellular domains with their ligands varies markedly between TLR paralogs.
Keywords: Toll-like receptor, innate immunity, inflammation, leucine-rich repeat, dsRNA, Pattern recognition receptor
Introduction
In the initial phase of an infection, the innate immune system generates a rapid inflammatory response that blocks the growth and dissemination of the infectious agent. This response is followed, in vertebrates, by the development of an acquired immune response in which highly specific B and T cell receptors recognize the pathogen and induce responses that lead to its elimination (Janeway, Jr. et al., 2002). The antigen receptors of the acquired immune system are well characterized. They consist of many structurally similar molecules with different binding specificities created by somatic rearrangements and mutations within the binding site regions of the B and T cell receptor variable domains (Jung et al., 2004;Schatz et al., 2005). By contrast, the receptors of the innate immune system are germline-encoded and have been selected by evolution to recognize pathogen-derived compounds that are essential for pathogen survival or endogenous molecules that are released by the host in response to infection (Matzinger, 1994;Yang et al., 2010;Erridge, 2010). Innate immune receptors, also known as pattern recognition receptors (PRRs), have been identified in the serum, on the cell surface, in endosomes, and in the cytoplasm (Medzhitov, 2007). The Toll-like receptors (TLRs) represent a particularly important group of PRRs (Gay et al., 2007). TLR paralogs are located on cell surfaces or within endosomes and have important roles in the host defense against pathogenic organisms throughout the animal kingdom. In humans, ten TLRs respond to a variety of Pathogen-Associated Molecular Patterns (PAMPs), including lipopolysaccharide (TLR4), lipopeptides (TLR2 associated with TLR1 or TLR6), bacterial flagellin (TLR5), viral dsRNA (TLR3), viral or bacterial ssRNA (TLRs 7 and 8) and CpG-rich unmethylated DNA (TLR9), among others (Kumar et al., 2009).
The TLRs are type I integral membrane receptors, each with an N-terminal ligand recognition domain, a single transmembrane helix, and a C-terminal cytoplasmic signaling domain (Bell et al., 2003). The signaling domains of TLRs are known as Toll IL-1 Receptor (TIR) domains because they share homology with the signaling domains of IL-1R family members (O'Neill et al., 2007). TIR domains are also found in many adaptor proteins that interact homotypically with the TIR domains of TLRs and IL-1 receptors as the first step in the signaling cascade. Remarkably, homologs of TIR domains are also found in some plant proteins that confer resistance to pathogens (Burch-Smith et al., 2007), suggesting that the TIR domain represents a very ancient motif that served an immune function before the divergence of plants and animals. The transmembrane domains of TLRs each contain a typical stretch of approximately 20 uncharged, mostly hydrophobic residues. TLRs that recognize nucleic acid PAMPs interact through their transmembrane domains with a multispan transmembrane protein known as UNC93B, which directs these TLRs to endocytic compartments (Brinkmann et al., 2007;Kim et al., 2008). The remaining TLR paralogs do not interact with UNC93B, and traffic directly to the cell surface. The N-terminal ectodomains (ECDs) of TLRs are glycoproteins with 550–800 amino acid residues (Bell et al., 2003). These ectodomains are either extracellular or in endosomes where they encounter and recognize molecules released by invading pathogens. Here we review our current understanding of the structural basis for ligand recognition and signal transduction by TLRs.
Leucine-Rich Repeats - the building blocks of TLRs
All TLR ECDs are constructed of tandem copies of a motif known as the leucine-rich repeat (LRR), which is typically 22–29 residues in length and contains hydrophobic residues spaced at distinctive intervals (Figure 1A). This motif is found in many proteins in animals, plants and microorganisms, including many proteins involved in immune recognition (Palsson-McDermott et al., 2007). In three dimensions, all LRRs adopt a loop structure, beginning with an extended stretch that contains three residues in the β-strand configuration that were recently reviewed (Bella et al., 2008) (Figure 1B). When assembled into a protein, multiple consecutive LRRs form a solenoid structure, in which the consensus hydrophobic residues point to the interior to form a stable core and the β strands align to form a hydrogen-bonded parallel β sheet. Because the β strands are more closely packed than the non-β portions of the LRR loops, the solenoid is forced into a curved configuration in which the concave surface is formed by the β sheet (Kajava, 1998) (Figure 1C). As a result, each LRR protein contains a concave surface, a convex surface, an ascending lateral surface that consists of loops connecting the β strand to the convex surface, and a descending lateral surface on the opposite side (Bella et al., 2008).
Figure 1.
The structure of Leucine-Rich Repeats. (A) LRR consensus sequences for TLR3 and Ribonuclease Inhibitor. Residues forming the β strand are highlighted in orange. (B) A LRR loop from hTLR3 and a LRR loop from RI, with the conserved residues forming a hydrophobic core. The boxed regions form the surfaces involved in ligand binding. (C) Ribbon diagram of TLR3 (Bell et al., 2005) (2A0Z) and Ribonuclease Inhibitor (Kobe et al., 1995) (1DFJ). (D) Ribonuclease inhibitor complexed with ribonuclease A (Kobe et al., 1995) (1DFJ). The LRR protein is shown in blue and the ligand in orange. (E) Lamprey VLR complexed with H-trisaccharide (Han et al., 2008) (3E6J). (F) Glycoprotein Ib alpha complexed with the von Willebrand factor A1 domain (Huizinga et al., 2002) (1M10). Figures generated with program Pymol (DeLano, 2002).
The first LRR protein structure described was ribonuclease inhibitor (RI) (Kobe et al., 1995). The LRRs in this protein are relatively long, typically 27–29 amino acids in length, and all LRRs have 3–4 turns of α-helix on their convex surfaces opposite the β sheet. RI contains 16 LRRs that do not form a complete circle, but form a “horseshoe” structure (Figure 1C). In the innate immune system, a family of cytoplasmic danger sensors known as “Nod-like receptors” (NLRs) contain nine or fewer contiguous RI-like LRRs at their C-terminal ends, which are thought to be involved in recognizing danger signals. Based on the RI structure, one would predict that the LRRs of the NLR proteins form banana-shaped structures with a β-sheet on their concave surfaces and α-helices on their convex surfaces.
The TLR-ECDs typically contain 19–25 LRRs which, like RI, form horseshoe structures. In contrast to RI, the consensus LRR of the TLRs is 24 residues in length (Figure 1A), which does not allow for the formation of multi-turn helices on their convex sides. Consequently, inter-strand distances on the convex side are shorter for TLRs than for RI, which gives rise to TLR-ECD structures with lower curvature and larger outer diameters (~90Å) than for RI (Figure 1C). The 24 residue consensus LRRs adopt a variety of configurations on their convex sides, often containing bits of secondary structure such as β-strands, 310 helices, and polyproline II helices. In other proteins that contain 24 residue LRRs, for example glycoprotein Ibα (Uff et al., 2002), Nogo receptor (He et al., 2003), and the variable lymphocyte receptors (VLRs) of jawless vertebrates among others, a slight twist relates each LRR to its neighbor, which generates a non-planar overall structure. The twist in these proteins is most likely due to the inherent twist found in β-sheets. By contrast, the known TLR-ECD structures are more planar, suggesting that structures on the convex and lateral sides of TLR LRRs counteract the twist tendency on the concave side. Planarity of TLR-ECDs may be important for ligand binding and activation (see below).
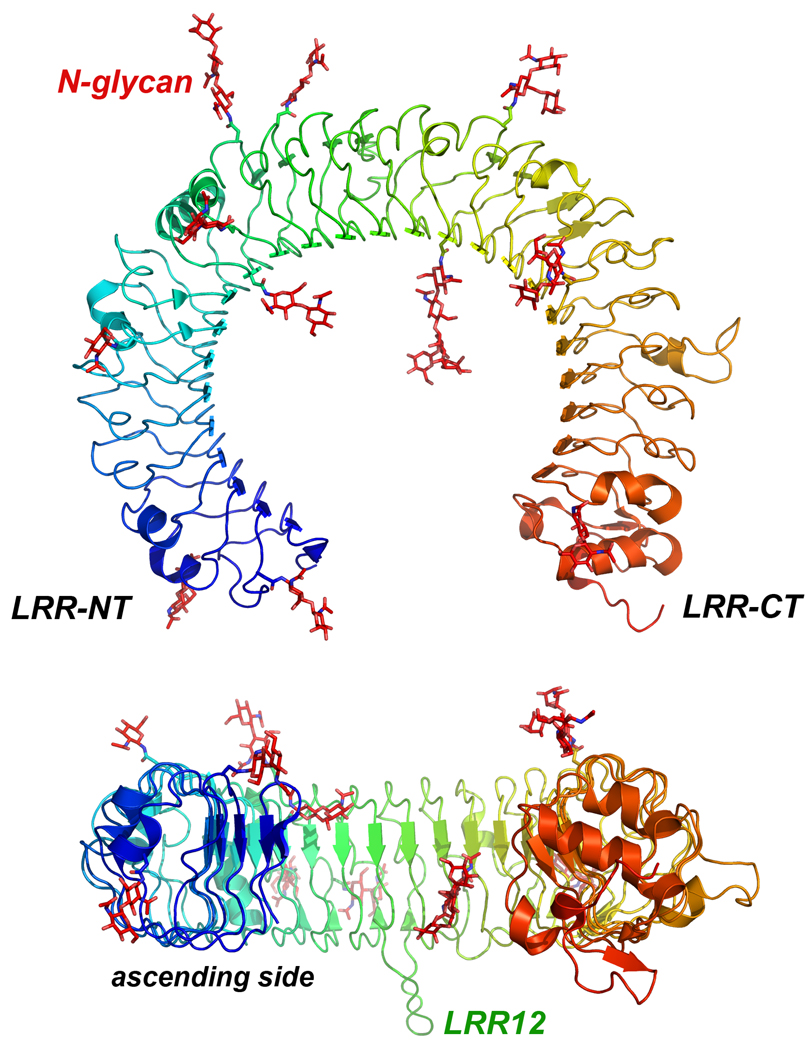
A characteristic feature of TLR-ECDs is the frequent occurrence of LRRs that are substantially larger than the consensus 24 residues, especially in TLRs 7, 8, and 9. These extra residues often produce loops that protrude from the TLR-ECD horseshoe, usually on the ascending or convex side of the LRR (Figure 1B). The TLR-ECDs also contain structures that cap the N and C-terminal ends known as the LRR-NT and LRR-CT motifs, respectively (Figure 2). The LRR-NTs are disulfide-linked β-hairpins, whereas the LRR-CTs are globular structures that contain two α-helices and are stabilized by two disulfide bonds. Similar capping motifs have been observed in several other proteins that contain 24 residue LRRs (He et al., 2003;Huizinga et al., 2002).
Figure 2.
The structure of a TLR-ECD (hTLR3). Top and side views of the TLR3-ECD, with the N-linked glycosyl moieties (Bell et al., 2005) (2A0Z). The LRRs are capped by the LRR-NT and LRR-CT motifs and the molecule is flat with one glycan-free side (ascending side) that is involved in receptor dimerization.
In most LRR proteins, ligand binding occurs on the concave surface (Figure 1D). For example, the VLRs of jawless vertebrates generate great diversity by randomly joining LRRs from a large pool of genomically encoded LRR cassettes into the mature VLR protein. In these molecules, the highest diversity in amino acid sequence occurs on the concave β surface. In the two ligand-VLR structures available, binding occurs on this surface (Deng et al., 2010;Han et al., 2008;Velikovsky et al., 2009)(Figure 1E). In addition, a large loop that interacts with the ligand protrudes from the LRR-CT in both structures, which is also seen in the glycoprotein Ibα-von Willebrand factor complex (Huizinga et al., 2002) (Figure 1F). By contrast, in the known TLR-ligand structures, ligand binding occurs most often on the ascending lateral surface of the TLR-ECD (Jin et al., 2007;Kang et al., 2009;Liu et al., 2008;Park et al., 2009)(Figure 2). This surface is the only portion of the molecule that completely lacks N-linked glycan and is free to interact with a ligand.
The structure of TLRs
Based on sequence homologies, vertebrate TLRs can be grouped into six subfamilies, TLR1/2/6/10, TLR3, TLR4, TLR5, TLR7/8/9 and TLR11/12/13/21/22/23 (Matsushima et al., 2007;Roach et al., 2005). Not all vertebrate species express all TLR paralogs; humans, for example lack all members of the TLR11 family. As indicated in Table 1, the ECDs of the ten human TLRs vary in LRR numbers and extent of N-linked glycosylation. To date, the structures of the ECDs of TLRs 1, 2, 3, 4 and 6 (human or mouse) have been reported (Table 2). All ECDs assume the typical horseshoe-shape, but their structures cannot be superimposed because of variations in curvature. In the known structures, the glycans are distributed throughout the molecule, except for the lateral face formed by the ascending loops of the LRRs (Figure 2). This glycan-free face is involved in dimerization upon ligand binding in the known TLR/ligand structures (see below).
Table 1.
Main features of the 10 human TLR molecules.
| TLR | Residues | LRRsa | N-linked glycosylation sitesb | Accession code |
|---|---|---|---|---|
| 1 | 786 | 19 | 4 (7) | Q15399 |
| 2 | 784 | 19 | 3 (4) | O60603 |
| 3 | 904 | 23 | 11 (15) | O15455 |
| 4 | 839 | 21 | 5 (10) | O00206 |
| 5 | 858 | 20 | (7) | O60602 |
| 6 | 796 | 19 | 8 (9) | Q9Y2C9 |
| 7 | 1049 | 25 | (14) | Q9NYK1 |
| 8 | 1041 | 25 | (18) | Q9NR97 |
| 9 | 1032 | 25 | (18) | Q9NR96 |
| 10 | 811 | 19 | (8) | Q9BXR5 |
The number of LRRs in the extracellular domain do not include the LRR-NT or LRR-CT motifs.
Number of N-glycosylation sites observed in the crystal structure or predicted by the NetNGlyc server 1.0 in parentheses (http://www.cbs.dtu.dk/services/NetNGlyc/)
Table 2.
TLR-ECD and TIR domain structuresa.
| Molecule | Residues TLR\VLR |
Resol. Å |
PDB code | Reference |
|---|---|---|---|---|
| TLR monomers | ||||
| hTLR2b | T1–284\V136–199 | 1.8 | 2Z80 | (Jin et al., 2007) |
| mTLR2/Pam3CSK4 | T27–506\V136–199 | 1.8 | 2Z81 | (Jin et al., 2007) |
| mTLR2/Pam2CSK4 | T27–506\V133–199 | 2.6 | 2Z82 | (Jin et al., 2007) |
| mTLR2/pnLTAc | T27–506\V133–200 | 2.5 | 3A7B | (Kang et al., 2009) |
| mTLR2/PE-DTPAd | T27–506\V133–200 | 2.4 | 3A7C | (Kang et al., 2009) |
| hTLR3 | T22–696 | 2.4 | 2A0Z | (Bell et al., 2005) |
| hTLR3 | T27–664 | 2.1 | 1ZIW | (Choe et al., 2005) |
| mTLR3 | T27–697 | 2.6 | 3CIG | (Liu et al., 2008) |
| hTLR4 | T27–228\V128–199 | 1.7 | 2Z62 | (Kim et al., 2007) |
| hTLR4 | T27–527\V133–199 | 2.0 | 2Z63 | (Kim et al., 2007) |
| hTLR4 | V24–82\ T228–383 | 1.9 | 2Z66 | (Kim et al., 2007) |
| hTLR4/hMD-2/Eritoran | T27–228\V19–158 | 2.7 | 2Z65 | (Kim et al., 2007) |
| mTLR4/mMD-2 | T27–625 | 2.8 | 2Z64 | (Kim et al., 2007) |
| hMD-2 | 2.0 | 2E56 | (Ohto et al., 2007) | |
| hMD-2/lipid IVa | 2.2 | 2E59 | (Ohto et al., 2007) | |
| TLR dimeric complexes | ||||
| hTLR1/hTLR2/Pam3CSK4 | T25–475\V133–199, T27–506\V133–199 | 2.1 | 2Z7X | (Jin et al., 2007) |
| mTLR3/dsRNA | T28–697 | 3.4 | 3CIY | (Liu et al., 2008) |
| hTLR4/MD-2/LPS | T27–631 | 3.1 | 3FXI | (Park et al., 2009) |
| mTLR2/mTLR6/Pam2CSK4 | T26–506\V133–200, T33–482\V157–232 | 2.9 | 3A79 | (Kang et al., 2009) |
| TIR domains | ||||
| hTIR1b | 2.9 | 1FYV | (Xu et al., 2000) | |
| hTIR2 | 3.0 | 1FYW | (Xu et al., 2000) | |
| hTIR2 P681H mutant | 2.8 | 1FYX | (Xu et al., 2000) | |
| hTIR2 C713S mutant | 3.2 | 1O77 | (Tao et al., 2002) | |
| hTIR10 | 2.2 | 2J67 | (Nyman et al., 2008) | |
| TIR (human IL-1RAPL) | 2.3 | 1T3G | (Khan et al., 2004) | |
| TIR (Arabidopsis) | 2.0 | 3JRN | (Chan et al., 2010) | |
| pdTIR (Paracoccus) | 2.5 | 3H16 | (Chan et al., 2009) | |
| TIR (MyD88) | NMR | 2Z5V | (Ohnishi et al., 2009) | |
| TIR (MyD88) | NMR | 2JS7 | unpublished | |
Available in the PDB as of 2010.
Human (h) and mouse (m) proteins with the residue numbers shown for the TLR part (T) and the VLR part (V) in case of the chimeric constructs. The length of the full TLR-ECDs including the signal peptide: TLR1 577 residues, TLR2 585, TLR3 696, TLR4 627 and TLR6 582.
pnLTA - S. pneumoniae lipoteichoic acid
PE-DTPA- 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine-N-diethylenetriaminepentaacetic acid (synthetic phospholipid derivative)
The TLR1/2/6/10 subfamily
TLR2 resides on the plasma membrane where it responds to lipid-containing PAMPs such as lipoteichoic acid and di- and tri-acylated cysteine-containing lipopeptides (Takeda et al., 2003). It does this by forming dimeric complexes with either TLR1 or TLR6 on the plasma membrane. The structures of TLR2/TLR1 and TLR2/TLR6 with lipopeptide ligands have been determined (Jin et al., 2007;Kang et al., 2009). To facilitate crystallization and structure determination the LRR-CT and the last one or two LRRs of TLRs 1, 2, and 6 were replaced by corresponding regions of a hagfish VLR (Table 2) (Kim et al., 2007). Structural analyses revealed that the ECDs of TLRs 1, 2, and 6 contain three distinctive subdomains: N-terminal, central and C-terminal (Figure 3A). The N-terminal subdomain contains the LRR-NT capping motif and LRRs 1–4, with typical 24 residue LRR modules. In this region, two features commonly found in the interiors of LRR structures are preserved: an asparagine ladder, formed by a continuous network of hydrogen bonds between consensus asparagines (Fig 1A) and neighboring backbone oxygens (Kajava, 1998); and a spine formed by consecutive consensus phenylalanine residues (Fig 1A) (Jin et al., 2007). By contrast, the central and C-terminal subdomains have LRR modules with atypical sequences, and their β sheet conformations deviate from those seen in standard LRRs. The central subdomain is also lacking the asparagine ladder and phenylalanine spine.
Figure 3.
Structure of the TLR1/TLR2/Pam3CSK4 complex. (A) The TLR2-ECD showing the position of the three subdomains: N-terminal, central and C-terminal. (B) hTLR1 and Pam3CSK4 ligand interactions mapped on the molecular surface. (C) hTLR2 and Pam3CSK4 interactions mapped on the surface. (D) Ribbon diagram of TLR1/TLR2 binding Pam3CSK4 (magenta) (Jin et al., 2007) (2Z7X).
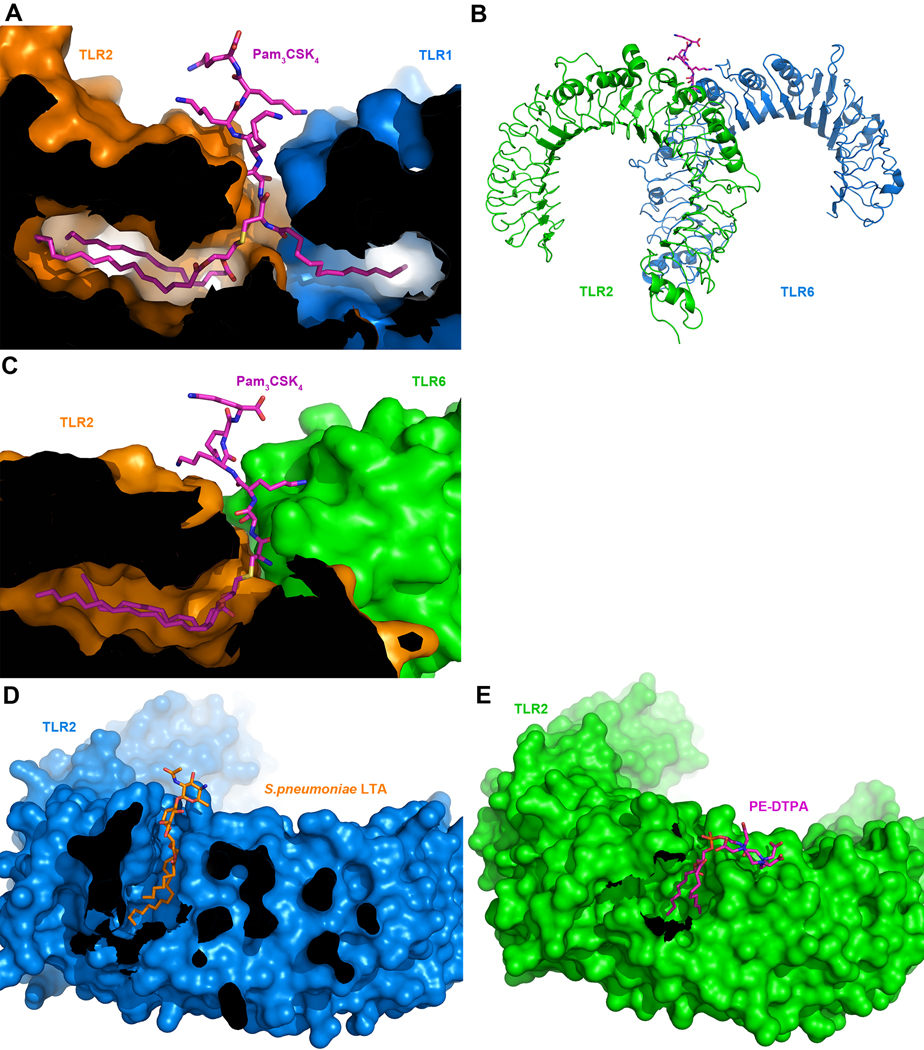
The border between the central and C-terminal subdomains (LRRs 9–12) harbors ligand-binding pockets on the convex side in both TLR1 and TLR2 ECDs (Figure 3B–C). These pockets are lined with hydrophobic residues and can accommodate fairly long lipid chains from lipopeptide ligands. The shapes of lipid binding pockets vary between species (Jin et al., 2007). For example, the mouse TLR2 binding pocket is shorter than the human. As a result, mouse TLR2 binds relatively short lauryl chains more efficiently than human TLR2, which is also reflected in the capacity of lauryl3CSK4 to activate mouse, but not human TLR2. In the TLR1/TLR2 complex (Figure 3D), the triacylated lipopeptide ligand, Pam3CSK4, bridges the two TLR-ECDs by inserting the two ester-bound palmitoyl groups into the TLR2 binding pocket, and the single amide-bound palmitoyl chain into the TLR1 pocket (Jin et al., 2007) (Figure 3B, 4A). TLRs 1 and 2 also interact with the head group of Pam3CSK4 by forming hydrogen bonds with the glycerol and peptide portions and by forming hydrophobic interactions with the sulfur atom. Importantly, TLRs 1 and 2 interact with each other on their dimerization surfaces via several hydrogen bonds and hydrophobic interactions in the vicinity of the binding pockets. The importance of these interactions to TLR1/2 function is suggested by the observation that a polymorphic mutation in this region, P315L, abolishes its ability to respond to lipopeptide ligands (Omueti et al., 2007). The ligand-protein and protein-protein interactions bring the C-terminal regions of the ECDs in close proximity, giving rise to an overall structure of the complex that has been described as having an “m”-shape (Figure 3D). The close apposition of the C-terminal regions likely facilitates the dimerization of the TIR domains on the cytoplasmic side of the plasma membrane.
Figure 4.
Ligand binding by TLR 1, 2 and 6. (A) Cross-section through the molecular surface of the TLR1/TLR2/Pam3CSK4 complex (2Z7X). (B) Ribbon diagram of TLR2/TLR6 binding Pam2CSK4 (magenta) (Kang et al., 2009)(3A79). (C) Cross-section through the molecular surface of the TLR2/TLR6/Pam2CSK4 complex (3A79). (D) Cross-section through the molecular surface of the TLR2/pnLTA complex (Kang et al., 2009)(3A7B). (E) Cross-section through the molecular surface of the TLR2/PE-DTPA complex (Kang et al., 2009)(3A7C).
Whereas the TLR1/2 complex recognizes tri-acylated lipopeptides such as Pam3CSK4, the TLR2/6 complex recognizes the di-acylated ligand, Pam2CSK4. The latter ligand lacks the lipid chain that binds in the hydrophobic pocket of TLR1, raising the question of how the di-acylated lipopepetide is able to stabilize a complex between TLRs 2 and 6. The crystal structure of the TLR2/TLR6/Pam2CSK4 complex (Kang et al., 2009) indicates that TLR6 and TLR1 have very similar, horseshoe structures and that the TLR2/6 complex adopts the same overall “m”-shaped structure as seen in the TLR1/2 complex (Figure 4B). However, TLRs 1 and 6 contain important structural differences in their ligand binding and dimerization regions. In TLR6, the side chains of two phenylalanine residues block the lipid binding channel, leading to a channel that is less than half the length as that of TLR1 (Figure 4C). This structural feature provides selectivity for diacylated over triacylated lipopeptides, as confirmed by the observation that mutation of the F343 and F365 residues in TLR6 to their TLR1 counterparts allows TLR6 to respond to the triacylated Pam3CSK4 (Kang et al., 2009). In the TLR2/6 complex, the two lipid chains of Pam2CSK4 are buried in the TLR2 hydrophobic pocket as in the TLR1/2 complex while the peptide part of Pam2CSK4 forms several hydrogen bonds with both TLR2 and TLR6 (Figure 4C). The LRR11 loop in TLR6 is slightly displaced relative to its position in TLR1 and forms a prominent hydrogen bond with the carbonyl oxygen of the first peptide bond of the ligand. In addition, the LRR11-LRR14 regions of TLR2 and TLR6 form a heterodimeric interface via hydrophobic and hydrophilic interactions of their surface-exposed residues. The area of hydrophobic interaction is 80% larger than in the hTLR1/hTLR2 complex, suggesting that this surface interaction together with the hydrogen bond between LRR11 and the ligand drives the heterodimerization of TLR6.
Lipoteichoic acid (LTA), a glycolipid from Gram+ bacterial membranes contains a diacylated glycerol group connected by an ether bond to a variable carbohydrate moiety. LTAs vary in their capacity to activate TLR2, for example S. aureus LTA is a much more potent activator than S. pneumoniae LTA. The crystal structure for S. pneumoniae LTA bound to the TLR2/VLR hybrid shows differences compared to lipopeptide bound to TLR2 (Kang et al., 2009) (Figure 4D). Curiously, this LTA failed to dimerize TLR2 or induce hetero-dimers with TLR1 or TLR6, although it was capable of activating TLR2 in cells, probably by forming a complex with TLR1 (Han et al., 2003). Another nonpeptidic ligand PE-DTPA (a synthetic derivative of phosphatidylethanolamine) binds mTLR2/VLR similar to LTA (Kang et al., 2009) (Figure 4E). These TLR2/ligand structures are missing the hydrogen bonding network present in TLR2-lipopeptide complexes, due to the shift of the carbohydrate head groups relative to the peptide head groups of lipopeptide ligands. This shift is also enhanced by the repulsion between the head group oxygen atom of the ligand and the hydrophobic sulfur binding site of TLR2, which is normally filled by the sulfur atom from the lipopeptide cysteine. The hydrogen bonding network is possibly required to maintain the correct conformation of the TLR2 LRR11 loop that makes crucial intermolecular contacts with TLR1 or TLR6 in the heterodimeric complexes.
There are four different types of protein-ligand interactions for the TLR 1, 2 and 6 complexes. These interactions can be ranked in order of their importance: hydrophobic interactions with the TLR2 pocket, hydrogen bonding of peptide head groups, hydrophobic interactions with the TLR1 channel and hydrophobic interaction of the conserved cysteine from peptide headgroups. The heterodimer is further stabilized by hydrophobic, hydrogen-bonding and ionic interactions between the TLR molecules.
Phylogenic analysis identifies TLR10 as part of the TLR-1 family (Roach et al., 2005). Based on the above TLR1/TLR2 and TLR2/TLR6 complex structures, homology models of hTLR10 complexes were constructed and refined through molecular dynamics simulations (Govindaraj et al., 2010). The hTLR2/hTLR10 and hTLR1/hTLR10 complex models were similar to the available TLR1 family complexes. However, the binding orientation of the hTLR10 homodimer was different due to the presence of negatively charged surfaces near LRR 11–14 that define a specific binding pocket. Docking experiments suggest that Pam3CSK4 might be a ligand for the hTLR2/hTLR10 complex and PamCysPamSK4 might bind to hTLR1/hTLR10 and the hTLR10 homodimer.
TLR3
TLR3 recognizes dsRNA, which is produced by most viruses at some stage in their lifecycles and is a potent indicator of viral infection. In contrast to several cytoplasmic receptors for dsRNA, TLR3 is localized to endosomes where it recognizes dsRNA. Studies using soluble TLR3-ECD protein (Leonard et al., 2008) or immobilized TLR3-GFP constructs (Wang et al., 2010) showed that TLR3-ECD is monomeric in solution but binds as dimers to 45 bp segments of dsRNA, the minimum length required for TLR3 binding and activation. In addition, binding is independent of base sequence and occurs only at pH 6.5 and below (Leonard et al., 2008). A crystal structure of the TLR3-ECD dimer complexed with a 46 bp dsRNA oligonucleotide explains these properties.
The first reported crystal structure of a TLR binding domain was that of the unliganded form of TLR3-ECD (Bell et al., 2005;Choe et al., 2005) (Figure 2). The TLR3-ECD horseshoe is largely uniform and flat, lacking the subdomain structure seen in TLRs 1, 2, 4 and 6. However, irregularities in the LRRs produce short alpha helices on the convex side of the horseshoe and two large, conserved loops that protrude from the lateral and convex faces of LRRs 12 and 20, respectively. TLR3-ECD is heavily glycosylated, with 15 predicted N-glycosylation sites, of which 11 are visible in the crystal structure. The glycans decorate all faces of the TLR3-ECD, except for the lateral surface on the C-terminal side of the β sheet. This face offers a large, planar surface for interaction with dsRNA (Figure 2). In the crystal structure of the TLR3-ECD-dsRNA complex, the glycan-free surfaces of two TLR3-ECDs sandwich the dsRNA molecule, generating an “m”-shaped structure (Liu et al., 2008) (Figure 5A). No conformational change in the TLR3-ECD occurs upon ligand binding. In the complex, the dsRNA interacts at two sites on each TLR3-ECD, one near the N-terminus (encompassing LRR-NT and LRRs 1–3), and one near the C-terminus (involving LRRs 19–23) (Figure 5B). In addition, the two ECDs interact with each other at their LRR-CT motifs. Mutational analyses (Wang et al., 2010) have established that the simultaneous interaction of all three sites is required for stable binding of dsRNA to TLR3.
Figure 5.
Structure of the TLR3/dsRNA complex. (A) Molecular surface of TLR3 dimer (green) with bound dsRNA (Liu et al., 2008)(3CIY). The interaction of the C-terminal capping motifs stabilizes the TLR3 dimer. (B) Top view.
Although the N and C-terminal dsRNA sites are separated by 55–60 Å in each ECD, the two N-terminal sites in the complex are separated by 110 Å. This latter distance correlates with a dsRNA length of ~45 base pairs and explains why dsRNA oligonucleotides less than about 40 bp cannot bind or activate TLR3 (Leonard et al., 2008). Because cells normally contain short (25 bp or less) stretches of dsRNA for example in miRNA and tRNA hairpins, the inability of TLR3 to bind dsRNA less than 40 bp most likely provides an important mechanism for preventing auto-reactive responses against self dsRNA. In binding dsRNA, the TLR3-ECD interacts only with the ribose-phosphate backbone, accounting for the lack of RNA sequence specificity in binding. This feature would prevent the viruses from escaping detection by mutation. All binding-site residues are located on the glycan-free surface of the ECD. Of a particular interest is the Asn-413 N-linked paucimanose moiety that reaches out from the concave surface and contacts the dsRNA helix, but the relevance of this interaction is unclear.
The majority of TLR3/dsRNA interactions are hydrophilic, involve hydrogen bonds and salt bridges, and account for a total buried area of 1103 Å2 (4.4% of the TLR3-ECD surface). Particularly important are electrostatic interactions between the phosphate groups from the dsRNA backbone and the imidazole rings of four histidine residues, three in the N-terminal site, and one in the C-terminal site. Mutation of two of these histidines (H39 and H60 in the N-terminal site) to alanine abolishes binding and responsiveness to dsRNA, indicating that the salt bridges formed by these residues are essential for complex formation. This could explain the pH-dependency of dsRNA binding because these side chains would be protonated only below pH 6.5 and able to interact with the RNA phosphates. The structure of the complex reveals the main reasons for the inability of TLR3 to interact with dsDNA. The helical structure of dsDNA is the B form, whereas dsRNA assumes the A form. The B helix would not be structurally compatible with the two terminal binding sites on the TLR3-ECD horseshoe. Also, there are several hydrogen bonds between TLR3-ECD and the 2’-hydroxyl groups of dsRNA that would be missing in dsDNA.
The only interaction between the two TLR3 molecules in the complex occurs at the interface between the two LRR-CT motifs, and therefore this site is responsible for dimerization. In this site, the two LRR-CTs are related by two-fold symmetry and the interactions between the LRR-CTs are mainly hydrophilic, consisting of hydrogen bonds and salt bridges. The dimerization site is essential for dsRNA binding (Wang et al., 2010) because it correctly positions the four dsRNA binding sites in the complex, but in addition it also brings the two C-terminal residues within ~25 Å of each other. In the cell, two LRR-CT motifs that interact on the luminal side of an endosome would presumably bring the two TIR domains together on the cytoplasmic side, forming a dimeric scaffold on which adaptor molecules could bind and initiate signaling.
TLR4 and MD-2
Lipopolysaccharide (LPS), an essential component of the outer membrane of Gram negative bacteria, induces a powerful inflammatory response that can lead to septic shock and death (Beutler et al., 2003). LPS signals through TLR4 by complexing co-receptor MD-2, which is anchored by several hydrogen bonds to the lateral and concave surface of TLR4-ECD and contacts residues from the LRR2-LRR10 area (Kim et al., 2007) (Figure 6A). LPS-binding protein (LBP) and CD14 deliver and load the LPS to the TLR4-bound MD-2.
Figure 6.
Structure of the TLR4/MD-2/LPS complex. (A) Ribbon diagram of TLR4 green, MD-2 (blue) binding LPS (magenta) (Park et al., 2009)(3FXI). (B) Cross-section through the molecular surface of the MD-2/LPS complex. (C) TLR4 and LPS interactions mapped on their molecular surface. (D) Cross-section through the molecular surface of the MD-2/lipid IVa complex (Ohto et al., 2007)(2E59). (E) Cross-section through the molecular surface of the MD-2/Eritoran complex (Kim et al., 2007) (2Z65).
One of the important factors that determine the inflammatory potential of an LPS molecule (Rietschel et al., 1994) is the number of lipid chains in the Lipid A portion of LPS. Six chains provide an optimal inflammatory activity, whereas Lipid A with five chains has approximately 100-fold less activity. Ligands with only four lipid chains, such as lipid IVa and Eritoran, have antagonistic activity (Teghanemt et al., 2005). Because the lipids interact with the MD-2 pocket through hydrophobic contacts, variations in the lipid chain structure can be accommodated by shifting the position of the chains in the pocket. With a smaller number of lipid chains, Lipid A can move deeper into the pocket and the phosphate groups cannot participate in ionic interactions with TLRs (Park et al., 2009). These two phosphate groups are also essential for the endotoxic activity of LPS, and deleting either of them reduces the endotoxic activity by ~100 fold (Rietschel et al., 1994).
TLR4-ECD has 21 LRRs capped by LRR-NT and LRR-CT motifs. The structures of the TLR4-ECD and several chimeric fragments of TLR4 capped with hagfish VLR domains (Kim et al., 2007) show a horseshoe structure that is much less planar than the TLR3-ECD. Three subdomains can be differentiated, N-terminal, central and C-terminal, with different degrees of twist and curvature. The central subdomain contains only one variable residue between the last leucine residue of a preceding LRR motif and the first leucine residue of the next LRR motif, in contrast with the standard two variable residues (Fig 1A). The length of these LRR modules also varies between 20–30 residues, conferring a smaller radius and greater twist angle to the subdomain. The central subdomains of human and mouse TLR4 differ most in structure, apparently due to the MD-2 binding to the mTLR4 LRR9 loop.
MD-2 is a co-receptor molecule that binds both the extracellular domain of TLR4 and the hydrophobic portion of LPS (Visintin et al., 2006). The crystal structure of hMD-2 complexed to lipid IVa has been determined (Ohto et al., 2007) (Figure 6D). Lipid IVa, a precursor of Lipid A, is an antagonist for hTLR4 but an agonist for mTLR4 (Means et al., 2000). The molecule adopts the β cup topology of the ML family of lipid-binding proteins that have sandwiched antiparallel β-sheets and conserved disulfide bridges (Derewenda et al., 2002). The β-sheets of hMD-2 form a very deep and narrow hydrophobic pocket, with a surface area of ~1000 Å that can bury the four lipid strands of lipid IVa. The opening of the pocket is lined by positively charged residues and three disulfide bridges stabilize the cup-like structure. Based on this structure, it seemed unlikely that the hMD-2 pocket would be able to accommodate more than four lipid chains without conformational changes, but another structure with bound LPS showed that additional room for the ligand is generated, not by conformational change, but by displacing the LPS glucosamine backbone upwards by ~ 5 Å (Park et al., 2009). In addition to the displacement, the glucosamine backbones are also rotated by ~180 degrees, interchanging the two phosphate groups. The crystal structure of the TLR4-ECD/MD-2/Eritoran complex was also reported (Kim et al., 2007) (Figure 6E). Eritoran is a synthetic antagonist with four lipid chains that occupy ~90% of the hMD-2 hydrophobic pocket, while the di-glucosamine sugars are fully exposed to solvent and the phosphate groups form essential ionic bonds with positively charged residues at the opening of the pocket. Binding of Eritoran does not induce significant structural changes in hMD-2. The available structures suggest that the MD-2 pocket evolved to bind large and structurally different ligands (Kim et al., 2007). Agonists and antagonists will bind in a similar fashion, but with their glucosamine backbones rotated by ~180 degrees.
The 3.1 Å resolution structure of the TLR4/MD-2/LPS complex (Park et al., 2009) shows two E.coli LPS molecules bound (Figure 6A). Five of the LPS lipid chains are buried in the large hydrophobic MD-2 pocket, while the sixth one is exposed and makes hydrophobic contacts with conserved phenylalanines on the other TLR4-ECD (Figure 6B). The LPS phosphate groups form ionic interactions with positively charged residues on MD-2 and TLR4-ECD (Figure 6C). In addition, the F126 and L87 loops of MD-2 interact with the second TLR4-ECD molecule. All these interactions bring the two copies of the TLR4-ECD/MD-2/LPS complex together into a typical “m”-shaped signaling complex.
Cells that express TLR4/MD-2 also express a homologous complex, RP105/MD-1, that regulates the LPS response (Akashi-Takamura et al., 2008;Divanovic et al., 2007). RP105 is a type I receptor with an ECD resembling that of TLR4. However, RP105 lacks a C-terminal TIR domain. MD-1, like MD-2, binds LPS and a crystal structure of a lipid IVa/MD-1 complex has been reported (Yoon et al., 2010). Although both MD-1 and MD-2 are members of the ML family of lipid binding proteins, they use different surfaces for ligand recognition. RP105/MD-1 binds directly to TLR4/MD-2, and on B cells RP105/MD-1 enhances the LPS response (Miyake et al., 1994;Miura et al., 1998;Miyake et al., 1998), whereas on macrophages it attenuates the response (Divanovic et al., 2005).
TLR5
TLR5 is one of the few TLRs that recognize a protein PAMP, bacterial flagellin (Hayashi et al., 2001). It is highly expressed in gut, especially in lamina propria dendritic cells (Uematsu et al., 2009) where it controls the composition of the microbiota (Vijay-Kumar et al., 2010). With no structure available, TLR5-ECD is predicted to contain 20 LRRs, with five loops extending from the ascending or convex surfaces (Bell et al., 2003). Mutational analyses of flagellin have located the site recognized by TLR5 as lying in the conserved D1 domain (Donnelly et al., 2002). This domain is exposed in monomeric flagellin, but is buried in the polymerized flagellin fiber, suggesting that TLR5 recognizes flagellin monomers that are released upon depolymerization of flagellin polymers (Smith et al., 2003). The portion of the TLR5-ECD that interacts with flagellin is less well defined. Human and mouse TLR5 differ in their ability to recognize WT or mutated flagellin molecules from different sources (Andersen-Nissen et al., 2007;Miao et al., 2007). Moreover, mouse TLR5 gains human specificity when one residue, Pro268, is mutated to its human counterpart, alanine and vice versa. Sequence analysis predicts that 19 Pro268 lies on the ascending/convex surface of LRR9, an atypical LRR motif (Bell et al., 2003). Thus it is likely that LRR9 plays an important role in flagellin recognition by TLR5.
The TLR 7, 8 and 9 family
No structure has yet been reported for any member of the TLR 7, 8, and 9 subfamily. Like TLR3, these TLRs are located in endosomes and recognize nucleic acid PAMPs. However, their amino acid sequences suggest that the structures of TLRs 7–9 ECDs are markedly different from TLR3 (Bell et al., 2003). TLRs 7–9 ECDs each comprise 25 LRRs and are heavily glycosylated. They all contain large insertions in LRRs 2, 5, and 8 that most likely give rise to structures that loop out from the dimerization surfaces of the ECDs. In addition, the ECDs of TLRs 7–9 contain stretches of approximately 40 residue between LRRs 14 and 15 that have undefined structure. Because these stretches are the only portions that show a high degree of species variability in each of the three paralogs, it is likely that these regions are relatively unstructured. TLR9, which recognizes single stranded DNA with unmethylated CpG sequences, especially in viral and bacterial DNA, has been the most extensively studied paralog of this family (Hemmi et al., 2000;Kumar et al., 2009). There is evidence (Latz et al., 2007) that TLR9 undergoes an extensive conformational change when binding CpG DNA, which would make it very different from TLR3. A ligand-induced conformational change could be dependent on the unstructured region if it served as a hinge within the horseshoe, thus allowing a large conformational transition to occur upon ligand binding. More recently, several studies have suggested that proteases are required for TLR9 function (Asagiri et al., 2008;Ewald et al., 2008;Matsumoto et al., 2008;Park et al., 2008), and that TLR9 is cleaved within the undefined region between LRRs 14 and 15 (Park et al., 2008), consistent with this being an unstructured, exposed stretch of amino acid residues. Another report suggests that the C-terminal fragment, lacking the first 14 LRRs and the undefined region, is active by itself implying that that the N-terminal fragment is not needed for ligand recognition (Park et al., 2008). This observation seems to be inconsistent with a more recent study (Peter et al., 2009) showing that mutations within the N-terminal fragment inactivate TLR9. Indeed it seems unlikely that a large portion of the TLR9-ECD would be highly conserved throughout vertebrate evolution, and yet not be essential for TLR9 function. Clearly, a detailed structural analysis of the TLR9-ECD is needed to clarify the mechanism of ligand binding and activation of TLR9 as well as TLRs 7 and 8.
Common features and differences between the signaling complexes
The signaling complex structures of the TLRs with their respective ligands reveal the diverse mechanisms of recognition of a wide variety of PAMPs. These PAMPs carry characteristic molecular signatures that are unique to different classes of pathogens and are detected by the TLRs. In all structures, the signaling complex consists of an “m”-shaped TLR dimer, in which the ECD N-termini extend to the opposite ends and the C-termini interact in the middle. Formation of the homo- or heterodimer supports the hypothesis that dimerization of the ECDs positions the cytoplasmic TIR domains close enough to dimerize and initiate a downstream signaling cascade (Jin et al., 2007;Kang et al., 2009;Liu et al., 2008;Park et al., 2009). In most cases, two ECDs cradle a single ligand molecule, except for TLR4 where two MD-2 molecules each bind a ligand. Most known TLR structures do not use the concave beta-sheet surface for ligand binding, a characteristic mode of binding for other LRR proteins. TLR4 is an exception because it binds its adaptor protein MD-2 in the concave surface.
In all TLR complexes, the TLR-ECDs are related by an approximate two-fold symmetry axis. In the TLR 1, 2, 3 and 6 complexes, the non-glycosylated surface of two TLR molecules sandwich a ligand molecule, while in the TLR4 complex the coreceptor molecule MD-2 binds the ligand directly. TLR3 binds its ligand mainly by hydrogen bonding and electrostatic interactions while hydrophobic interactions dominate the ligand binding in other known TLR structures. The buried surface area in the TLR1/TLR2 interaction is similar to that in the TLR3 homodimer. In the TLR3/dsRNA complex the protein-protein interactions occur only at the LRR-CT, while direct interactions between TLR1/TLR2 or TLR2/TLR6 occur near the binding pockets. In the TLR3/dsRNA and TLR4/MD-2/LPS complexes the two C-terminal residues are ~25 Å apart, whereas in the TLR1/2 and TLR2/6 complexes they are ~40 Å apart. This larger distance may be due to the fact that the native LRR-CT motifs were replaced with hagfish VLRs, however. In the TLR4/MD-2/LPS complex, despite the close proximity of the C-termini there is no direct interaction between the two molecules in this region. In the known TLR-ligand complexes, ligands bind TLRs by different mechanisms, but in each case, the ligand bridges two TLR-ECDs on the same glycan-free surface and forms dimers with similar overall architecture. Whether this paradigm will apply to the TLR7–9 family remains to be determined.
The TIR domain
The TLRs signal pathogen attack by dimerization of their cytoplasmic TIR domains in response to ligand-induced dimerization of the ectodomains (Figure 7A). TIR dimerization of TLRs is recognized by TIR domains on the adaptor proteins MyD88, MAL, TRIF, and TRAM, which then trigger downstream signaling pathways, leading to the expression of inflammatory cytokines, various anti-viral and anti-pathogen proteins and to initiation of the adaptive immune response (Figure 7B). The TIR domain is one of many evolutionarily conserved building block of the immune system (Figure S1) (Palsson-McDermott et al., 2007).
Figure 7.
(A) Structural model of the full-length TLR3/dsRNA signaling complex. The model is based on the mTLR3/dsRNA structure (3CIY) and a TLR3 TIR domain homology model based on the TLR10 TIR structure (2J67). The transmembrane portions have been modeled as α-helices. (B) TLR signaling pathways. TLR3 signals exclusively through TRIF, while TLR4 can use both the TRIF and the MyD88 pathways. All other TLRs use the MyD88 pathway.
The crystal structures of isolated TIR domains have been reported for TLRs 1, 2 (Xu et al., 2000) and 10 (Nyman et al., 2008) (Table 2), IL-1RAPL (Khan et al., 2004) and for TIR domains from Arabidopsis thaliana (Chan et al., 2010) and Paracoccus denitrificans (Chan et al., 2009). In addition, an NMR structure has been reported for the MyD88 TIR domain (Ohnishi et al., 2009). In all TIR domains, alternating β strands and α-helices are arranged as a central five-stranded parallel β-sheet surrounded by five α-helices.
The TIR domains from both TLR1 and TLR2 exist as monomers in the crystal. Despite sharing 50% sequence identity there are important conformational differences between the two structures. By contrast, the 2.2 Å structure for the TIR domain of human TLR10 revealed a two-fold symmetric dimer which has been taken to represent the signaling dimer for the TIRs (Figure 8) (Nyman et al., 2008). The BB-loop that joins strands βB and αB is one of the essential points of interaction in the dimeric interface, which also contains residues from the DD-loop, and the αC-helix. In addition to being involved in TIR dimer formation, the BB-loop is sufficiently exposed to interact with adaptor proteins during signal transduction. In TLR4, the P681H polymorphism, which lies in the BB-loop, abolishes signaling in response to LPS (Poltorak et al., 1998), indicating that the BB loop plays an essential role in TLR signaling. The structures of the plant (Chan et al., 2010) and bacterial TIR domains (Chan et al., 2009) are similar to their mammalian counterparts. However, in the latter a dimer is formed that is very different from the TLR10 dimer. The BB loops are not involved in dimerization contacts but are highly exposed on the surface. An important question left in TLR structural biology is the orientation of TIR domains that activate the signaling cascade. Available TIR domain structures lack the region immediately following their transmembrane (TM) segment, making it harder to predict their exact orientation. In the TLR10 TIR dimer, the N-termini of both molecules have to simultaneously point towards the membrane, to accommodate the linker between the TM segment and the first N-terminal residue from the TIR.
Figure 8.
Structure of the TIR domain dimer from TLR10. (A) Ribbon diagram of the dimeric TIR10, with the two interacting BB-loops highlighted in gold and red (Nyman et al., 2008) (2J67). (B) Molecular surface of dimeric TIR10.
Purified TIR domains from the MAL and MyD88 adaptor proteins have been shown to form stable heterodimers in solution (Dunne et al., 2003). Dunne et al. also produced models of the TIR domains from human TLR4, MAL and MyD88 and used these to model the interactions between pairs of these TIR domains. In these models, the BB loop does not seem to be involved in either TIR dimerization or in heterotypic interactions with TIRs from the adaptor proteins, which is contrary to expectation. Clearly, more structures of interacting TIR domains will be required to better understand how TIR domains function. In addition to its TIR domain, MyD88 contains a death domain. Following binding to TLR TIR domains, MyD88 death domains can interact with the death domains of members of the IRAK family of Ser/Thr kinases, triggering their activation and initiating downstream signaling cascades. A structure of a ternary complex formed by the death domains of human MyD88, IRAK4 and IRAK2 (Lin et al., 2010) showed that these molecules form a left handed helical structure containing in order 6 MyD88, 4 IRAK4 and 4 IRAK2 death domains. The MyD88-IRAK4 oligomeric assembly – also called the myddosome - was also confirmed by cryo EM and small angle X-ray scattering experiments (Motshwene et al., 2009). There are three types of interactions observed between the death domains in the myddosome. SNPs in the human MyD88 death domain, S34Y and R98C interfere with the myddosome assembly and may drastically contribute to susceptibility to infection (George et al., 2011). If each TLR-TIR dimer binds two MyD88 TIR domains then the large myddosome superhelix could possibly bridge several activated receptor dimers into a network (Gay et al., 2011). These scaffolds might include more than one type of MyD88-dependent TLR, enabling synergistic responses to microbial stimuli. There are other cases where death domains assemble in superhelical oligomeric signaling complexes like the Death-Inducing Signaling Complex (DISC) (Scott et al., 2009) or the PIDDosome (Park et al., 2007).
Concluding remarks
TLRs are germ-line encoded pattern recognition receptors that initiate defensive responses against a wide variety of pathogens. TLRs are type I membrane receptors with mainly planar, horseshoe-shaped LRR ECDs. TLR-ECD-ligand complex structures are known for TLR1/TLR2 and TLR6/TLR2 with lipopeptides, TLR3 with dsRNA, and TLR4/MD-2 with LPS. Although the TLR-ligand interactions are very different, they all produce an “m”-shaped dimeric complex with C termini in the middle, and N-termini on the outside. Pairing of the ECD C-termini can lead to dimerization of the cytoplasmic TIR domains, which is recognized homotypically by adapter molecule TIR domains. MyD88, the most commonly used adapter, also has death domains that interact with IRAK kinase death domains to form a superhelical myddosome, which presumably initiates the signaling cascade.
Despite substantial progress in our understanding of the structural basis of TLR recognition and signaling, several questions need to be addressed. There is still no structural data for TLRs 5, 7, 8, and 9. Amino acid sequences of the TLR 7–9 family suggest that they may differ from other TLRs in both structure and mode of ligand recognition. How TLR-TIR domains interact with each other or with TIRs from adapter molecules is still unclear. Structures of adapter molecules would provide insight into how they translate TIR recognition into IRAK activation. Better understanding of the structural basis for PAMP recognition and signaling by TLRs could lead to the development of adjuvants that specifically bind to TLR-ECDs and activate TLRs or of anti-inflammatory drugs that block TLR mediated signaling.
Supplementary Material
Acknowledgements
This work was support by the Intramural Research Program of the National Institutes of Health (NIDDK and NCI) and by a National Institutes of Health/Food and Drug Administration intramural biodefense award from NIAID.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akashi-Takamura S, Miyake K. TLR accessory molecules. Curr. Opin. Immunol. 2008;20:420–425. doi: 10.1016/j.coi.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Andersen-Nissen E, Smith KD, Bonneau R, Strong RK, Aderem A. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. J. Exp. Med. 2007;204:393–403. doi: 10.1084/jem.20061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asagiri M, Hirai T, Kunigami T, Kamano S, Gober HJ, Okamoto K, Nishikawa K, Latz E, Golenbock DT, Aoki K, Ohya K, Imai Y, Morishita Y, Miyazono K, Kato S, Saftig P, Takayanagi H. Cathepsin K-dependent toll-like receptor 9 signaling revealed in experimental arthritis. Science. 2008;319:624–627. doi: 10.1126/science.1150110. [DOI] [PubMed] [Google Scholar]
- Bell JK, Botos I, Hall PR, Askins J, Shiloach J, Segal DM, Davies DR. The molecular structure of the Toll-like receptor 3 ligand-binding domain. Proc. Natl. Acad. Sci. U. S. A. 2005;102:10976–10980. doi: 10.1073/pnas.0505077102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JK, Mullen GE, Leifer CA, Mazzoni A, Davies DR, Segal DM. Leucine-rich repeats and pathogen recognition in Toll-like receptors. Trends Immunol. 2003;24:528–533. doi: 10.1016/s1471-4906(03)00242-4. [DOI] [PubMed] [Google Scholar]
- Bella J, Hindle KL, McEwan PA, Lovell SC. The leucine-rich repeat structure. Cell Mol. Life Sci. 2008;65:2307–2333. doi: 10.1007/s00018-008-8019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutler B, Rietschel ET. Innate immune sensing and its roots: the story of endotoxin. Nat. Rev. Immunol. 2003;3:169–176. doi: 10.1038/nri1004. [DOI] [PubMed] [Google Scholar]
- Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim YM. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, nesh-Kumar SP. The functions of plant TIR domains. Sci. STKE. 2007;2007:e46. doi: 10.1126/stke.4012007pe46. [DOI] [PubMed] [Google Scholar]
- Chan SL, Low LY, Hsu S, Li S, Liu T, Santelli E, Le NG, Reed JC, Woods VL, Jr, Pascual J. Molecular mimicry in innate immunity: crystal structure of a bacterial TIR domain. J. Biol. Chem. 2009;284:21386–21392. doi: 10.1074/jbc.C109.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SL, Mukasa T, Santelli E, Low LY, Pascual J. The crystal structure of a TIR domain from Arabidopsis thaliana reveals a conserved helical region unique to plants. Protein Sci. 2010;19:155–161. doi: 10.1002/pro.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- Deng L, Velikovsky CA, Xu G, Iyer LM, Tasumi S, Kerzic MC, Flajnik MF, Aravind L, Pancer Z, Mariuzza RA. A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13408–13413. doi: 10.1073/pnas.1005475107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derewenda U, Li J, Derewenda Z, Dauter Z, Mueller GA, Rule GS, Benjamin DC. The crystal structure of a major dust mite allergen Der p 2, and its biological implications. J. Mol. Biol. 2002;318:189–197. doi: 10.1016/S0022-2836(02)00027-X. [DOI] [PubMed] [Google Scholar]
- Divanovic S, Trompette A, Atabani SF, Madan R, Golenbock DT, Visintin A, Finberg RW, Tarakhovsky A, Vogel SN, Belkaid Y, Kurt-Jones EA, Karp CL. Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat. Immunol. 2005;6:571–578. doi: 10.1038/ni1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divanovic S, Trompette A, Petiniot LK, Allen JL, Flick LM, Belkaid Y, Madan R, Haky JJ, Karp CL. Regulation of TLR4 signaling and the host interface with pathogens and danger: the role of RP105. J. Leukoc. Biol. 2007;82:265–271. doi: 10.1189/jlb.0107021. [DOI] [PubMed] [Google Scholar]
- Donnelly MA, Steiner TS. Two nonadjacent regions in enteroaggregative Escherichia coli flagellin are required for activation of toll-like receptor 5. J. Biol. Chem. 2002;277:40456–40461. doi: 10.1074/jbc.M206851200. [DOI] [PubMed] [Google Scholar]
- Dunne A, Ejdeback M, Ludidi PL, O'Neill LA, Gay NJ. Structural complementarity of Toll/interleukin-1 receptor domains in Toll-like receptors and the adaptors Mal and MyD88. J. Biol. Chem. 2003;278:41443–41451. doi: 10.1074/jbc.M301742200. [DOI] [PubMed] [Google Scholar]
- Erridge C. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J. Leukoc. Biol. 2010;87:989–999. doi: 10.1189/jlb.1209775. [DOI] [PubMed] [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ, Gangloff M. Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- Gay NJ, Gangloff M, O'Neill LA. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol. 2011 doi: 10.1016/j.it.2010.12.005. [DOI] [PubMed] [Google Scholar]
- George J, Motshwene PG, Wang H, Kubarenko AV, Rautanen A, Mills TC, Hill AV, Gay NJ, Weber AN. Two human MYD88 variants, S34Y and R98C, interfere with MyD88-IRAK4-myddosome assembly. J. Biol. Chem. 2011;286:1341–1353. doi: 10.1074/jbc.M110.159996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindaraj RG, Manavalan B, Lee G, Choi S. Molecular modeling-based evaluation of hTLR10 and identification of potential ligands in Toll-like receptor signaling. PLoS. One. 2010;5:e12713. doi: 10.1371/journal.pone.0012713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han BW, Herrin BR, Cooper MD, Wilson IA. Antigen recognition by variable lymphocyte receptors. Science. 2008;321:1834–1837. doi: 10.1126/science.1162484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SH, Kim JH, Martin M, Michalek SM, Nahm MH. Pneumococcal lipoteichoic acid (LTA) is not as potent as staphylococcal LTA in stimulating Toll-like receptor 2. Infect. Immun. 2003;71:5541–5548. doi: 10.1128/IAI.71.10.5541-5548.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, Eng JK, Akira S, Underhill DM, Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- He XL, Bazan JF, McDermott G, Park JB, Wang K, Tessier-Lavigne M, He Z, Garcia KC. Structure of the Nogo receptor ectodomain: a recognition module implicated in myelin inhibition. Neuron. 2003;38:177–185. doi: 10.1016/s0896-6273(03)00232-0. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K, Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Huizinga EG, Tsuji S, Romijn RA, Schiphorst ME, de Groot PG, Sixma JJ, Gros P. Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297:1176–1179. doi: 10.1126/science.107355. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu. Rev. Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, Lee H, Lee JO. Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Jung D, Alt FW. Unraveling V(D)J recombination; insights into gene regulation. Cell. 2004;116:299–311. doi: 10.1016/s0092-8674(04)00039-x. [DOI] [PubMed] [Google Scholar]
- Kajava AV. Structural diversity of leucine-rich repeat proteins. J. Mol. Biol. 1998;277:519–527. doi: 10.1006/jmbi.1998.1643. [DOI] [PubMed] [Google Scholar]
- Kang JY, Nan X, Jin MS, Youn SJ, Ryu YH, Mah S, Han SH, Lee H, Paik SG, Lee JO. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- Khan JA, Brint EK, O'Neill LA, Tong L. Crystal structure of the Toll/interleukin-1 receptor domain of human IL-1RAPL. J. Biol. Chem. 2004;279:31664–31670. doi: 10.1074/jbc.M403434200. [DOI] [PubMed] [Google Scholar]
- Kim HM, Park BS, Kim JI, Kim SE, Lee J, Oh SC, Enkhbayar P, Matsushima N, Lee H, Yoo OJ, Lee JO. Crystal structure of the TLR4-MD-2 complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem. J. 2009;420:1–16. doi: 10.1042/BJ20090272. [DOI] [PubMed] [Google Scholar]
- Latz E, Verma A, Visintin A, Gong M, Sirois CM, Klein DC, Monks BG, McKnight CJ, Lamphier MS, Duprex WP, Espevik T, Golenbock DT. Ligand-induced conformational changes allosterically activate Toll-like receptor 9. Nat. Immunol. 2007;8:772–779. doi: 10.1038/ni1479. [DOI] [PubMed] [Google Scholar]
- Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The TLR3 signaling complex forms by cooperative receptor dimerization. Proc. Natl. Acad. Sci. U. S. A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–890. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, Davies DR. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto F, Saitoh S, Fukui R, Kobayashi T, Tanimura N, Konno K, Kusumoto Y, kashi-Takamura S, Miyake K. Cathepsins are required for Toll-like receptor 9 responses. Biochem. Biophys. Res. Commun. 2008;367:693–699. doi: 10.1016/j.bbrc.2007.12.130. [DOI] [PubMed] [Google Scholar]
- Matsushima N, Tanaka T, Enkhbayar P, Mikami T, Taga M, Yamada K, Kuroki Y. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC. Genomics. 2007;8:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzinger P. Tolerance, danger, and the extended family. Annu. Rev. Immunol. 1994;12:991–1045. doi: 10.1146/annurev.iy.12.040194.005015. [DOI] [PubMed] [Google Scholar]
- Means TK, Golenbock DT, Fenton MJ. The biology of Toll-like receptors. Cytokine Growth Factor Rev. 2000;11:219–232. doi: 10.1016/s1359-6101(00)00006-x. [DOI] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Miao EA, Andersen-Nissen E, Warren SE, Aderem A. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 2007;29:275–288. doi: 10.1007/s00281-007-0078-z. [DOI] [PubMed] [Google Scholar]
- Miura Y, Shimazu R, Miyake K, Akashi S, Ogata H, Yamashita Y, Narisawa Y, Kimoto M. RP105 is associated with MD-1 and transmits an activation signal in human B cells. Blood. 1998;92:2815–2822. [PubMed] [Google Scholar]
- Miyake K, Shimazu R, Kondo J, Niki T, Akashi S, Ogata H, Yamashita Y, Miura Y, Kimoto M. Mouse MD-1, a molecule that is physically associated with RP105 and positively regulates its expression. J. Immunol. 1998;161:1348–1353. [PubMed] [Google Scholar]
- Miyake K, Yamashita Y, Hitoshi Y, Takatsu K, Kimoto M. Murine B cell proliferation and protection from apoptosis with an antibody against a 105-kD molecule: unresponsiveness of X-linked immunodeficient B cells. J. Exp. Med. 1994;180:1217–1224. doi: 10.1084/jem.180.4.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, Sandercock AM, Robinson CV, Latz E, Gay NJ. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009;284:25404–25411. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyman T, Stenmark P, Flodin S, Johansson I, Hammarstrom M, Nordlund P. The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J. Biol. Chem. 2008;283:11861–11865. doi: 10.1074/jbc.C800001200. [DOI] [PubMed] [Google Scholar]
- O'Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- Ohnishi H, Tochio H, Kato Z, Orii KE, Li A, Kimura T, Hiroaki H, Kondo N, Shirakawa M. Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc. Natl. Acad. Sci. U. S. A. 2009;106:10260–10265. doi: 10.1073/pnas.0812956106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto U, Fukase K, Miyake K, Satow Y. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science. 2007;316:1632–1634. doi: 10.1126/science.1139111. [DOI] [PubMed] [Google Scholar]
- Omueti KO, Mazur DJ, Thompson KS, Lyle EA, Tapping RI. The polymorphism P315L of human toll-like receptor 1 impairs innate immune sensing of microbial cell wall components. J. Immunol. 2007;178:6387–6394. doi: 10.4049/jimmunol.178.10.6387. [DOI] [PubMed] [Google Scholar]
- Palsson-McDermott EM, O'Neill LA. Building an immune system from nine domains. Biochem. Soc. Trans. 2007;35:1437–1444. doi: 10.1042/BST0351437. [DOI] [PubMed] [Google Scholar]
- Park B, Brinkmann MM, Spooner E, Lee CC, Kim YM, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- Park HH, Logette E, Raunser S, Cuenin S, Walz T, Tschopp J, Wu H. Death domain assembly mechanism revealed by crystal structure of the oligomeric PIDDosome core complex. Cell. 2007;128:533–546. doi: 10.1016/j.cell.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J. Immunol. 2009;182:7690–7697. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van HC, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Rietschel ET, Kirikae T, Schade FU, Mamat U, Schmidt G, Loppnow H, Ulmer AJ, Zahringer U, Seydel U, Di PF, Schreier M, Brade H. Bacterial endotoxin: molecular relationships of structure to activity and function. FASEB J. 1994;8:217–225. doi: 10.1096/fasebj.8.2.8119492. [DOI] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:9577–9582. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG, Spanopoulou E. Biochemistry of V(D)J recombination. Curr. Top. Microbiol. Immunol. 2005;290:49–85. doi: 10.1007/3-540-26363-2_4. [DOI] [PubMed] [Google Scholar]
- Scott FL, Stec B, Pop C, Dobaczewska MK, Lee JJ, Monosov E, Robinson H, Salvesen GS, Schwarzenbacher R, Riedl SJ. The Fas-FADD death domain complex structure unravels signalling by receptor clustering. Nature. 2009;457:1019–1022. doi: 10.1038/nature07606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, ndersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu. Rev. Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Tao X, Xu Y, Zheng Y, Beg AA, Tong L. An extensively associated dimer in the structure of the C713S mutant of the TIR domain of human TLR2. Biochem. Biophys. Res. Commun. 2002;299:216–221. doi: 10.1016/s0006-291x(02)02581-0. [DOI] [PubMed] [Google Scholar]
- Teghanemt A, Zhang D, Levis EN, Weiss JP, Gioannini TL. Molecular basis of reduced potency of underacylated endotoxins. J. Immunol. 2005;175:4669–4676. doi: 10.4049/jimmunol.175.7.4669. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Immune responses of TLR5(+) lamina propria dendritic cells in enterobacterial infection. J. Gastroenterol. 2009;44:803–811. doi: 10.1007/s00535-009-0094-y. [DOI] [PubMed] [Google Scholar]
- Uff S, Clemetson JM, Harrison T, Clemetson KJ, Emsley J. Crystal structure of the platelet glycoprotein Ib(alpha) N-terminal domain reveals an unmasking mechanism for receptor activation. J. Biol. Chem. 2002;277:35657–35663. doi: 10.1074/jbc.M205271200. [DOI] [PubMed] [Google Scholar]
- Velikovsky CA, Deng L, Tasumi S, Iyer LM, Kerzic MC, Aravind L, Pancer Z, Mariuzza RA. Structure of a lamprey variable lymphocyte receptor in complex with a protein antigen. Nat. Struct. Mol. Biol. 2009;16:725–730. doi: 10.1038/nsmb.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin A, Iliev DB, Monks BG, Halmen KA, Golenbock DT. MD-2. Immunobiology. 2006;211:437–447. doi: 10.1016/j.imbio.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu L, Davies DR, Segal DM. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J. Biol. Chem. 2010;285:36836–36841. doi: 10.1074/jbc.M110.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Tao X, Shen B, Horng T, Medzhitov R, Manley JL, Tong L. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- Yang D, Tewary P, de la RG, Wei F, Oppenheim JJ. The alarmin functions of high-mobility group proteins. Biochim. Biophys. Acta. 2010;1799:157–163. doi: 10.1016/j.bbagrm.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SI, Hong M, Han GW, Wilson IA. Crystal structure of soluble MD-1 and its interaction with lipid IVa. Proc. Natl. Acad. Sci. U. S. A. 2010;107:10990–10995. doi: 10.1073/pnas.1004153107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.