Abstract
Apolipoprotein B-editing enzyme, catalytic polypeptide-1 (APOBEC1) is a cytidine deaminase, initially identified by its activity in converting a specific cytidine (C) to uridine (U) in apolipoprotein B (apoB) mRNA transcripts in the small intestine. Editing results in translation of a truncated apoB isoform with distinct functions in lipid transport. To address the possibility that APOBEC1 edits additional mRNAs, we developed a transcriptome-wide comparative RNA-Seq screen. We identified and validated 32 previously undescribed mRNA targets of APOBEC1 editing, all of which are located in AU-rich segments of transcript 3′ untranslated regions (3′ UTRs). Further analysis established several characteristic sequence features of editing targets, which were predictive for the identification of additional APOBEC1 substrates. The transcriptomics approach to RNA editing presented here dramatically expands the list of APOBEC1 mRNA editing targets and reveals a novel cellular mechanism for the modification of transcript 3′ UTRs.
RNA editing refers to processes in which the base sequence of polynucleotide RNA is modified at specific sites. RNA editing events introduce molecular diversity to sequences “hard coded” in genomic DNA and contribute to numerous and varied biological functions. Adenosine-to-inosine conversion in tRNA anticodons impacts coding specificity (reviewed in ref. 1). Mitochondrial RNA editing occurs in many diverse species by different mechanisms 2. Editing of mRNA can alter protein coding sequences (reviewed in ref. 3) and modulate gene expression 4-7. In higher eukaryotes, two enzyme families mediate mRNA editing: the Adenosine Deaminases Acting on RNA (ADARs) convert adenosine to inosine, and the polynucleotide cytidine deaminases (AID/APOBECs) convert cytidine to uridine.
The apolipoprotein B (apoB) transcript was the first mRNA editing target identified in mammals 8,9. The apoB protein product exists as two isoforms, both important in lipid metabolism. The full-length isoform, apoB-100, is produced by the liver and forms the principle lipoprotein component of LDL particles. The shorter apoB-48 isoform is produced by the small intestine, in which it is essential for chylomicron lipoprotein particle formation and the absorption and transport of dietary lipid. Both isoforms of apoB originate from an identical primary transcript but are not regulated by differential splicing or post-translational processing. In the small intestine, cytidine 6666 of the apoB mRNA is deaminated to uridine, thereby converting a glutamine codon (CAA) to a stop codon (UAA)8,9. Upon translation, this results in production of the truncated apoB-48 protein. The site-specific mRNA modification is mediated by a multiprotein “editosome” complex, the catalytic component of which is APOBEC1 (ref. 10).
The first member of the APOBEC/AID family of enzymes to be identified, APOBEC1 is a zinc-dependent cytidine deaminase11 present only in mammals12. In vitro , purified APOBEC1 binds polynucleotide RNA 13, with a preference for AU-rich sequences 14. APOBEC1 associates with APOBEC1 Complementation Factor (ACF), a RNA binding component of the editosome necessary for apoB mRNA editing 15,16. ACF selectively binds an 11-nt mooring sequence several bases downstream of the edited cytidine in apoB mRNA 15. This sequence motif is required for the site-specific editing of the apoB transcript 17, and is necessary and sufficient to induce C-to-U conversion in AU-rich heterologous RNA in vitro 18. Apobec1−/− mice lack detectable apoB mRNA editing in small intestine and have no apoB-48 in serum 19,20. Though hepatic overexpression of APOBEC1 is oncogenic in mice and rabbits 21, at present apoB mRNA is the only known physiological editing target of APOBEC1 in healthy tissue.
The identification of novel RNA editing targets has proven challenging, in large part due to the technical difficulty of detecting single nucleotide alterations over entire transcriptomes. The development of ultra-high throughput sequencing technologies provides powerful tools for more comprehensive investigation of RNA editing. One recent study used target capture and ultra-high throughput sequencing to detect A-to-I editing in numerous computationally predicted RNA targets 22. Whole transcriptome sequencing (RNA-Seq) represents another promising option for broad characterization of RNA editing. Though RNA-Seq is frequently used for transcriptome mapping and quantification 23-25, it has also been successfully applied to the analysis of single nucleotide polymorphisms (SNPs) in expressed genes 26,27.
We reasoned that, given sufficient transcript coverage and read depth, the single nucleotide resolution of RNA-Seq could be used to identify candidate mRNA editing sites throughout a transcriptome. Applying a novel comparative RNA-Seq screening approach to murine small intestine enterocytes, we have identified and validated numerous, previously unknown APOBEC1 mRNA editing targets. Unlike the well-characterized site in the apoB coding sequence, these newly recognized editing sites are located in the 3′UTRs of diverse transcripts. These sites share several characteristic sequence features, including a downstream (3′) motif similar to the mooring sequence in apoB mRNA. Bioinformatics analysis based on these features was used to predict additional APOBEC1 editing targets, which were subsequently validated by standard sequencing techniques. Finally, many of the APOBEC1 editing sites identified here were found to be located within transcript regions conserved in mammalian evolution, implying functional importance. Our findings demonstrate the feasibility and utility of a novel transcriptomics approach to RNA editing studies and reveal numerous additional mRNA targets of APOBEC1 editing, thereby suggesting functions for this enzyme beyond its previously characterized role.
RESULTS
A comparative RNA-Seq screen for mRNA editing targets
To identify candidate mRNA editing sites, we developed a comparative RNA-Seq screen that distinguishes single nucleotide variations between two transcriptomes (Fig. 1). Jejunal epithelial cells were isolated from the small intestine of C57/BL6 wild-type and congenic Apobec1−/− mice (Supplementary Fig. 1). RNA-Seq libraries were prepared from poly-A+ mRNA and deep sequenced, generating 76,766,760 (wild-type) and 50,509,000 (Apobec1−/−) 36-nt reads. Reads were aligned to the mouse reference genome (mm9, NCBI 37.1), allowing for up to two mismatches per sequence. As accurate mapping and individual base content of the sequences is critical in identifying single nucleotide variants associated with editing, only those reads with sufficient quality scores that mapped to unique sites in the genome were used for analysis (42,770,803 and 28,877,750 reads for wild-type and Apobec1−/−, respectively, Supplementary Table 1). Though this stringent alignment policy likely reduced coverage of orthologous and repetitive transcript regions, it eliminated a potential source of false positive hits from the incorrect mapping of mismatch-containing reads. Read coverage of transcripts at single base resolution was extensive, particularly for genes expressed at moderate to high levels (Supplementary Fig. 2).
FIGURE 1.
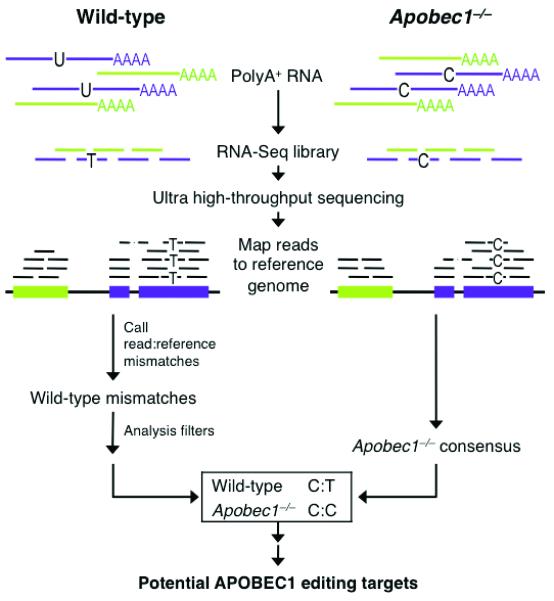
Comparative RNA-Seq Screen for APOBEC1 mRNA Editing Targets. Schematic workflow of RNA-Seq Screen.
The strategy for detecting potential RNA editing events involved identifying sample-specific single nucleotide mismatches in RNA-Seq reads relative to reference sequence (Fig. 1 and Supplementary Table 2). On account of APOBEC1’s cytidine deaminase activity, we used a modified SNP-calling algorithm to find those sites within RefSeq exons at which the reference genome contained a C and wild-type reads contained Ts (or reference G and RNA-Seq read As for (−)-strand transcripts, in genomic context). These sites were then compared to Apobec1−/− reads. If the corresponding position in Apobec1−/− reads also contained the mismatch, the site was discarded as a likely genomic polymorphism or non-APOBEC1 modification. However, if the corresponding location in Apobec1−/− reads matched the reference sequence, the site was considered for additional analysis (example in Supplementary Fig. 3). After filtering out those sites with insufficient read coverage (< 5 reads for wild-type, < 3 for Apobec1−/−) and/or mismatch probability scores (Supplementary Table 2), we were left with a set of 39 candidate APOBEC1 mRNA editing targets. When candidate targets were ranked by mismatch probability score (described in Methods), the top hit was the well-characterized site in apoB mRNA, which served as an internal positive control for the screening method. This result confirmed that our sequencing methodology and analysis pipeline can successfully detect single nucleotide editing events on a transcriptome scale.
Validation of candidate APOBEC1 mRNA editing sites
To validate the potential editing events identified by our iiRNA-Seq screen, we used standard dideoxynucleotide Sanger sequencing to examine the sites in genomic DNA and RNA (cDNA) isolated from intestinal epithelium. All validation samples were independently prepared from different mice than those used for RNA-Seq libraries. Sanger sequencing results for several sites are presented in Figure 2 (a–d). We observed clear evidence of C-to-U(T) RNA editing at 33 of the 39 candidate sites. The C/T chromatogram peaks in wild-type cDNA were of varied intensity, indicating differences in editing levels. At one site (chr3:73442586(−), Fig. 2d), we observed additional editing at a cytidine adjacent to the location identified by the screen. To further validate APOBEC1-specific editing, we selected several sites for subcloning and additional Sanger sequencing. C/T mismatches at candidate editing sites were observed only in subclones derived from wild-type cDNA; none were present in wild-type genomic DNA, Apobec1−/− genomic DNA or Apobec1−/− cDNA (Supplementary Fig. 4, a–e). Additionally, low level “hyperediting” of C residues in close proximity to the primary editing site was observed in a minority of subclones for several targets, including apoB (Supplementary Fig. 4f). This phenomenon has been previously described for apoB mRNA and is of unknown functional significance 28-30.
FIGURE 2.
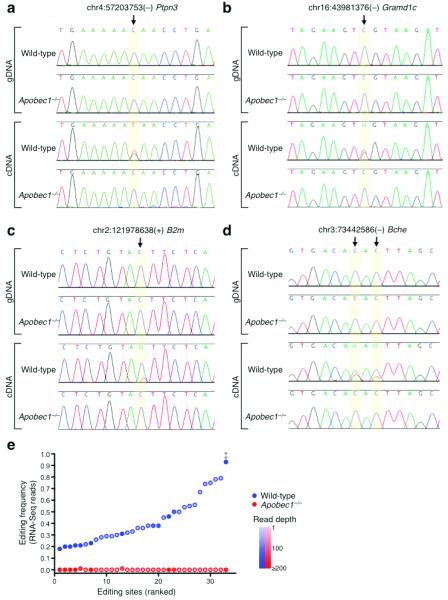
Validation of APOBEC1 mRNA Editing Targets. (a, b, c and d) Representative examples of conventional Sanger sequencing chromatograms for wild-type and Apobec1−/− genomic DNA and cDNA at editing sites. (a) chr4:57203753(−) in the Ptpn3 transcript, (b) chr16:43981376(−) in the Gramd1c transcript, (c) chr2:121978638(+) in the B2m transcript, and (d) chr3:73442586(−) in the Bche transcript. (e) Editing frequency of ranked APOBEC1 sites. ‡ indicates apoB editing site.
A list of validated APOBEC1 mRNA editing targets appears in Supplementary Table 3. Unlike the edited coding sequence of apoB mRNA, all of the newly identified APOBEC1 sites are located in 3′ UTRs. We used our RNA-Seq read data to estimate the editing level of each site ([# of T reads] / [# of C reads + # of T reads]). ApoB mRNA displayed the most pronounced editing (0.92), while editing frequency of 3′ UTR sites ranged from 0.18 to 0.79 (Fig. 2e). Editing frequencies calculated from RNA-Seq reads were very similar to those determined by cDNA amplification, subcloning, and Sanger sequencing (examples in Supplementary Fig. 4e).
APOBEC1 mRNA editing targets share characteristic features
Target recognition by RNA editing enzymes is typically determined by the sequence and/or structural context of the edited base 31,32. Although features contributing to apoB mRNA editing have been previously characterized, it was unclear whether similar attributes would apply to APOBEC1 editing of 3′ UTR targets. To determine if the identified sites share common features that might designate them for APOBEC1 editing, we examined the sequences around the target cytidines.
APOBEC1 has RNA binding activity with a preference for sequences rich in A and U 13,14. The sequence region (101-nt) surrounding the apoB mRNA editing site is also particularly AU-rich (0.70 AU content). We determined the AU content of the APOBEC1 edit site 3′ UTRs (0.63) and found them to be significantly (P < 0.0001) more AU-rich than comparable sets of 3′ UTRs chosen at random (Fig. 3a). We also found that, within these AU-rich 3′ UTRs, the local regions (101-nt centered on site) containing the edit sites were further enriched for A and U bases (0.69, P = 0.0006, Fig. 3b–c). These results are consistent with a model in which APOBEC1 targets require a high AU sequence context for efficient editing.
FIGURE 3.
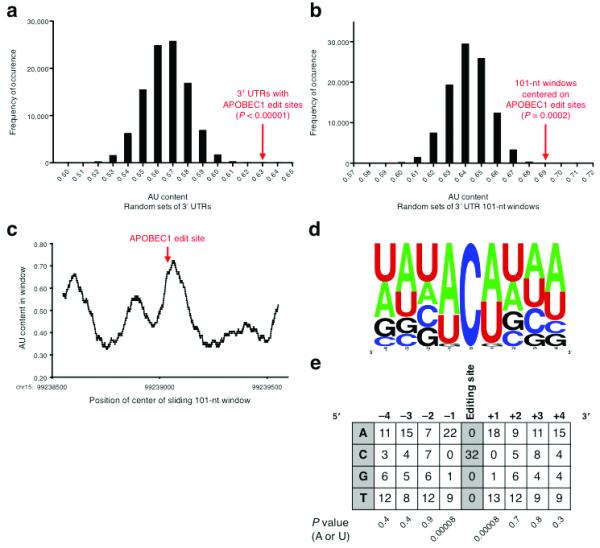
Sequence Features of APOBEC1 Editing Sites. (a) AU content of random sets of 3′ UTRs. The value for the APOBEC1 editing site-containing 3′ UTRs is well to the right of the distribution. (b) AU content of random 101-nt windows within APOBEC1 editing site-containing 3′ UTRs. The value for the windows centered on the editing sites is well to the right of the distribution. (c) AU content in a sliding 101-nt window in the Tmbim6 3′ UTR. The AU content peaks near the editing site, chr15:99239051(+). (d) Frequency plot of bases flanking the APOBEC1 editing sites, aligned on the target cytidine. (e) Base counts of nucleotides flanking the APOBEC1 editing sites. The nucleotides immediately adjacent to the target cytidine tend overwhelmingly to be A or U. P values were computed using the binomial test. For a given column, the P value is the probability that the skew towards A or U is a random occurrence.
Aside from regional sequence content, other cytidine deaminases in the AID/APOBEC family exhibit strong preferences for particular bases immediately neighboring their editing targets 33. Such preferences have not been described for APOBEC1, likely due to its perceived specificity for a single substrate. By aligning the 3′ UTR edit sites, we found that almost all of the edited cytidines were immediately flanked by A or U bases at the −1 and +1 position (P = 8 × 10−5, Fig. 3d–e). There were no apparent nucleotide preferences at the −4,−3,−2 and +2,+3,+4 positions relative to the editing site.
DNA and RNA binding proteins often recognize and bind to sequence motifs in their molecular targets. To ascertain whether the APOBEC1 editing targets identified here share a common sequence element potentially important for editosome recognition, we used the Multiple Em for Motif Elicitation (MEME) algorithm 34 to analyze the sequence regions (101-nt centered on target C) surrounding the editing sites. MEME analysis revealed a significant (log likelihood ratio = 157, E value = 8.8 ×10−1, compared to 65 and 3 × 102 for shuffled sequence control) 10-nt motif in regions adjacent to most (21/31) editing sites (Fig. 4a). Next, we used the motif consensus sequence, WRAUYANUAU, to manually align the edit site-containing sequences (Fig. 4b). We found close or exact consensus motif matches downstream (3′) of almost every editing site, with most (24/32) appearing 4–6 nt from the target cytidine. Of note, the consensus motif also matches the first 10-nt of the apoB mooring sequence, which is 5-nt downstream of its editing site 17,35. These results suggest that most of the 3′ UTR sites are edited by a similar mechanism as the apoB transcript.
FIGURE 4.
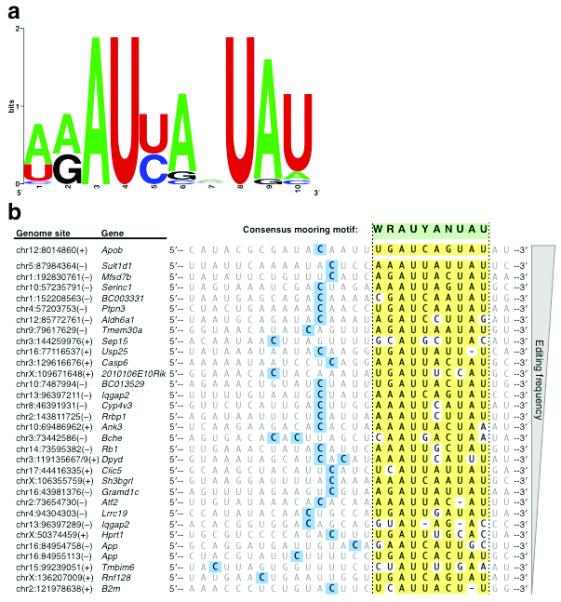
APOBEC1 mRNA Editing Targets Share a Characteristic Sequence Motif. (a) Frequency plot of sequence motif identified by MEME analysis of regions flanking APOBEC1 editing site. Log likelihood ratio (157) and E value (8.8 ×10−1) are significant as compared to the “best” motif of a shuffled sequence control (Log likelihood ratio 65, E value 3 × 102). (b) Alignment of APOBEC1 target sequences by consensus sequence motif. Edited cytidines are shaded in blue. Yellow shading indicates a match to the consensus sequence motif, represented in green.
Predicted APOBEC1 targets are not edited in coding sequences
With the set of newly identified target sites and their characteristic sequence features described above, we now have a refined list of criteria for sequences edited by APOBEC1. Based on these findings, we derived an APOBEC1 editing “sequence pattern,” consisting of a cytidine flanked on both sides by either A or U and followed by an appropriately spaced mooring motif (WCWN2–4WRAUYANUAU, Fig. 5a). In order to evaluate the distribution of potential APOBEC1 editing targets throughout the transcriptome, we searched for this sequence pattern in all RefSeq exons. As enumerated in Figure 5b, we found nearly 400 examples of this pattern in mouse mRNAs (Supplementary Table 4), 181 of which occurred in transcripts expressed in small intestine enterocytes (RNA-Seq analysis, RPKM ≥ 1.0). Bypassing the comparative editing screen workflow, we directly examined the wild-type RNA-Seq read sequences at these sites for evidence of RNA editing (Fig. 5c). Of the 74 patterns located in 3′ UTRs with read coverage (≥ 3 wild-type reads), we detected C/T mismatches indicative of editing at 32 sites. Of the 34 patterns present in coding exons covered by RNA-Seq reads, only the apoB site displayed evidence of editing. A subset of these sites (7 in coding sequences, 14 in 3′ UTR sequences) was additionally examined by Sanger sequencing, which confirmed C-to-U editing in 9 of the 3′ UTR sites but none of the coding sequence sites (Fig. 5d–f). Though many of these sites were not detected in the RNA-Seq screen due to insufficient read coverage in the Apobec1−/− library and/or relatively low editing frequencies, the APOBEC1 sequence pattern derived from the initially identified targets was clearly predictive for additional APOBEC1 3′ UTR editing targets.
FIGURE 5.
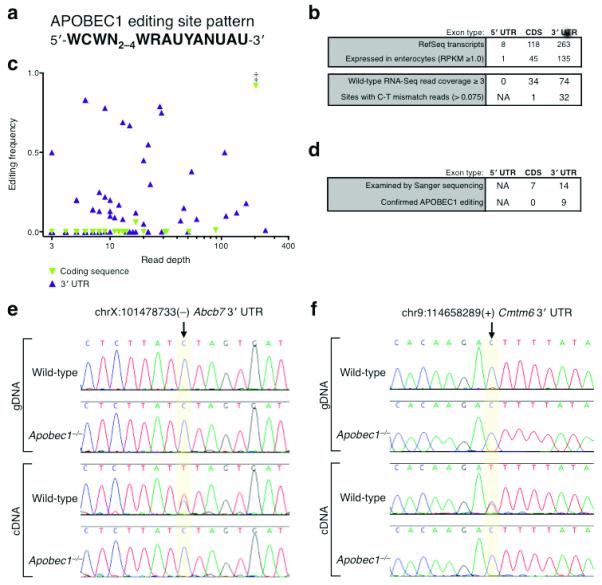
Sequence Pattern Prediction of APOBEC1 mRNA Editing Targets. (a) APOBEC1 editing site pattern used to search for additional targets in RefSeq transcripts. (b) Occurrences of APOBEC1 editing site pattern in RefSeq transcripts by type, listed by intestine epithelium expression level and wild-type RNA-Seq read coverage. (c) Editing frequency at predicted APOBEC1 target sites as evaluated by wild-type read content. ‡ indicates apoB editing site. (d) Sanger sequencing validation of predicted APOBEC1 editing sites in RefSeq transcripts by type. (e and f) Representative examples of conventional Sanger sequencing chromatograms for wild-type and Apobec1−/− genomic DNA and cDNA at predicted editing sites. (e) chrX:101478733(−) in the Abcb7 transcript, (f) chr9:114658289(+) in the Cmtm6 transcript.
These results suggest that while the APOBEC1 sequence pattern supports editing at numerous sites in transcript 3′ UTRs, it is not targeted when present in coding sequences. The pronounced exception of apoB raises questions about the mechanism of APOBEC1 sequence recognition and localization as well as the role for editing in 3′ UTRs.
APOBEC1 editing occurs in conserved mRNA sequence regions
Compared to other non-coding sequences, functional elements within 3′ UTRs are more likely to be conserved through evolution 36. When inspecting the APOBEC1 editing sites, we observed that many seemed to occur within regions of considerable phylogenetic conservation. Two examples are presented in Supplementary Figure 5a and 5b. In order to systematically assess the conservation of sequence regions containing APOBEC1 editing sites, we compared the conservation scores (phastCons scores for placental mammals 37) of 101-nt windows centered on the initially identified editing sites to random 101-nt windows within the same 3′ UTRs. As a set, the regions containing APOBEC1 editing sites are significantly (P = 0.01) more conserved (Supplementary Fig. 5c), suggesting that these sequences may be of functional importance.
Taken together, these results indicate that APOBEC1 site-specifically edits many mRNA transcripts other than apoB in small intestine enterocytes. They also provide the first reported examples of C-to-U mRNA editing in 3′ UTRs, a molecular mechanism that suggests additional roles for APOBEC1 beyond its function in apolipoprotein regulation.
DISCUSSION
Recent advances in ultra-high throughput DNA sequencing technologies have redefined the scale at which we can study transcriptomes. Early RNA-Seq experiments in yeast 23, mouse 24 and human 25 cells revealed numerous novel genes, splicing events and transcript untranslated regions, demonstrating a level of transcriptome complexity not previously appreciated. Despite the massive scope of whole transcriptome datasets, RNA-Seq also provides mRNA sequence information at single nucleotide resolution. Though such information can be used to examine “expressed SNPs” 26,27, it also provides a powerful tool with which to study RNA editing in an unbiased manner as described here.
As is the case for many RNA editing enzymes, an edited target (apoB mRNA) of APOBEC1 was identified prior to discovery of the enzyme responsible. Following the demonstration of C-to-U editing in apoB mRNA 8,9, numerous biochemical and genetic strategies were employed to eventually trace back the activity to an unidentified gene subsequently characterized as a cytidine deaminase, APOBEC1 10,11,38,39. Despite extensive study of APOBEC1 sequence preferences, as well as observation of NF1 mRNA editing in tumor cells 40,41, physiologic editing of mRNAs other than apoB has not been described. As studies examining limited sets of candidate transcripts did not reveal C-to-U alterations, APOBEC1 editing has been thought to be primarily specific for apoB mRNA.
In contrast to working from single substrate to enzyme, the comparative RNA-Seq screen described here has allowed for an unbiased and dramatically more comprehensive search for editing targets. In addition to detecting the well characterized editing site in apoB mRNA, we identified and validated 32 previously unknown APOBEC1 editing sites in the 3′ UTRs of diverse mRNA transcripts. This set of targets revealed several characteristic features of APOBEC1 editing sites, including a strong preference for A/U bases immediately neighboring the edited cytidine and a 3′ mooring motif encompassing that previously described for apoB. Using these sequence features as a guide, we found that while similar patterns are present in coding and untranslated sequences throughout the transcriptome, APOBEC1 editing is primarily constrained to 3′ UTRs. Though they do not result in protein coding changes, many of the editing sites identified here are located within evolutionarily conserved sequences, arguing for functional relevance.
The localization of all newly identified editing sites to transcript 3′ UTRs raises questions about the mechanism of apoB coding sequence editing by APOBEC1. Despite the presence of motifs consistent with APOBEC1 editing within coding and untranslated sequences throughout the transcriptome, our RNA-Seq data suggest that APOBEC1 only acts on those targets located in 3′ UTRs. Thus, with regard to APOBEC1 targeting, apoB coding sequence editing appears to be the exception rather than the rule. It is interesting to note that upon apoB mRNA editing by APOBEC1 the downstream coding sequence becomes a 3′ UTR. This may represent an important link regarding the relationship of this well-known mRNA target to the 3′ UTR editing sites.
The distribution of APOBEC1 sites in coding sequence (apoB) as well as transcript 3′ UTRs is reminiscent of editing by ADARs, another family of RNA deaminases. ADARs deaminate adenosine to inosine in numerous RNAs at diverse tissue sites 3. Much like C-to-U deamination in apoB, some initial examples of A-to-I modification in mRNA were observed in tissue-specific transcripts (i.e. GluR, in the brain), which have protein coding sequences modified as a consequence of editing 42. Similar coding changes have been observed in several other neuronal transcripts (reviewed in ref. 32). Additional A-to-I editing events have been found to impact protein sequence by initiating alternative RNA splicing events (reviewed in ref. 3). However, recent bioinformatic 43 and ultra high-throughput sequencing analyses 22 have demonstrated that most A-to-I RNA editing occurs in non-coding RNA sequences, especially transcript 3′ UTRs. As is the case for APOBEC1 (Fig. 2e), ADAR editing varies in efficiency between target transcripts 22. Though the functional consequences for most of these editing events remain largely unknown, a small number of targets have been examined in some detail. In these cases, A-to-I editing in 3′ UTR sequences has been shown to induce nuclear retention of transcripts 7, target mRNA cleavage 44, and potentially modify miRNA target sites to modulate gene expression 45,46. Though some remain controversial, these findings provide illustrative examples of how nucleotide changes in non-coding sequence can impact genetic output.
Might APOBEC1 editing of 3′ UTRs have similar functional consequences? Based on the sequence context of many sites described here, we can speculate on several possible functional outcomes for APOBEC1 editing: (1) 3′ UTRs contain sequence and structural motifs that are recognized by RNA binding proteins. APOBEC1 editing events in 4 transcript 3′ UTRs are predicted to generate new AU-rich elements (AREs, AUUUA pentamers), which could contribute to transcript instability via their interaction with various RNA-BPs (reviewed in 47). (2) 3′ UTRs represent the principle targets of transcript regulation by miRNAs. More than 35% of APOBEC1 editing sites are located within sequences that match the seed targets of known miRNAs (Supplementary Table 5). Cytidine deamination at these sites would modify target sequences and potentially abolish miRNA binding. Conversely, APOBEC1 editing could introduce new miRNA seed target sequences, or shift existing targets to sequences that recruit different miRNAs. It should be noted that miRNA targeting is enhanced within regions rich in A and U nucleotides 48, a prominent feature of APOBEC1 editing sites (Fig. 3a–c). (3) Finally, APOBEC1-mediated alterations in 3′ UTRs could impact additional post-transcriptional processes including transcript polyadenylation, subcellular localization, and translational efficiency.
Without flexible mouse enterocyte models that can manipulated In vitro, direct experimental evidence for the functional and physiological relevance of these editing events would require detection of altered translational outcomes in APOBEC1-expressing enterocytes in vivo. Furthermore, as Apobec1−/− epithelial cells accumulate triacylglycerol lipids due to apoB-related deficiencies in chylomicron formation 49, direct regulatory effects due to the absence of 3′ UTR editing of various target transcripts are difficult to evaluate, as they may be obscured by the indirect cellular effects of the absence of apoB editing on lipid metabolism. For these technical reasons, experimental evidence for the biological effects of APOBEC1 editing has been elusive. However, the localization of many APOBEC1 edit sites within regions conserved in mammalian evolution (Supplementary Fig. 5) implies functional relevance.
Though often considered an intestine-specific protein, APOBEC1 is expressed at diverse tissue sites in numerous mammals 50,51 (Rosenberg and Papavasiliou, unpublished data). As many of these tissues do not express apoB, the function of APOBEC1 and its editing activity has remained unclear. The identification of multiple, previously unknown physiologic editing substrates for APOBEC1 raises the possibility of functionally significant mRNA editing in these tissues. Thus, our results suggest additional functions for APOBEC1 beyond its characterized role in lipid transport, both in small intestine enterocytes as well as other cell types.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Charles Rice, Dr. Harold Smith, Dr. Donny Licatalosi and Eric Fritz for critical reading of this manuscript. We wish to extend additional gratitude to Dr. Smith and Dr. Robert Darnell for helpful discussion and suggestions. We thank Geulah Livshits for immunofluorescence assistance. This work was supported by the Cancer Research Institute (training grants to BRR and CEH), by NIH MSTP grant GM07739 (BRR and CEH) and by a pilot grant to FNP through the Rockefeller CTSA program (NIH/NCRR grant # UL1RR024143).
Appendix
METHODS
Mice
All C57/BL6 wild-type and congenic Apobec1−/− mice were used at 6–8 weeks of age. Apobec1−/− mice 19 were generously provided by N. Davidson (Washington University School of Medicine, St Louis, MO).
Isolation of small intestine enterocytes
Mouse small intestines were removed and washed in Hanks Buffered Saline Solution (HBSS) with Ca2+ and Mg2+. Jejunum segments were dissected, everted and cut into pieces of approximately 5 cm. Enterocytes were isolated with a protocol adapted from 52. Briefly, jejunum segments were washed 5 times in HBSS (with Ca2+ and Mg2+) containing 1% (v/v) fetal bovine serum (FBS) and then washed once in HBSS (without Ca2+ and Mg2+) containing 2% (w/v) glucose and 2% bovine serum albumin (BSA). Jejunum segements were transferred to enterocyte isolation buffer (HBSS without Ca2+ and Mg2+, 1.5 mM EDTA, 0.5 mM DTT) and incubated at 37°C with agitation for 30 minutes. Enterocyte suspensions were collected, washed and resuspended in TRI Reagent (Ambion) for RNA preparation or processed for immunolabeling and flow cytometry.
Preparation of RNA-Seq libraries
Enterocyte total RNA was prepared by TRI Reagent (Ambion) extraction and treated with TURBO DNase (Ambion). RNA-Seq libraries were prepared with a protocol adapted from 24 as follows. Isolation of poly-A+ mRNA from total RNA was performed on polyT resin (MicroPoly(A)Purist Kit, Ambion). Poly-A+ mRNA was fragmented in fragmentation buffer (40 mM Tris acetate pH 8.2, 100 mM potassium acetate, 30 mM magnesium acetate) at 94°C for exactly 4 minutes 30 seconds. Fragmented RNA was reverse transcribed (Superscript III, Invitrogen) and cDNA was prepared with the Double-stranded cDNA Synthesis Kit (Invitrogen). Sequencing adapter oligonucleotides (Illumina) were added with T4 DNA Ligase (Quick Ligation Kit, New England Biolabs). Double-stranded cDNA libraries were size-separated by agarose gel electrophoresis and fragments ranging from approximately 175–225 nt were excised and PCR amplified with linker specific primers (Illumina). Integrity and quality of RNA and cDNA were monitored throughout on the Agilent Bioanalyzer 2100.
Ultra high-throughput sequencing was performed on the Illumina Genome Analyzer II (GAII) by standard sequencing-by-synthesis reaction for 36-nt reads.
RNA-Seq read mapping and identification of read:reference mismatches
In order to reliably identify potential APOBEC1 editing events, we incorporated an RNA-Seq read mapping strategy focused on the accurate identification of single nucleotide read:reference mismatches. Reads were mapped to the C57/BL6 reference mouse genome (NCBI37/mm9) using the Tuxedo Tools software packages, Bowtie (short read alignment) 53 and TopHat (spliced read mapping for RNA-Seq) 54. To ensure subsequent identification of mismatches resulted in minimal false detection of editing sites, several conditions were applied to read mapping: (1) Up to 2 mismatches were permitted in the seed region (first 28-nt) of each read. (2) Mismatches required a minimum base quality score. (3) Only reads mapping to unique sites in the genome were analyzed; all alignments to multiple sites or ambiguous positions were suppressed.
Once suitable RNA-Seq alignments were generated, single nucleotide read:reference mismatches were identified with SAMTools software 55. The standard SAMTools workflow was used to generate a “pileup” output that contained all mapped RNA-Seq reads and their quality scores on a reference base-by-base scale and a corresponding consensus base call at each position. This information was used to identify single nucleotide variants in the wild-type dataset with a quality-conscious algorithm 56. Each variant is assigned a mismatch probability score (also referred to as SNP quality score), defined as the Phred-scaled probability that the read consensus is identical to the reference.
The index of read:reference variant sites was filtered on several criteria to restrict analysis to those mismatches related to APOBEC1 editing. First, as many mismatch sites are the result of off-target mapping to intergenic and intronic sites, only those sites that mapped to RefSeq exons were retained. Next, to identify editing consistent with APOBEC1 cytidine deaminase activity, only sites at which the reference base was a C and the read consensus call included T were selected for additional consideration. Mismatch sites annotated as SNPs (dbSNP build 128, ftp.ncbi.nih.gov/snp) were also discarded. Finally, the remaining sites were compared to read consensus base calls in the Apobec1−/− dataset. Only those sites at which wild-type read consensus contained C:T mismatches and Apobec1−/− read consensus contained a high-confidence C:C match were deemed potential editing sites. This list was further reduced by removing those sites with insufficient read depth (<5 reads for wild-type, < 3 reads for Apobec1−/−) and/or insufficient confidence scores (Phred-scaled mismatch probability < 45 for wild-type, Phred-scaled consensus probability < 30 for Apobec1−/−; detailed in 56).
Unless otherwise described, all filters and database queries were performed with standard shell scripts or Galaxy tools (http://g2.bx.psu.edu)57.
APOBEC1 editing site validation
Genomic DNA and RNA were prepared from wild-type and Apobec1−/− small intestine enterocytes by standard methods. cDNA was prepared from total RNA by Superscript III reverse transcriptase (Invitrogen) and random hexamer priming and/or 3′RACE oligo(dT) priming (Invitrogen). Sequences containing potential APOBEC1 editing sites were PCR amplified using TurboPfu high-fidelity polymerase (Stratagene). Primer sequences appear in Supplementary Table 6. Primer extension sequencing was performed by GENEWIZ, Inc. (South Plainfield, NJ) using Applied Biosystems BigDye version 3.1 and 3730xl DNA Analyzer.
Analysis of APOBEC1 editing site sequence features
For the evaluation of the AU content of the 3′ UTRs that contain the APOBEC1 editing sites, the histogram (Fig. 3a) was generated by computing the AU content for each of 100,000 random sets of suitably chosen 3′ UTRs.
For the evaluation of the AU content of the 101-nt windows centered on the APOBEC1 editing sites, the histogram (Fig. 3b) was generated by computing the AU content for each of 100,000 random sets of suitably chosen 101-nt windows from within the same 3′ UTRs.
Base identity at positions immediately flanking the edited cytidine was assessed by aligning sequences on the editing site. P values for the occurrence of A or U nucleotides were computed by the binomial test. The background frequencies for the nucleotides were computed from the 101-nt windows centered on the edit sites.
Sequence motif analysis was performed with the MEME algorithm (http://meme.nbcr.net/meme4_3_0/cgi-bin/meme.cgi34 on a set 101-nt sequences centered on the edit sites. Statistical significance was approximated by comparing the log likelihood ratio and E-value of the best reported hit to those of the top hit returned by an identical analysis of randomly-shuffled input sequence.
All logo and frequency plots were generated with WebLogo (http://weblogo.berkeley.edu/)58.
Additional information and detailed explanations for all calculations appears in the Supplementary Methods.
APOBEC1 sequence pattern analysis
SequenceSearcher software (http://athena.bioc.uvic.ca/tools/SequenceSearcher) 59 was used to perform regular expression searches for the APOBEC1 consensus sequence pattern within a compiled collection of all RefSeq exon sequences. The list of predicted sites was filtered on gene expression level (RPKM ≥ 1.0, data not shown) in wild-type small intestine enterocytes. When sufficient coverage was available (≥ 3 reads), wild-type RNA-Seq reads mapped to each site were examined for evidence of editing (C:T mismatches above a background frequency of 0.075).
Assessment of phylogenetic conservation
PhastCons scores 37 for placental mammals were used to evaluate evolutionary conservation in 101-nt windows centered on the editing sites. For a set of windows in 3′ UTR genomic intervals, mean phastCons scores were computed by dividing the sum of scores for all nucleotides in all windows by the total number of nucleotides. The mean phastCons score for APOBEC1 edit site-containing windows was compared to each of 10,000 sets of random windows derived from the same 3′ UTRs.
Additional information and detailed explanations for calculations appears in the Supplementary Methods.
Footnotes
ACCESSION CODES: NCBI Gene Expression Omnibus (GEO); RNA-Seq read and alignment data: GSE24958
References
- 1.Grosjean H, et al. Enzymatic conversion of adenosine to inosine and to N1-methylinosine in transfer RNAs: a review. Biochimie. 1996;78:488–501. doi: 10.1016/0300-9084(96)84755-9. [DOI] [PubMed] [Google Scholar]
- 2.Gray MW. Diversity and evolution of mitochondrial RNA editing systems. IUBMB Life. 2003;55:227–33. doi: 10.1080/1521654031000119425. [DOI] [PubMed] [Google Scholar]
- 3.Pullirsch D, Jantsch MF. Proteome diversification by adenosine to inosine RNA-editing. RNA biology. 2010;7 doi: 10.4161/rna.7.2.11286. [DOI] [PubMed] [Google Scholar]
- 4.Kumar M, Carmichael GG. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci USA. 1997;94:3542–7. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–63. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–75. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 7.Chen L-L, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen SH, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science. 1987;238:363–6. doi: 10.1126/science.3659919. [DOI] [PubMed] [Google Scholar]
- 9.Powell LM, et al. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell. 1987;50:831–40. doi: 10.1016/0092-8674(87)90510-1. [DOI] [PubMed] [Google Scholar]
- 10.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 11.Navaratnam N, et al. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–12. [PubMed] [Google Scholar]
- 12.Conticello SG, Thomas CJF, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–77. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 13.Navaratnam N, et al. Evolutionary origins of apoB mRNA editing: catalysis by a cytidine deaminase that has acquired a novel RNA-binding motif at its active site. Cell. 1995;81:187–95. doi: 10.1016/0092-8674(95)90328-3. [DOI] [PubMed] [Google Scholar]
- 14.Anant S, MacGinnitie AJ, Davidson NO. apobec-1, the catalytic subunit of the mammalian apolipoprotein B mRNA editing enzyme, is a novel RNA-binding protein. J Biol Chem. 1995;270:14762–7. [PubMed] [Google Scholar]
- 15.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000;20:1846–54. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lellek H, et al. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem. 2000;275:19848–56. doi: 10.1074/jbc.M001786200. [DOI] [PubMed] [Google Scholar]
- 17.Shah RR, et al. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem. 1991;266:16301–4. [PubMed] [Google Scholar]
- 18.Backus JW, Smith HC. Apolipoprotein B mRNA sequences 3′ of the editing site are necessary and sufficient for editing and editosome assembly. Nucleic Acids Res. 1991;19:6781–6. doi: 10.1093/nar/19.24.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirano K, et al. Targeted disruption of the mouse apobec-1 gene abolishes apolipoprotein B mRNA editing and eliminates apolipoprotein B48. J Biol Chem. 1996;271:9887–90. doi: 10.1074/jbc.271.17.9887. [DOI] [PubMed] [Google Scholar]
- 20.Morrison JR, et al. Apolipoprotein B RNA editing enzyme-deficient mice are viable despite alterations in lipoprotein metabolism. Proc Natl Acad Sci USA. 1996;93:7154–9. doi: 10.1073/pnas.93.14.7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamanaka S, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci USA. 1995;92:8483–7. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–3. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 23.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–9. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–8. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 25.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–60. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 26.Heap GA, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum Mol Genet. 2010;19:122–34. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamanaka S, Poksay KS, Driscoll DM, Innerarity TL. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J Biol Chem. 1996;271:11506–10. doi: 10.1074/jbc.271.19.11506. [DOI] [PubMed] [Google Scholar]
- 29.Sowden M, Hamm JK, Smith HC. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J Biol Chem. 1996;271:3011–7. doi: 10.1074/jbc.271.6.3011. [DOI] [PubMed] [Google Scholar]
- 30.Sowden MP, Eagleton MJ, Smith HC. Apolipoprotein B RNA sequence 3′ of the mooring sequence and cellular sources of auxiliary factors determine the location and extent of promiscuous editing. Nucleic Acids Res. 1998;26:1644–52. doi: 10.1093/nar/26.7.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davidson NO. The challenge of target sequence specificity in C-->U RNA editing. J Clin Invest. 2002;109:291–4. doi: 10.1172/JCI14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–46. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beale RCL, et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol. 2004;337:585–96. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 34.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 35.Backus JW, Smith HC. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res. 1992;20:6007–14. doi: 10.1093/nar/20.22.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duret L, Dorkeld F, Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–22. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadjiagapiou C, Giannoni F, Funahashi T, Skarosi SF, Davidson NO. Molecular cloning of a human small intestinal apolipoprotein B mRNA editing protein. Nucleic Acids Res. 1994;22:1874–9. doi: 10.1093/nar/22.10.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau PP, Zhu HJ, Baldini A, Charnsangavej C, Chan L. Dimeric structure of a human apolipoprotein B mRNA editing protein and cloning and chromosomal localization of its gene. Proc Natl Acad Sci USA. 1994;91:8522–6. doi: 10.1073/pnas.91.18.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukhopadhyay D, et al. C-->U editing of neurofibromatosis 1 mRNA occurs in tumors that express both the type II transcript and apobec-1, the catalytic subunit of the apolipoprotein B mRNA-editing enzyme. Am J Hum Genet. 2002;70:38–50. doi: 10.1086/337952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res. 1996;24:478–85. doi: 10.1093/nar/24.3.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–13. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 43.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–5. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 44.Osenberg S, Dominissini D, Rechavi G, Eisenberg E. Widespread cleavage of A-to-I hyperediting substrates. RNA. 2009;15:1632–9. doi: 10.1261/rna.1581809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Borchert GM, et al. Adenosine deamination in human transcripts generates novel microRNA binding sites. Hum Mol Genet. 2009;18:4801–7. doi: 10.1093/hmg/ddp443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang H, Landweber LF. Hypothesis: RNA editing of microRNA target sites in humans? RNA. 2007;13:463–7. doi: 10.1261/rna.296407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–50. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kendrick JS, Chan L, Higgins JA. Superior role of apolipoprotein B48 over apolipoprotein B100 in chylomicron assembly and fat absorption: an investigation of apobec-1 knock-out and wild-type mice. Biochem J. 2001;356:821–7. doi: 10.1042/0264-6021:3560821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirano K, Min J, Funahashi T, Davidson NO. Cloning and characterization of the rat apobec-1 gene: a comparative analysis of gene structure and promoter usage in rat and mouse. J Lipid Res. 1997;38:1103–19. [PubMed] [Google Scholar]
- 51.Nakamuta M, et al. Alternative mRNA splicing and differential promoter utilization determine tissue-specific expression of the apolipoprotein B mRNA-editing protein (Apobec1) gene in mice. Structure and evolution of Apobec1 and related nucleoside/nucleotide deaminases. J Biol Chem. 1995;270:13042–56. doi: 10.1074/jbc.270.22.13042. [DOI] [PubMed] [Google Scholar]
- 52.Xie Y, Nassir F, Luo J, Buhman K, Davidson NO. Intestinal lipoprotein assembly in apobec-1−/− mice reveals subtle alterations in triglyceride secretion coupled with a shift to larger lipoproteins. Am J Physiol Gastrointest Liver Physiol. 2003;285:G735–46. doi: 10.1152/ajpgi.00202.2003. [DOI] [PubMed] [Google Scholar]
- 53.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor J, Schenck I, Blankenberg D, Nekrutenko A. Using galaxy to perform large-scale interactive data analyses. Curr Protoc Bioinformatics. 2007 doi: 10.1002/0471250953.bi1005s19. Chapter 10, Unit 10.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marass F, Upton C. Sequence Searcher: A Java tool to perform regular expression and fuzzy searches of multiple DNA and protein sequences. BMC Res Notes. 2009;2:14. doi: 10.1186/1756-0500-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


