Human infertility is a major worldwide health problem for individuals and their partners (1). Because genetic defects are thought to underlie many of the unexplained pathologies in infertility cases, animal models are expected to provide valuable clues to the underlying defects. More than 200 infertile or subfertile genetic mouse models have already been generated, defining key DNA repair and signaling pathways and other processes involved in mammalian reproduction (2). Many of these models were generated by targeted disruption of known genes or were fortuitously identified as spontaneously arising mutants. Because of the complexity of reproduction, however, this number almost certainly represents only a small fraction of the genes controlling this process (3).
To discover new genes in gametogenesis, John Schimenti and colleagues have undertaken an ambitious phenotypebased screen in mice based on chemical mutagenesis of either embryonic stem cells (ethylmethanesulfonate) or whole animals (ethylnitrosourea). In a pilot study, 11 mouse fertility mutants have thus far been generated that affect several different stages of gametogenesis in one or both sexes, including meiosis (4). In this issue of PNAS, Libby et al. (5) report the positional cloning of the first of the mutant genes, Mei1 (meiosis defective 1). Mei1 is expressed almost exclusively in the gonads, in particular, in the testis of prepubertal and adult males and in the ovary of late embryonic females (i.e., embryonic day 17.5), the time of meiotic prophase. The human MEI1 gene is predicted to encode a protein with 79% identity to the mouse protein, and other vertebrate homologs have also been identified. However, the encoded protein contains no significant homology to previously described proteins, and homologs are not evident in yeast, worms, or flies. Thus, with this forward genetic approach, a novel vertebrate meiosis gene has been identified, highlighting the power of the chemical mutagenesis screen for infertility studies.
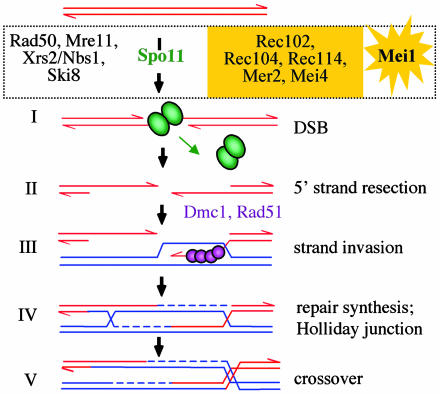
Despite being a novel gene, precise characterization of the Mei1 mutant phenotype in relation to that of other mouse meiotic mutants has provided insight into its function (5, 6). Central to meiosis is recombination between homologous chromosomes resulting in crossover recombinants. In conjunction with sister-chromatid cohesion, crossovers maintain the physical connections between homologs necessary for chromosome congression at the metaphase plate to permit segregation of homologs at the first meiotic division. The events of meiotic recombination, as described for Saccharomyces cerevisiae (7), are as follows (Fig. 1): double-strand breaks (DSBs) are introduced to initiate recombination (step I); 5′ terminal strands at the DSBs are degraded to yield 3′ single-stranded tails (step II); the tails invade the intact, homologous chromosome (step III); repair synthesis ensues and double-Holliday junction intermediates are formed (step IV); and resolution results in mature crossover products (step V).
Fig. 1.
Meiotic recombination resulting in crossover recombinants, as proposed for S. cerevisiae. DSBs, which initiate recombination, are introduced by the Spo11 protein, which forms a covalent complex with the DNA ends (7). In addition to Spo11, a number of other proteins are required for DSB formation, including those that have roles in mitotic DSB repair (i.e., Rad50/Mre11/Xrs2) and those that have meiotic-specific roles (proteins in yellow box). Although homologs have not been identified in mammals for this latter group of proteins, Mei1 mutation in the mouse leads to a similar phenotype as Spo11 mutation, implying that Mei1 is required with Spo11 for DSB formation. See text for further details. Note: recombination is between replicated homologous chromosomes, but only one sister chromatid is shown for each homolog.
The catalytic activity for DSB formation appears to reside in the Spo11 protein, which presumably acts as a transesterase rather than as an endonuclease (Fig. 1, step I) (7). In the mouse, ≈250 DSBs are inferred to be introduced by the Spo11 protein (see, e.g., ref. 8). Because there are only ≈24 crossovers, most DSBs do not give rise to crossovers but may instead be repaired by recombination without crossing over. DSB formation leads to a DNA damage response, including phosphorylation of histone H2AX (γH2AX) (9), and repair of the Spo11-generated DSBs involves the strand invasion proteins Rad51 and Dmc1, which form foci at the DSB sites (Fig. 1, step III), and other repair proteins (7, 8, 10, 11).
Spo11 does not act alone, however, because at least nine other proteins are required in S. cerevisiae for DSB formation (Fig. 1, step I), such that null mutations in any one of those genes confer the same meiotic phenotype as the Spo11 null mutation (7). Four of these proteins, like Spo11, are conserved in other organisms: Three have mitotic roles in DSB repair (i.e., the Rad50 complex) and one is cytoplasmic in vegetative yeast cells, but has a nuclear meiotic role (i.e., Ski8) (C. Arora and S. Keeney, personal communication). However, the other five proteins, which like Spo11 are expressed exlusively during meiosis, show limited or no sequence conservation with proteins in other species. Conversely, some genes required for DSB formation in other organisms, e.g., rec6 in Schizosaccharomyces pombe (12) and mei-P22 in Drosophila melanogaster (13), are also novel proteins, demonstrating the importance of phenotype-based screens for identifying genes involved in meiotic recombination.
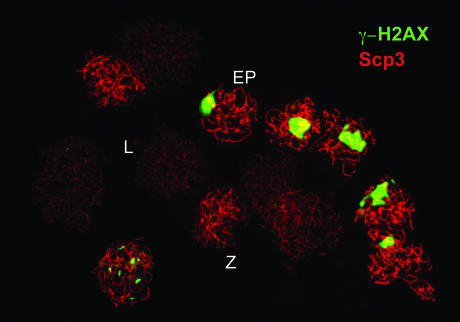
Phenotypically, the mouse Mei1 mutant shows striking similarities to Spo11-deficient mice (10, 11), suggesting that it, too, fails to introduce DSBs into meiotic chromosomes. Notably, in both mutants γH2AX staining (Fig. 2) and Rad51 foci are nearly eliminated in early meiotic prophase (5, 9–11), although Rad51 foci can be induced by cisplatin treatment, indicating a defect in DSB formation rather than in focus formation per se. At later stages, i.e., at late zygotene/early pachytene in spermatocytes, the sex body containing the X and Y chromosomes stains brightly for γH2AX in both mutants (Fig. 2) (ref. 9 and L. Reinholdt and J. Schimenti, personal communication), indicating that this histone modification on the sex body is independent of Spo11-generated DSBs (9). The similarity of the phenotypes suggests that Mei1 assists Spo11 in DSB formation, possibly as an ortholog to one of the yeast DSB formation genes; alternatively, it may have a new function that arose during vertebrate evolution.
Fig. 2.
Chromosomes from Spo11–/– spermatocytes are nearly devoid of γH2AX staining (green), except in the brightly staining sex body that contains the sex chromosomes. Spermatocytes at different stages of meiotic progression are shown. In leptotene (L), short stretches of axial elements of the synaptonemal complex are formed as evidenced by Scp3 staining (red). In the Spo11 mutant, little γH2AX staining is apparent, such that nuclei are faint. Wild-type (WT) spermatocytes are usually strongly stained for γH2AX at this stage as a result of DSB formation (e.g., see figure 6D in ref. 5). Nuclei that proceed to zygotene (Z) have longer axial elements that are readily visible with Scp3 staining; γH2AX staining is only rarely seen in the Spo11 mutant, as shown. WT spermatocytes experience a decline of γH2AX staining at zygotene, as DSBs begin to be repaired (data not shown). At late zygotene and early pachytene (EP), the sex chromosomes stain heavily for γH2AX in both mutant (as shown) and WT nuclei (data not shown), whereas the autosomes do not stain. WT pachytene nuclei have complete synapsis of homologous chromosomes (data not shown), whereas full synapsis is never achieved in the Spo11 mutant (see refs. 10 and 11). (Magnification: ×400.)
The apparent lack of DSB formation and subsequent impairment of meiotic recombination in both the Spo11 and Mei1 mutants leads to meiotic catastrophe and subsequent germ cell loss, although the response is sexually dimorphic (6, 10, 11). In both sexes, meiotic chromosome structures begin to assemble normally as evidenced by deposition of Scp3 into axial elements, but homologous chromosomes fail to synapse properly (e.g., Fig. 2). In males, an immediate apoptotic loss is seen at the late zygotene/early pachytene stages. In females, oocytes can progress further to form follicles, although they are profoundly defective in chromosome congression at the metaphase I spindle, such that a normal division cannot be completed (ref. 6 and M. Di Giacomo, F. Baudat, S. Keeney, and M.J., unpublished results).
As well as mutants in DSB formation, other meiotic mutants have been generated in the mouse that affect downstream steps in recombination. Like Spo11, several of the genes have been identified based on their homology to yeast genes involved in the process (e.g., Dmc1) (7). In other cases, meiotic roles have been found for genes originally identified to have roles in mitotic recombination in mammalian cells. Examples of the latter include Brca1 and Brca2 (14). In Brca1 and Brca2 hypomorphic mutants, DSBs are introduced by Spo11 as gauged by γH2AX staining, but subsequent steps are defective as evidenced by impaired recruitment of Rad51 into foci (15, 16).
Genes with roles in meiotic recombination will likely continue to be identified through homology searches or through their role in mitotic recombination. Nevertheless, the phenotype-based screen that led to the discovery of Mei1 is particularly suited for the identification of novel meiosis-specific genes that have little homology to genes in other organisms because of sequence divergence or the evolution of vertebrate-specific functions. This approach will likely be valuable for the identification of novel genes involved in other aspects of gametogenesis as well. The emerging application of gene profiling to germ cells by using microarray technology is also expected to lead to the discovery of mammalian genes involved in gametogenesis (see refs. 3, 17, and 18 and references therein). Once the phenotype-based screen is expanded it will be interesting to determine what percentage of genes are novel and which were identified through other approaches, to gauge the degree of saturation achieved by the various approaches. Importantly for human infertility, in individuals where the precise characterization of the cellular and chromosomal defects in germ cells is feasible, it may be possible in the future to discern the underlying genetic defect by using the growing catalog of mouse mutants.
Acknowledgments
We thank Monica Di Giacomo, Charanjit Arora, Scott Keeney, Mary Ann Handel, Shyam Sharan, Pellegrino Rossi, Susanna Dolci, and Claudio Sette for communication of unpublished results; Scott Keeney for critical reading of the manuscript; and Peter Moens, Mary Ann Handel, and William Bonner for antibodies and assistance. M.B. was supported by an award from The Lalor Foundation and an American-Italian Cancer Foundation Fellowship, and M.J. was supported by National Institutes of Health Grant HD40916.
See companion article on page 15706.
References
- 1.De Kretser, D. M. & Baker, H. W. (1999) J. Clin. Endocrinol. Metab. 84, 3443–3450. [DOI] [PubMed] [Google Scholar]
- 2.Matzuk, M. M. & Lamb, D. J. (2002) Nat. Cell Biol. 4, Suppl., s41–s49. [DOI] [PubMed] [Google Scholar]
- 3.Schultz, N., Hamra, F. K. & Garbers, D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 12201–12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward, J. O., Reinholdt, L. G., Hartford, S. A., Wilson, L. A., Munroe, R. J., Schimenti, K. J., Libby, B. J., O'Brien, M., Pendola, J. K., Eppig, J. & Schimenti, J. C. (2003) Biol. Reprod. 69, 1615–1625. [DOI] [PubMed] [Google Scholar]
- 5.Libby, B. J., Reinholdt, L. G. & Schimenti, J. C. (2003) Proc. Natl. Acad. Sci. USA 100, 15706–15711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Libby, B. J., De La Fuente, R., O'Brien, M. J., Wigglesworth, K., Cobb, J., Inselman, A., Eaker, S., Handel, M. A., Eppig, J. J. & Schimenti, J. C. (2002) Dev. Biol. 242, 174–187. [DOI] [PubMed] [Google Scholar]
- 7.Keeney, S. (2001) Curr. Top. Dev. Biol. 52, 1–53. [DOI] [PubMed] [Google Scholar]
- 8.Moens, P. B., Kolas, N. K., Tarsounas, M., Marcon, E., Cohen, P. E. & Spyropoulos, B. (2002) J. Cell Sci. 115, 1611–1622. [DOI] [PubMed] [Google Scholar]
- 9.Mahadevaiah, S. K., Turner, J. M., Baudat, F., Rogakou, E. P., de Boer, P., Blanco-Rodriguez, J., Jasin, M., Keeney, S., Bonner, W. M. & Burgoyne, P. S. (2001) Nat. Genet. 27, 271–276. [DOI] [PubMed] [Google Scholar]
- 10.Baudat, F., Manova, K., Yuen, J. P., Jasin, M. & Keeney, S. (2000) Mol. Cell 6, 989–998. [DOI] [PubMed] [Google Scholar]
- 11.Romanienko, P. J. & Camerini-Otero, R. D. (2000) Mol. Cell 6, 975–987. [DOI] [PubMed] [Google Scholar]
- 12.Davis, L. & Smith, G. R. (2001) Proc. Natl. Acad. Sci. USA 98, 8395–8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu, H., Jang, J. K., Kato, N. & McKim, K. S. (2002) Genetics 162, 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jasin, M. (2002) Oncogene 21, 8981–8993. [DOI] [PubMed] [Google Scholar]
- 15.Xu, X., Aprelikova, O., Moens, P., Deng, C. X. & Furth, P. A. (2003) Development (Cambridge, U.K.) 130, 2001–2012. [DOI] [PubMed] [Google Scholar]
- 16.Sharan, S. K., Pyle, A., Coppola, V., Babus J., Swaminathan, S., Benedict, J., Swing D., Martin, B. K., Tessarollo, L., Evans J. P., et al. (2004) Development (Cambridge, U.K.) 131, 131–142. [DOI] [PubMed] [Google Scholar]
- 17.Rossi, P., Dolci, S., Sette, C., Capolunghi, F., Pellegrini, M., Loiarro, M., Di Agostino, S., Paronetto, M. P., Grimaldi, P., Merico, D., et al. (2004) Gene Expression Patterns, in press. [DOI] [PubMed]
- 18.Schlecht, U. & Primig, M. (2003) Reproduction 125, 447–456. [DOI] [PubMed] [Google Scholar]