Abstract
The facial branchiomotor neurons undergo a characteristic tangential migration in the vertebrate hindbrain. Several signaling mechanisms have been implicated in this process, including the non-canonical Wnt/planar cell polarity (PCP) pathway. However, the role of this signaling pathway in controlling the dynamics of these neurons is unclear. Here, we describe the cellular dynamics of the facial neurons as they migrate, focusing on the speed and direction of migration, extension of protrusions, cell shape and orientation. Furthermore, we show that the PET/LIM domain protein Prickle1b (Pk1b) is required for several aspects of these migratory behaviors, including cell orientation. However, we find that centrosome localization is not significantly affected by disruption of Pk1b function, suggesting that polarization of the neurons is not completely lost. Together, our data suggest that Pk1b function may be required to integrate the multiple migratory cues received by the neurons into polarization instructions for proper posterior movement.
Keywords: facial branchiomotor neurons, prickle1b, planar cell polarity/non-canonical Wnt, tangential neuronal migration, neuronal polarity
Introduction
Neuronal migration is an essential step in the development of organized neural circuitry. During vertebrate development all neurons undergo one or more forms of migration from their birthplace to their final destination. Modes of migration include glial-associated radial migration as well as glia-independent tangential migration. While the cell behaviors involved in radial migration have been studied extensively (for reviews, see Marín and Rubinstein 2003; Métin et al. 2008), much less is known about tangential migration. To understand more about tangential migration we have investigated the cell behaviors of facial branchiomotor neurons during their tangential migration through the zebrafish hindbrain.
The facial neurons are a subset of the cranial branchiomotor neurons common to all vertebrates. Facial branchiomotor neurons (FBMNs) are born in the ventralmost portion of rhombomere (r) 4 of the hindbrain, and shortly thereafter their cell bodies commence a characteristic posterior migration from r4 to r6-7. In the zebrafish (Danio rerio), this tangential migration begins as soon as the first neurons are born, at 16 hours post-fertilization (hpf), and is completed by 48 hpf. After the facial neurons have completed their posterior movement, they migrate dorsolaterally within r6-7 to their final destination in the neural tube. Meanwhile, the axons of the FBMNs exit the neural tube from r4 and innervate the derivatives of the second branchial arch.
Multiple genes are required for the normal tangential migration of zebrafish FBMNs (reviewed by Chandrasekhar 2004; Song 2007). These include several components of the non-canonical Wnt/planar cell polarity (PCP) pathway (reviewed by Wada and Okamoto 2009a, 2009b), including vangl2 (Jessen et al. 2002; Bingham et al. 2002); frizzled3a (fz3a), cadherin EGF-like seven-pass G-type receptor 1a (celsr1a), celsr1b, celsr2 (Wada et al. 2006); scribble1 (Wada et al. 2005); prickle1a (pk1a, Carreira-Barbosa et al. 2003); and prickle1b (pk1b, Rohrschneider et al. 2007). Recently, similar roles in FBMN migration have been demonstrated for the murine PCP components wnt5a, fz3, and vangl2 (Vivancos et al. 2009), suggesting that findings in the zebrafish will be broadly applicable to vertebrates. Loss of function of any of these PCP genes results in a partial or complete disruption of facial neuron tangential migration. Instead, most or all neurons remain in r4, and although they can still migrate dorsally, they do so in close proximity to the fourth ventricle.
The PCP pathway has well-demonstrated roles in organizing epithelial cell layers in organisms from Drosophila to humans (reviewed by Montcouquiol et al. 2006; Simons and Mlodzik 2008). However, it is not clear how PCP components work together to bring about facial neuron migration, in which small numbers of neurons move through the neuroepithelium. Also puzzling is the fact that transplantation experiments in zebrafish have demonstrated primarily cell-nonautonomous requirements for several PCP components during tangential migration (Jessen et al. 2002; Wada et al. 2006). It has been proposed by Wada et al. (2006) that fz3a functions in the surrounding neuroepithelium to keep the facial neurons in a ventral position; loss of fz3a results in dorsal movement of FBMNs that is perhaps incompatible with tangential migration. Previous work in our lab has demonstrated that the proposed PCP gene pk1b is also required for zebrafish FBMN migration (Rohrschneider et al. 2007). However, we have shown that pk1b functions primarily cell-autonomously during facial neuron migration. This is the only PCP component known to act principally within the facial neurons during their tangential migration. Although cell-nonautonomous roles for pk1b cannot be ruled out, its unique expression and localization of function among other PCP components is intriguing.
Very little is known about the cellular dynamics of the facial neurons as they migrate from r4 to r6-7. Studies of migrating neurons in the mammalian cortex and cerebellum have demonstrated the importance of extension of protrusions, cell shape, and centrosome positioning (Tabata and Nakajima 2003; Ward et al. 2005; Komuro and Rakic 1998; Tanaka et al. 2004; Solecki et al. 2004). In this study, we have sought to broaden our understanding of tangential migration by studying the cellular dynamics of FBMN migration. To enable detailed cellular analysis we generated a new transgenic zebrafish line in which membrane-localized red fluorescent protein is expressed specifically in FBMNs, allowing us to image changes in cell shape and protrusions during directed neuronal migration. Further, we sought to determine how loss of Pk1b function affects these processes. We demonstrate that Pk1b-deficient FBMNs in r4 take on cell shapes characteristic of control FBMNs in r6. However, we show that the position of the centrosome, which has been reported to be coupled to cell soma movement during neuronal migration, is comparable in wildtype and Pk1b-deficient neurons in r4. Based on these data, we conclude that some aspects of the orientation of facial neurons in r4 are disrupted in response to Pk1b deficiency, resulting in the inability of these neurons to migrate tangentially out of r4.
Results
zCREST1:membRFP transgenic line allows visualization of facial neuron cellular dynamics
We have generated a new transgenic line in order to efficiently visualize the cellular and membrane dynamics of the migrating facial neurons. The zCREST1 enhancer of the islet1 promoter was previously shown to drive gene expression in the branchiomotor neurons (Uemura et al. 2005). We used a construct containing the zCREST1 enhancer, the minimal promoter of hsp70l, and membRFP (kind gift from Dr. Cecilia Moens; Figure 1A), to generate a stable transgenic line expressing the zCREST1:membRFP transgene in branchiomotor neurons (Figure 1B). This line also shows expression in a few cells of the otic vesicle (arrowhead in Figure 1D), but this did not detract from our ability to track FBMNs. In comparison to the islet1:GFP transgenic line (Figure 1C; Higashijima et al. 2000), the new zCREST1:membRFP line enabled more effective imaging of cell protrusions and cell shape changes (Figure 1B–D). We generated time-lapse movies of migrating FBMNs in both wildtype embryos (uninjected controls) and embryos rendered Pk1b-deficient by morpholino injection (Pk1b morphants) (for examples, see Supplemental Movies 1–4). For each movie we collected images over approximately a 1.5–2 hour period, commencing at one of two stages: 20 hpf, the earliest stage at which RFP was reliably detectable in live specimens without significant photobleaching, and 24 hpf, a stage at which many cells are undergoing active migration through r5 while neurons continue to be born in r4. We have focused our analysis on FBMNs that migrate from r4 to r6. Although neurons can also be observed migrating through r6 to r7, their origin is unclear and their identity as purely facial neurons cannot be determined (Chandrasekhar 2004). Furthermore, it is not clear whether these neurons receive the same migratory cues as those neurons which stop posterior migration in r6. We have therefore excluded these neurons from our analysis. As we previously reported (Rohrschneider et al. 2007), loss of Pk1b function completely disrupts tangential migration of facial neurons out of r4, although they are still able to move dorsolaterally through r4. Beyond this disruption of facial neuron migration Pk1b morphants show only minimal alteration in gross morphology; very mild convergent extension defects lead to a slightly shortened body axis (Supplemental Figure 1), but we have not observed any additional phenotypes.
Figure 1. zCREST1:membRFP transgenic line allows enhanced visualization of cell shape and protrusions in FBMNs.
A: zCREST1:membRFP transgene. Expression of membrane-targeted RFP was driven by branchiomotor neuron-specific zCREST1 enhancer of the islet1 promoter. B–D Dorsal views of live zCREST1:membRFP; islet1:GFP double-transgenic 24 hpf embryos. B: zCREST1:membRFP transgene expression is limited to branchiomotor neurons, as well as a subset of cells in the otic vesicle (yellow arrowheads). C: islet1:GFP transgene. D: Merged images. D’: Higher magnification view of D, highlighting protrusions visible with membRFP expression (white arrowheads).
Pk1b deficiency alters speed of migration of FBMNs
At both stages observed (20–22 hpf and 24–26 hpf), FBMNs in control embryos undergo a cumulative posterior migration from r4 to r6 (Supplemental Movies 1, 3; Figure 2A–B, E–F). This migration is saltatory in nature, as neurons frequently pause and extend protrusions. Neurons that are born in r4 within the duration of imaging appear to follow the track of axons laid out by neurons that have previously migrated. However, we frequently observed neurons undergoing movements toward and away from the midline i.e. perpendicular to the tangential direction. In general, these neurons extend protrusions toward the midline, briefly move toward the midline, and then reverse course and move back towards the track of axons. We discuss migratory behaviors in more detail below.
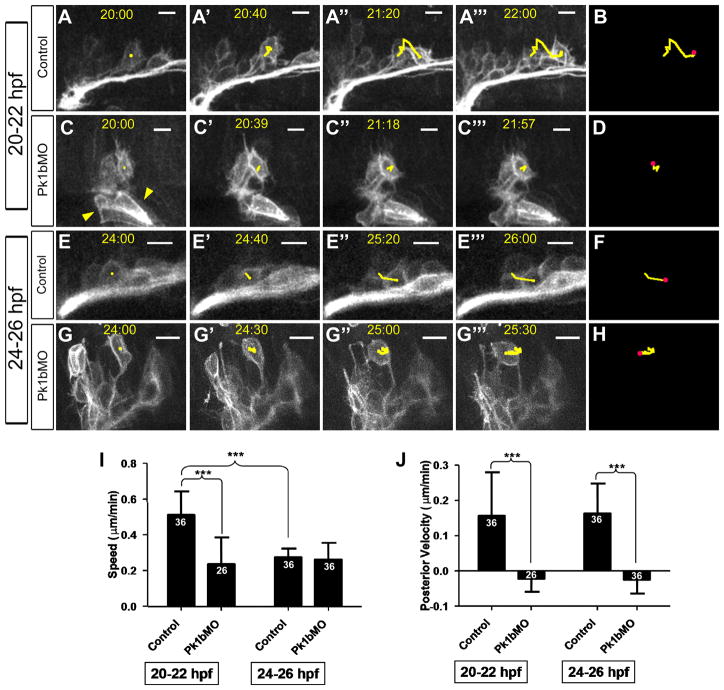
Figure 2. Pk1b deficiency alters both overall speed and posterior-directed velocity of FBMN migration.
A, C, E, G: Frames from time-lapse movies of zCREST1:membRFP embryos, showing movements of FBMNs. A-A’’’: Control, 20-22 hpf. C-C’’’: Pk1b morphant, 20–22 hpf. E-E’’’: Control, 24-26 hpf. G-G’’’: Pk1b morphant, 24–26 hpf. Scale bars = 10 μm. Note that there are membRFP-positive cells in otic vesicle (yellow arrowheads). B, D, F, H: Yellow lines represent cumulative movements of cells tracked in A, C, E, and G, respectively. Pink dots correspond to final position of tracked neurons during the course of the time-lapse analysis. I–J: Average FBMN speed (I) and posterior-directed velocity (J) was measured over the course of time-lapse movies. Numbers above each bar indicate the number of cells tracked (n). N = 3 embryos for each class. Scale bars: 10 μm. * – p<0.05; ** – p<0.01; *** – p<0.001.
Pk1b deficiency could affect FBMN migration in one of two ways: either the overall movement of the neurons could be disrupted, or the posterior-directed migration out of r4 may be specifically lost. We distinguished between these possibilities by comparing cell movements in time-lapse movies of Pk1b morphants (Supplemental Movies 2, 4; Figure 2C–D, G–H) to those in wildtype (WT) specimens. Importantly, although FBMNs do not exit r4 in Pk1b morphants, the neurons still move significantly within r4. Moreover, the morphant neurons often migrate to a more anterior position within r4, shortly before undergoing dorsal movement. Therefore, morphant neurons retain motile capacity, but are not able to move in a directed fashion out of r4.
In order to quantify the movements of control and Pk1b morphant neurons, we tracked individual cells in control and morphant embryos and compared both their overall speeds (movement in any direction) and posterior-directed velocities, relative to the point of facial nerve exit from r4 of the hindbrain, which represents the most reliable fixed reference point. The average overall speed of control neurons between 20 and 22 hpf was 0.514 μm/min, compared to 0.239 μm/min for morphants (Figure 2I; Table 1). Therefore, although Pk1b morphant neurons are not stationary, they move at a slower speed than control neurons at early stages of migration. However, by later stages (24–26 hpf), the speeds of control and Pk1b morphant neurons are more comparable (0.276 μm/min and 0.264 μm/min, respectively). We hypothesize that the decrease in speed of WT neurons at later stages of migration is indicative of a reduction in exploratory behaviors.
Table 1.
Analysis of time-lapse movies. Averages shown represent mean ± standard deviation. n refers to the number of individual neurons or protrusions analyzed.
| Speed | Velocity | Protrusion Angle | Number of Protrusions | Protrusion Length | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Average (μm/min) | n | Average (μm/min) | n | Average (°) | n | Medial Alignment (90° ± 30) | Average protrusions/cell/time point | n | Average (μm) | n | |
| Control, 20–22 hpf | 0.512 ±0.133 | 36 | 0.156 ±0.123 | 36 | 94 | 368 | 59% | 1.61 ± 1.16 | 228 | 4.89 ±2.75 | 368 |
| Pk1bMO, 20–22 hpf | 0.238 ±0.149 | 26 | −0.023 ±0.037 | 26 | 81 | 202 | 59% | 1.62 ± 1.32 | 141 | 4.76 ±2.81 | 202 |
| Control, 24–26 hpf | 0.276 ±0.049 | 36 | 0.163 ±0.085 | 36 | 108 | 147 | 66% | 0.72 ± 0.90 | 207 | 3.81 ±2.07 | 147 |
| Pk1bMO, 24–26 hpf | 0.264 ±0.094 | 36 | −0.026 ±0.039 | 36 | 123 | 188 | 50% | 1.06 ± 1.03 | 161 | 4.89 ±3.04 | 188 |
We also find substantial differences in the posterior-directed velocities of control and morphant neurons. Between 20 and 22 hpf, the average velocity of control neurons was 0.156 μm/min, while that of Pk1b morphant neurons was −0.023 μm/min (indicative of anterior movement within r4). The velocities at later stages of migration are not significantly different to those at earlier stages, either in control or in Pk1b morphant embryos (0.163 μm/min and −0.026 μm/min, respectively).
Together these data allow us to conclude that FBMNs in WT embryos undergo more exploratory movements at earlier stages of migration, and migrate more efficiently at later stages. Furthermore, Pk1b deficiency disrupts the ability of the facial neurons to move in a directed fashion, and also appears to hinder their overall ability to undergo exploratory behaviors in the surrounding neuroepithelium.
Protrusive behavior of facial neurons is altered by Pk1b deficiency
Previous studies in mammalian systems have described the protrusive behaviors of migrating neurons (Tabata and Nakajima 2003, Ward et al. 2005). Specifically, neurons send out protrusions in several directions in search of migratory cues. These protrusions are then stabilized or retracted, depending on the type of cue encountered (attractive or repulsive). While the nature of our time-lapse studies does not allow for a thorough measurement of protrusive stability, we have quantified the direction of protrusions extended (protrusion angle) as well as the average number and length of protrusions extended by the FBMNs (Figure 3A). For this analysis, we have focused on filopodial protrusions.
Figure 3. Protrusive behaviors of FBMNs are altered in Pk1b morphants.
A: Schematic illustrating how measurements were performed. A line was drawn from the base of each protrusion to its tip. The length of this line was defined as protrusion length. The angle between this line and the A–P axis, was defined as the protrusion angle. B–E: Rose diagrams illustrating range of FBMN protrusion angles. Numbers along right side indicate scale of y-axis (i.e. percentage of total protrusions for each angle subset). Yellow lines through diagrams indicate mean protrusion angle. F: Quantification of the average number of protrusions extended per cell, per time point in time-lapse movies. Numbers in each bar indicate the number of instances counted (n). N = 3 embryos for each class. G: Quantification of the average length of protrusions in time-lapse movies. Numbers in each bar indicate the number of protrusions scored (n). N = 3 embryos for each class. M – medial (90°), L – lateral (−90°), A – anterior (180°), P – posterior (0°). * – p<0.05; ** – p<0.01; *** p<0.001.
At both stages of migration analyzed, control FBMNs send the majority of their protrusions in the general direction of the midline (Figure 3B, D; Table 1). At earlier stages of migration, 59% of protrusions are sent towards the midline (90° ± 30), and the mean protrusion angle is 94°. By later stages, a higher percentage of protrusions (66%) are sent towards the midline, suggesting that signals from the midline may continually act to guide migrating neurons. In addition, the mean protrusion angle is 81°, signifying a slight but significant shift towards the direction of migration. In Pk1b-deficient embryos, the majority of protrusions are still extended towards the midline at both earlier and later stages of migration (Figure 3C, E). However, the mean angles are shifted away from the normal direction of migration (108° between 20 and 22 hpf, 123° between 24 and 26 hpf), consistent with the observed anteriorward movements of these neurons. Moreover, while the number of medially-directed protrusions is similar to WT at earlier stages of migration (59%), Pk1b-deficient neurons show a decrease in these types of protrusions at later stages of migration (50%). These data suggest that Pk1b morphant neurons may continue to search for migratory cues in several directions.
In control neurons, the average number of protrusions extended per cell per time point is 1.61 between 20 and 22 hpf (Figure 3F). This number decreases to 0.72 between 24 and 26 hpf. In addition, the average length of protrusions decreases over time (4.89 μm at earlier stages vs. 3.81 μm at later stages; Figure 3G). These data are further indications of the decrease in exploratory behaviors at later stages of migration. Both the number and length of protrusions extended in Pk1b morphant neurons are comparable to control embryos between 20 and 22 hpf (averages of 1.62 protrusions per cell per time point, 4.76 μm; Figure 3F, G). At 24–26 hpf, the average number of protrusions decreases but not to the same extent as in WT neurons (1.06; Figure 3F). Also in contrast to WT neurons, the average length of protrusions extended from Pk1b-deficient FBMNs does not decrease at later stages of migration (4.89 μm; Figure 3G). Our data on protrusions extended by FBMNs suggest that Pk1b deficiency causes the neurons to continue their exploratory behaviors at later stages of migration, while control neurons decrease these behaviors.
Facial neurons are improperly elongated in Pk1b-deficient embryos
Our analysis of FBMN dynamics thus far has demonstrated that Pk1b function is not absolutely required for FBMN movement or extension of protrusions. Rather, our data suggest that Pk1b morphant neurons may not be able to properly respond to migratory cues, which normally guide posterior-directed movement out of r4. We hypothesized that the loss of directed migration in Pk1b-deficient embryos may be due to changes in the polarity of the FBMNs. To test this hypothesis, we first analyzed the length-width ratios (LWR) of FBMNs in fixed embryos as a measure of cell shape (Figure 4, Table 2). A more elongated cell shape has been shown to be indicative of changes in polarity during migration (Komuro and Rakic 1998). In addition, loss of PCP signaling has been associated with changes in LWR in gastrulating embryos (Jessen et al. 2002). We observed that individual FBMNs rapidly change their shape as they migrate, both at early and later migration stages, yet certain tendencies are clear.
Figure 4. FBMN length-width ratio (LWR) is altered in Pk1b morphants.

A–D: Dorsal views of fixed embryos highlighting differences in cell shape and orientation along migratory path. (A) Control, 18 hpf; (B) Pk1b morphant, 18 hpf; (C) Control, 24 hpf; (D) Pk1b morphant, 24 hpf. Flattened confocal z-stacks. Scale bars = 20 μm. Dashed lines represent rhombomere boundaries, which were assessed by EphA4 immunostaining. E: Schematic explaining LWR measurement. A bisecting line was drawn through the longest axis of each neuron, and another bisecting line was drawn perpendicular to the first. The line that most closely aligned with the A–P axis was defined as the width, while the perpendicular line was defined as the length. If the value of LWR 1, we considered these cells to be “round”. F: Bar graphs summarizing average LWR in each rhombomere at both early and later stages of migration, in control and Pk1b morphant embryos. * – p<0.05; ** – p<0.01; *** – p<0.001.
Table 2.
Analysis of fixed embryos. Averages shown represent mean ± standard deviation. n refers to the number of individual neurons analyzed.
| LWR | Elongation Axis | |||
|---|---|---|---|---|
| Average | n | Medial Alignment (±90° ± 30) | n | |
| Control r4, 18 hpf | 0.881 ±0.464 | 53 | 51% | 53 |
| Control r5, 18 hpf | 0.967 ±0.466 | 39 | 38% | 39 |
| Control r6, 18 hpf | 0.780 ±0.462 | 31 | 61% | 31 |
| Pk1bMO r4, 18 hpf | 0.749 ± 0.437 | 118 | 70% | 118 |
| Control r4, 24 hpf | 1.020 ± 0.299 | 28 | 36% | 28 |
| Control r5, 24 hpf | 1.078 ± 0.471 | 42 | 36% | 42 |
| Control r6, 24 hpf | 0.957 ± 0.453 | 98 | 49% | 98 |
| Pk1bMO r4, 24 hpf | 0.812 ± 0.336 | 148 | 64% | 148 |
As we used fixed embryos for this analysis, we were able to detect membRFP at the earlier stage of 18 hpf, and compared embryos at this stage to 24 hpf embryos. At 18 hpf, control and Pk1b morphant neurons have very similar cell shapes (Figures 4A–B, F). In control neurons, cells are slightly elongated in the mediolateral axis in r4 (LWR = 0.881), but they become more round as they move into r5 (LWR = 0.967). Once the neurons reach r6, they once again become elongated in the mediolateral axis, as they undergo dorsolateral movement (LWR = 0.780). In Pk1b morphants, neurons are slightly more elongated in r4 compared to control neurons in r4, although this difference is not statistically significant (LWR = 0.749; Figure 4F).
At 24 hpf, differences in cell shape become more pronounced (Figure 4C–D, F). Control neurons in r4 and r5 are generally more round than at 18 hpf (LWR = 1.020 and 1.078, respectively), and in r6 become slightly more elongated in the mediolateral axis (LWR = 0.957). Importantly, the neurons in Pk1b morphants, all of which remain in r4, are significantly more elongated in the mediolateral axis (LWR = 0.811), and most closely resemble WT neurons in r6 that are undergoing dorsolateral migration. Together, these data suggest that while the shapes of WT and morphant FBMNs are largely similar at 18 hpf, Pk1b morphant cells are inappropriately elongated by 24 hpf, perhaps indicative of mispolarization of the neurons.
Pk1b function is required for proper orientation of facial neurons
To test further our hypothesis that Pk1b morphant neurons are incorrectly polarized, we determined the orientation of the neurons by measuring their axis of elongation at both early and later stages of migration (Figure 5, Table 2). Approximately 66% of control neurons in r4 are oriented in the mediolateral axis (90° ± 30) at 18 hpf (Figure 5B). As cells migrate into r5, they change shape more frequently, and thus display a wider range of elongation axes (Figure 5C). Once control cells reach r6, they once again elongate in the mediolateral axis as they undergo dorsolateral migration (Figure 5D). In Pk1b morphants, approximately 75% of cells are elongated in the mediolateral axis in r4 at 18 hpf (Figure 5E).
Figure 5. Pk1b-deficient FBMNs are elongated in the incorrect axis.
A: Schematic explaining elongation axis measurement. A bisecting line was drawn through the longest axis of each neuron. The angle of this line, relative to the A–P axis, was defined as the elongation axis. Values were measured between −90° and 90°. B–I: Rose diagrams illustrating range of elongation axes. Due to the nature of the measurement, diagrams are symmetrical. Numbers along right side indicate scale of y-axis (i.e. percentage of total neurons for each angle subset). n values indicated represent total number of neurons scored for each class. N = 5 embryos for each class. M – medial (90°), L – lateral (−90°), A – anterior (180°), P – posterior (0°). ‡, †, §, # – p<0.001
At 24 hpf, there is a larger variation in elongation axes, particularly in r4 and r5 of control embryos, where only about 35% of neurons are elongated in the mediolateral axis (Figure 5F–G). This number increases to approximately 50% in control r6 neurons (Figure 5H). In contrast, about 65% of Pk1b morphant neurons are mediolaterally elongated (Figure 5I). We conclude from these data that Pk1b function is required to properly orient the FBMNs in r4, and these results support our hypothesis that facial neurons in Pk1b morphant neurons are incorrectly polarized.
Centrosome positioning in r4 is not altered in Pk1b morphants
Proper position of the centrosome during neuronal migration in mammalian models is essential for organizing and polarizing the cellular cytoskeleton, and is also reported to be coupled to nucleokinesis (Solecki et al. 2004; Bellion et al. 2005; reviewed by Higganbotham and Gleeson 2007). Previous studies have demonstrated that the centrosome is positioned ahead of the nucleus (in the direction of migration) in tangentially-migrating neurons. A network of microtubules subsequently pulls the nucleus towards this microtubule organizing center (Tanaka et al. 2004; Solecki et al. 2004). This model has recently been disputed in radially-migrating granule cells (Umeshima et al. 2007), although this may reflect different modes of migration in different regions of the brain. While our data suggest that Pk1b deficiency perturbs the polarity of the facial neurons, the extent of this mispolarization is unclear. It is possible that this abnormal polarization disrupts the proper orientation of the facial neuron centrosomes in r4. Conversely, the centrosome could remain properly localized in spite of the other consequences of mispolarization. We therefore examined the position of the centrosome in the migrating facial neurons by immunolabeling with an antibody targeted against γ-tubulin.
The γ-tubulin protein is expressed at elevated levels in centrosomes (Fig. 6A–D). At both 18 hpf and 24 hpf the centrosomes of those FBMNs within r4 are indeed generally positioned posterior to the nucleus (i.e. ahead of the nucleus in the direction of migration; Figure 6F, J). For FBMNs in r5 the position of the centrosome varies more widely, but is still generally posterior to the nucleus (Figure 6G, K). By contrast, the centrosomes of FBMNs in r6 are often positioned medially in the cell i.e. behind the nucleus (Figure 6H, L). This shift in centrosome position could potentially correspond with the transition from tangential to dorsolateral migration of the neurons. In Pk1b morphant facial neurons the centrosomes remain positioned posterior to the nucleus, similar to the situation in WT FBMNs in r4 (Figure 6I, M). This result suggests that although the Pk1b morphant facial neurons are incorrectly elongated and oriented in r4, the position of the centrosome remains comparable to controls. In an effort to corroborate these results we also examined the expression of the neuronal polarity markers PKCζ and PIP3 (Ménager et al. 2004; Jiang et al. 2005; Sakakibara and Horwitz 2006; Higginbotham et al. 2006; for a review, see Marín et al. 2006). Unfortunately this data was uninformative because although asymmetric localization of these molecules was reported in amniote neurons, we did not detect any asymmetry of these markers in either WT or Pk1b-deficient zebrafish FBMNs (Supplemental Figure 2). Nevertheless, our data suggest that Pk1b deficiency does not perturb the ability of the centrosome to properly localize in facial neurons. Instead, the centrosome remains primed for tangential migration, but the neurons are unable to undergo directed migration into r5-6.
Figure 6. Centrosome remains primed for tangential migration in Pk1b-deficient neurons.
A–D: Dorsal views of fixed embryos illustrating positioning of centrosome in migrating neurons. (A) Control, 18 hpf; (B) Pk1b morphant, 18 hpf; (C) Control, 24 hpf; (D) Pk1b morphant, 24 hpf. Red: zCREST1:membRFP transgene expression. Blue: EphA4 immunostaining, to label r3 and r5. Green: γ-tubulin immunostaining, which allows visualization of centrosomes (bright puncta; white arrowheads) as well as the microtubule network (fainter green staining). Region of each cell with no green staining corresponds to nucleus. Flattened confocal z-stacks. Scale bars = 50 μm. E: Schematic demonstrating measurement of centrosome position. A bisecting line was drawn from the centrosome (bright green puncta) through each neuron. The angle of this line, relative to the A–P axis was defined as the centrosome position. F–M: Rose diagrams illustrating the range of centrosome positions in migrating FBMNs. Numbers along right side indicate scale of y-axis (i.e. percentage of total neurons for each angle subset). n values indicated represent total number of neurons scored for each class. N = 5 embryos for each class.
Discussion
Previous to this study, little was known about the cellular dynamics of FBMNs during their tangential migration in zebrafish. Here we have performed a comprehensive analysis of FBMN dynamics, characterizing several aspects of cell behaviors during migration in both WT and Pk1b-deficient embryos. In Figure 7 we summarize our data and present a model of FBMN migration that incorporates previous results as well as our new findings. According to the model several guidance molecules are present in the neuroepithelium surrounding the FBMNs, including cues that normally guide the tangential migration of the neurons, as well as others that the neurons do not normally migrate toward (black and grey plus signs, respectively). Our imaging studies also suggest that there are migratory cues (black question marks) present at the midline (dashed line). Reported zebrafish neuroepithelial guidance molecules that influence facial neuron migration include chemokines Sdf1a and Hgf1/2, the receptors for which, Cxcr4b and Met respectively, are expressed in the migrating neurons (Sapède et al. 2005; Cubedo et al. 2009; Elsen et al. 2009). In addition, roles for the extracellular matrix molecule lamininα1 and the adhesion molecule Tag-1 have been reported (Sittaramane et al. 2009). Within the facial neurons, the transcription factors Hoxb1a (McClintock et al. 2002) and Tbx20 (Pocock et al. 2008), as well as the transcription elongation factor Spt5 (Cooper et al. 2005) are required cell-autonomously for migration. The requirement of numerous molecules suggests that many signals must be integrated in order for the facial neurons to effectively migrate.
Figure 7. Summary of cellular dynamics of wildtype and Pk1b morphant FBMNs.
A–B: Schematics illustrating the dynamics of FBMNs during tangential migration at early (A) and later (B) stages of migration. C–D: Schematics illustrating the effects of Pk1b loss-of-function on FBMNs at early (C) and later (D) stages of migration. See text for details. M – medial (90°), L – lateral (−90°), A – anterior (180°), P – posterior (0°).
At early stages of normal migration (Fig. 7A) WT facial neurons extend many protrusions in the general direction of the midline. Neurons actively migrating in the posterior direction are primarily round. A subset of neurons elongates and transiently migrates in the direction of the midline. The centrosome is positioned in the posterior of the cell (i.e. ahead of the nucleus in the direction of migration). At later stages of migration (Fig. 7B) the neurons reduce their exploratory behaviors. The number and length of protrusions decreases and the neurons decrease their average speed while maintaining posterior-directed velocity. As neurons migrate through r5 the centrosome remains localized to the posterior, but as neurons in r6 begin dorsolateral movement, the position of the centrosome shifts. In Pk1b morphants (Fig. 7C) facial neurons are slightly more elongated than their WT counterparts, and incorrectly aligned with the mediolateral axis. The number and length of protrusions is comparable to controls, however they are oriented slightly more towards the anterior. The centrosome remains properly positioned in the posterior, primed for tangential migration. At later stages (Fig. 7D) protrusions remain longer than those of controls and are oriented yet more anteriorly. Aberrant cell shape and orientation of Pk1b-deficient FBMNs are amplified at this later stage, and cells migrate to more anterior positions within r4. As we already know that Pk1b functions primarily cell-autonomously within the FBMNs, our new results support the hypothesis that the facial neurons are improperly polarized in Pk1b-deficient embryos.
Pk1b regulates exploratory behaviors of migrating facial neurons
Our results illustrate the range of migratory behaviors in early- and later-migrating facial neurons. The relatively high ratio of speed to posterior velocity in early-migrating control neurons suggests that these cells undergo numerous exploratory movements at 20–22 hpf. By 24 hpf, neurons begin to migrate at a slower speed yet with similar velocity. In addition, control neurons significantly decrease the number of protrusions extended after 24 hpf. Together, our results reveal a transition from an initial exploratory phase towards a phase of highly directed migration, supporting the hypothesis that later-migrating neurons may follow the path of axons laid down by early-migrating neurons. We and others (Wada et al. 2005) have noted that the FBMNs migrate in very close apposition not only to their own axons, but also to axons of other neurons, which together comprise the medial longitudinal fasciculus (MLF). Although we have not detected any significant protrusive behavior towards the MLF in transverse hindbrain explants (data not shown), the mere proximity of these axon tracts may be sufficient to guide the neurons following their initial exploratory phase.
We have shown that blocking Pk1b function results in prolongation of FBMN exploratory behaviors, including extension of protrusions, into later stages of development. Cell culture studies have demonstrated that Prickle isoforms are upregulated in post-mitotic neurons and play a role in extension of axons and dendrites (Okuda et al. 2007; Fujimura et al. 2009). However, we do not find any overt changes in axon pathfinding in response to Pk1b-deficiency (Rohrschneider et al. 2007). We do find that a significant number of protrusions in both WT and Pk1b morphant neurons are extended in the general direction of the midline. From our analysis of transverse hindbrain explants as well as fixed specimens (data not shown), it is apparent that these midline-directed protrusions are oriented more dorsally than medially. Similar dorsal-midline protrusive behavior has been described in islet1:GFP transgenic fz3a mutants (Wada et al. 2006). Surprisingly, Wada et al. did not detect these protrusions in WT embryos; we attribute our differing results to our improved ability to visualize cell protrusions using the zCREST1:membRFP transgene (Figure 1B–D). Our finding of midline-directed protrusions suggests that the FBMNs receive guidance cues from the midline and/or are capable of responding to molecules at the midline; at present, we do not know the identity of this cue(s).
One possible candidate is the chemokine Sdf1a, which shows elevated expression along the midline of the neuroepithelium at both early and later stages of facial neuron migration (Cubedo et al. 2009); as mentioned above Sdf1a and its receptor Cxcr4b are required for facial neuron migration.
Our time-lapse imaging has demonstrated that FBMNs in Pk1b morphants are able to move, but they do not migrate in a directed fashion out of r4. Previous studies in embryos deficient in the PCP component Vangl2 have similarly shown that FBMN movement is not impaired, although directed migration is lost (Jessen et al. 2002; Sittaramane et al. 2009). In addition to a loss of posterior-directed migration, we find that Pk1b morphant facial neurons migrate at significantly slower speeds than WT neurons during the early exploratory stages. Nevertheless, our data reveal that Pk1b-deficient neurons display a similar range of behaviors to WT cells at this stage. The decrease in overall speed of Pk1b morphant neurons may be due to a direct effect on the migratory machinery of the facial neurons, or it may be a more indirect consequence of the inability of the facial neurons to respond to migratory cues in the surrounding neuroepithelium.
Pk1b function is required for some aspects of facial neuron polarization
We also demonstrate that Pk1b function plays a role in regulating several aspects of the orientation and shape of facial neurons. The decrease in LWR, plus the reduced range of both direction of protrusions and axis of elongation suggest that the facial neurons may be mispolarized in Pk1b morphants. However, we found the centrosome was properly positioned in facial neurons within r4 of Pk1b morphants. These results suggest that there are multiple aspects of polarity that must be regulated in order for facial neurons to migrate, based on both asymmetric localization of proteins as well as shape and orientation. Pk1b may regulate a subset of these aspects of polarity; alternatively, Pk1b may function to integrate intracellular polarity signals, and to translate these into appropriate cell shape and orientation.
Does Pk1b function with other PCP components during facial neuron migration?
Prickle homologues have traditionally been associated with PCP signaling. However, several notable differences between facial neuron migration and other PCP-dependent processes exist. For example, Pk1b is expressed at elevated levels within the facial neurons, while other PCP components examined are expressed more broadly in the neuroepithelium (Jessen et al. 2002; Wada et al. 2005, 2006). Overexpression of Pk1b mRNA does not cause a facial neuron migration phenotype, nor can overexpression rescue disrupted migration in Pk1b morphants (Mapp and Prince, in prep), suggesting that localized expression of Pk1b is key to its function. Furthermore, mosaic analyses have demonstrated that Pk1b functions primarily in the facial neurons (Rohrschneider et al. 2007), while vangl2, scribble1, fz3a, and celsr2 all function primarily in the neuroepithelium (Jessen et al. 2002; Wada et al. 2005, 2006). Specifically, 61% of WT neurons are able to migrate out of r4 in a Pk1b-deficient environment, while WT neurons cannot effectively migrate out of r4 in vangl2-, scribble1-, fz3a-, or celsr2-deficient environments (0%, 0%, 0%, 1.5% respectively). Conversely, 22% of Pk1b-deficient neurons are able to migrate out of r4 in a WT environment, while neurons deficient in other PCP components fare significantly better in this assay (33% of vangl2-deficient, ≥ 45% of fz3a-deficient, and ≥ 56% of celsr2-deficient neurons). We have recently demonstrated a genetic interaction between pk1b and vangl2; in addition, exogenous Vangl2 and Pk1b are able to interact in vivo (Mapp and Prince, in prep). However, we cannot exclude the possibility that Pk1b functions, at least in part, independent of other PCP components.
The Pk1b homologue Pk1a is also required for facial neuron migration (Carreira-Barbosa et al. 2003). These two zebrafish Pk1 proteins show significant homology, with the PET and LIM domains sharing 86% and 84% identity, respectively (Rohrschneider et al., 2007). However, these duplicated genes do not show equivalent expression domains: while pk1b is expressed at elevated levels in the migrating facial neurons, pk1a is not expressed within the neurons (Carreira-Barbosa et al. 2003; V. Sittaramane and A. Chandrasekhar, personal communication). Although cell-autonomy of action has not yet been determined for Pk1a, its expression pattern predicts that it will prove to function beyond the facial neurons. Furthermore, depletion of either Pk1a or Pk1b results in a complete disruption of facial neuron migration, a finding inconsistent with redundant function of these two proteins.
Wada et al. (2006) previously showed that defective facial neuron migration in fz3a mutant embryos was primarily due to a failure of the surrounding neuroepithelium to restrict dorsal movement of facial neurons. The authors hypothesized that PCP-mediated contacts between neuroepithelial cells may be decreased or eliminated, resulting in the ability of facial neurons to prematurely enter more dorsal regions. Our results illustrate that facial neurons in Pk1b morphant embryos also improperly undergo dorsal movement (Rohrschneider et al. 2007). As Pk1b functions primarily cell-autonomously, it is likely that the basic cellular dynamics of the neurons themselves are affected by Pk1b deficiency. While our experiments cannot exclude a role for the surrounding neuroepithelium in regulating the cellular dynamics of facial neurons, data presented here suggest that disruption of dynamics within the neurons can also result in their abnormal dorsal movement.
In this study we have described the dynamic cell behavior of facial neurons during their migration through the hindbrain. We have also uncovered new details of the role Pk1b plays in controlling FBMN cellular behaviors. However, the precise mechanisms through which Pk1b interacts with other PCP players, as well as non PCP-pathway molecules, remain to be elucidated.
Experimental Procedures
Fish lines and husbandry
zCREST1:membRFP transgenic fish were generated as follows: the zCREST1 enhancer of the islet1 promoter (Uemura et al. 2005), plus the hsp70l minimal promoter was used to drive expression of a membrane-targeted red fluorescent protein (membRFP) specifically in the branchiomotor neurons. This construct was kindly provided by Dr. Cecilia Moens. Plasmid DNA for this construct was injected into 1-cell stage *AB embryos at a concentration of 50 μg/ml, and injected embryos raised to adulthood. The progeny of these fish were screened for membRFP expression, and a single adult founder used to generate the transgenic line utilized throughout this study. To also visualize rhombomeres 3 and 5 the zCREST1:membrane-RFP fish were crossed to pGFP5.3 transgenic fish (Picker et al. 2002). All embryos were maintained at 28.5°C and staged as previously described (Kimmel et al. 1995).
Imaging
Fixed embryos were imaged on an LSM510 confocal microscope (Carl Zeiss Microimaging Inc., Thornwood, NY). For analysis of real-time migration, time-lapse movies were generated from zCREST1:membRFP; pGFP5.3 transgenic embryos. Live embryos were mounted dorsal-side down in glass-bottom Petri dishes and imaged on an inverted SP5 confocal microscope (Leica Microsystems Inc., Bannockburn, IL). Z-stacks were captured every 3–8 minutes for up to 2.5 hours. A total of 6 movies from WT specimens and 6 movies from Pk1b-deficient specimens were used for the analyses presented in this study.
Cell measurements
All cell measurements were made using ImageJ software (W.S. Rasband, http://rsb.info.nih.gov/ij/), and analyzed in Microsoft Excel, GraphPad Prism (GraphPad, La Jolla, CA), and/or Oriana (Kovach Computing Services, Anglesey, Wales). Statistical significance was determined by one-way ANOVA, with a p value of 0.05 or less considered significant. Circular data were plotted on rose diagrams using RockWorks (RockWare, Golden, CO), and statistical significance was determined using Watson’s U2 test and/or Watson-Williams F-test.
To measure the number, length, and direction of cell protrusions, protrusions were traced from flattened time-lapse Z-stacks in ImageJ. Direction was defined as the angle relative to the embryonic anterior-posterior axis. To measure the movements of individual cells, individual cells were tracked in flattened time-lapse Z-stacks using ImageJ. For each time point, the center of the cell body was marked (protrusions were ignored in determining the cell body center), and its position was measured relative to the point of facial nerve exit from r4. Average speed and posterior-directed velocity were calculated.
Fixed embryos were analyzed at 18 hpf and 24 hpf, and the axis of elongation and the length-width ratio (LWR) were determined. To measure the elongation axis, a line was drawn through the cell body, bisecting it along its longest axis. The angle of that line, relative to the anterior-posterior axis of the embryo, was determined. To measure LWR, a line was drawn through the cell body, bisecting it along its longest axis. A bisecting line was also drawn, perpendicular to the first. The line that most closely aligned with the anterior-posterior axis was defined as the “length,” while the perpendicular line was defined as the “width.”
To determine MTOC position, Z-stacks of γ-tubulin stained, fixed embryos were analyzed. A bisecting line was drawn from the centrosome to the opposite end of the cell, and the angle relative to the anterior-posterior axis was determined.
Morpholinos and Antibodies
Splice-blocking morpholinos against Pk1b were injected as previously described (Rohrschneider et al. 2007). The following primary antibodies were used for this study: mouse anti-γ-tubulin (Sigma-Aldrich #T6557, St. Louis, MO), rabbit anti-RFP (Millipore #AB3216, Billerica, MA), mouse anti-PIP3 (Invitrogen #A-21328; Carlsbad, CA), rabbit anti-PKCζ (Santa Cruz Biotechnology #sc-216; Santa Cruz, CA) and rabbit anti-EphA4 (kindly provided by Dr. David Wilkinson). For immunolabeling, embryos were fixed in 4% paraformaldehyde for 1–2 hours at room temperature, and processed as previously described (Prince et al. 1998).
Supplementary Material
A, B:Lateral views of 24 hpf WT (A) and Pk1b morphant (B) embryos.
A, B, E, F: Dorsal views of fixed zCREST1:membRFP transgenic embryos antibody stained for PKCζ (A, E) and PIP3 (B, F). C, D, G, H: Transverse views of fixed -zCREST1:membRFP transgenic embryos antibody stained for PKCζ (C, G) and PIP3 (D, H). Embryos were sectioned through r5 (C, D) or r4 (G, H). Scale bar = 20 μm.
Time-lapse movie of control zCREST1:membRFP transgenic embryo, beginning at 20 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of Pk1b morphant zCREST1:membRFP transgenic embryo, beginning at 20 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of control zCREST1:membRFP transgenic embryo, beginning at 24 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of Pk1b morphant zCREST1:membRFP transgenic embryo, beginning at 24 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Acknowledgments
We thank Dr. Cecilia Moens for providing the zCREST1:membRFP construct, Dr. David Wilkinson for providing EphA4 antibody, Dr. Edwin Ferguson for other reagents, as well as Ru Yi Teow and Dr. Vytas Bindokas for assistance with microscopy. We are indebted to Dr. Richard Fehon for comments on the manuscript, Dr. Anand Chandrasekhar for sharing unpublished data, and members of the Prince lab for discussion and fish care.
Funding
This work was funded by NSF Graduate Research Fellowships to OMM and MRR, and a March of Dimes grant (FY07-410) to VEP, and an NIH training grant to the University of Chicago (5T32GM007197). This research was also supported by a grant from the University of Chicago Imaging Institute and the University of Chicago CTSA (National Center for Research Resources #UL1RR024999).
References
- Bellion A, Baudoin JP, Alvarez C, Bornens M, Metin C. Nucleokinesis in tangentially migrating neurons comprises two alternating phases: Forward migration of the Golgi/centrosome associated with centrosome splitting and myosin contraction at the rear. J Neurosci. 2005;25:5691–5699. doi: 10.1523/JNEUROSCI.1030-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. The zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Concha ML, Takeuchi M, Ueno N, Wilson SW, Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: Development of vertebrate branchiomotor neurons. Dev Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Armstrong J, Moens CB. Zebrafish foggy/spt 5 is required for migration of facial branchiomotor neurons but not for their survival. Dev Dyn. 2005;234:651–658. doi: 10.1002/dvdy.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubedo N, Cerdan E, Sapede D, Rossel M. CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons. Mol Cell Neurosci. 2009;40:474–484. doi: 10.1016/j.mcn.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Elsen GE, Choi LY, Prince VE, Ho RK. The autism susceptibility gene met regulates zebrafish cerebellar development and facial motor neuron migration. Dev Biol. 2009;335:78–92. doi: 10.1016/j.ydbio.2009.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura L, Watanabe-Takano H, Sato Y, Tokuhisa T, Hatano M. Prickle promotes neurite outgrowth via the dishevelled dependent pathway in C1300 cells. Neurosci Lett. 2009;467:6–10. doi: 10.1016/j.neulet.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham H, Tanaka T, Brinkman BC, Gleeson JG. GSK3beta and PKCzeta function in centrosome localization and process stabilization during slit-mediated neuronal repolarization. Mol Cell Neurosci. 2006;32:118–132. doi: 10.1016/j.mcn.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–283. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Jessen JR, Topczewski J, Bingham S, Sepich DS, Marlow F, Chandrasekhar A, Solnica-Krezel L. Zebrafish trilobite identifies new roles for strabismus in gastrulation and neuronal movements. Nat Cell Biol. 2002;4:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Guo W, Liang X, Rao Y. Both the establishment and the maintenance of neuronal polarity require active mechanisms: Critical roles of GSK-3beta and its upstream regulators. Cell. 2005;120:123–135. doi: 10.1016/j.cell.2004.12.033. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Komuro H, Rakic P. Distinct modes of neuronal migration in different domains of developing cerebellar cortex. J Neurosci. 1998;18:1478–1490. doi: 10.1523/JNEUROSCI.18-04-01478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- Menager C, Arimura N, Fukata Y, Kaibuchi K. PIP3 is involved in neuronal polarization and axon formation. J Neurochem. 2004;89:109–118. doi: 10.1046/j.1471-4159.2004.02302.x. [DOI] [PubMed] [Google Scholar]
- Metin C, Vallee RB, Rakic P, Bhide PG. Modes and mishaps of neuronal migration in the mammalian brain. J Neurosci. 2008;28:11746–11752. doi: 10.1523/JNEUROSCI.3860-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montcouquiol M, Crenshaw EB, 3rd, Kelley MW. Noncanonical wnt signaling and neural polarity. Annu Rev Neurosci. 2006;29:363–386. doi: 10.1146/annurev.neuro.29.051605.112933. [DOI] [PubMed] [Google Scholar]
- Okuda H, Miyata S, Mori Y, Tohyama M. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth. FEBS Lett. 2007;581:4754–4760. doi: 10.1016/j.febslet.2007.08.075. [DOI] [PubMed] [Google Scholar]
- Picker A, Scholpp S, Bohli H, Takeda H, Brand M. A novel positive transcriptional feedback loop in midbrain-hindbrain boundary development is revealed through analysis of the zebrafish pax2.1 promoter in transgenic lines. Development. 2002;129:3227–3239. doi: 10.1242/dev.129.13.3227. [DOI] [PubMed] [Google Scholar]
- Pocock R, Mione M, Hussain S, Maxwell S, Pontecorvi M, Aslam S, Gerrelli D, Sowden JC, Woollard A. Neuronal function of Tbx20 conserved from nematodes to vertebrates. Dev Biol. 2008;317:671–685. doi: 10.1016/j.ydbio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Prince VE, Joly L, Ekker M, Ho RK. Zebrafish hox genes: Genomic organization and modified colinear expression patterns in the trunk. Development. 1998;125:407–420. doi: 10.1242/dev.125.3.407. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Sakakibara A, Horwitz AF. Mechanism of polarized protrusion formation on neuronal precursors migrating in the developing chicken cerebellum. J Cell Sci. 2006;119:3583–3592. doi: 10.1242/jcs.03080. [DOI] [PubMed] [Google Scholar]
- Sapède D, Rossel M, Dombly-Chaudière C, Ghysen A. Role of Sdf1 chemokine in the development of lateral line efferent and facial motor neurons. Proc Natl Acad Sci U S A. 2005;102:1714–1718. doi: 10.1073/pnas.0406382102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons M, Mlodzik M. Planar cell polarity signaling: From fly development to human disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittaramane V, Sawant A, Wolman MA, Maves L, Halloran MC, Chandrasekhar A. The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev Biol. 2009;325:363–373. doi: 10.1016/j.ydbio.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial- guided neuronal migration. Nat Neurosci. 2004;7:1195–1203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- Song MR. Moving cell bodies: Understanding the migratory mechanism of facial motor neurons. Arch Pharm Res. 2007;30:1273–1282. doi: 10.1007/BF02980268. [DOI] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Multipolar migration: The third mode of radial neuronal migration in the developing cerebral cortex. J Neurosci. 2003;23:9996–10001. doi: 10.1523/JNEUROSCI.23-31-09996.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Serneo FF, Higgins C, Gambello MJ, Wynshaw-Boris A, Gleeson JG. Lis1 and doublecortin function with dynein to mediate coupling of the nucleus to the centrosome in neuronal migration. J Cell Biol. 2004;165:709–721. doi: 10.1083/jcb.200309025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura O, Okada Y, Ando H, Guedj M, Higashijima S, Shimazaki T, Chino N, Okano H, Okamoto H. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev Biol. 2005;278:587–606. doi: 10.1016/j.ydbio.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Umeshima H, Hirano T, Kengaku M. Microtubule-based nuclear movement occurs independently of centrosome positioning in migrating neurons. Proc Natl Acad Sci U S A. 2007;104:16182–16187. doi: 10.1073/pnas.0708047104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos V, Chen P, Spassky N, Qian D, Dabdoub A, Kelley M, Studer M, Guthrie S. Wnt activity guides facial branchiomotor neuron migration, and involves the PCP pathway and JNK and ROCK kinases. Neural Dev. 2009;4:7. doi: 10.1186/1749-8104-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Iwasaki M, Sato T, Masai I, Nishiwaki Y, Tanaka H, Sato A, Nojima Y, Okamoto H. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 2005;132:2273–2285. doi: 10.1242/dev.01810. [DOI] [PubMed] [Google Scholar]
- Wada H, Okamoto H. Roles of noncanonical Wnt/PCP pathway genes in neuronal migration and neurulation in zebrafish. Zebrafish. 2009;6:3–8. doi: 10.1089/zeb.2008.0557. [DOI] [PubMed] [Google Scholar]
- Wada H, Okamoto H. Roles of planar cell polarity pathway genes for neural migration and differentiation. Dev Growth Differ. 2009;51:233–240. doi: 10.1111/j.1440-169X.2009.01092.x. [DOI] [PubMed] [Google Scholar]
- Wada H, Tanaka H, Nakayama S, Iwasaki M, Okamoto H. Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development. 2006;133:4749–4759. doi: 10.1242/dev.02665. [DOI] [PubMed] [Google Scholar]
- Ward ME, Jiang H, Rao Y. Regulated formation and selection of neuronal processes underlie directional guidance of neuronal migration. Mol Cell Neurosci. 2005;30:378–387. doi: 10.1016/j.mcn.2005.08.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A, B:Lateral views of 24 hpf WT (A) and Pk1b morphant (B) embryos.
A, B, E, F: Dorsal views of fixed zCREST1:membRFP transgenic embryos antibody stained for PKCζ (A, E) and PIP3 (B, F). C, D, G, H: Transverse views of fixed -zCREST1:membRFP transgenic embryos antibody stained for PKCζ (C, G) and PIP3 (D, H). Embryos were sectioned through r5 (C, D) or r4 (G, H). Scale bar = 20 μm.
Time-lapse movie of control zCREST1:membRFP transgenic embryo, beginning at 20 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of Pk1b morphant zCREST1:membRFP transgenic embryo, beginning at 20 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of control zCREST1:membRFP transgenic embryo, beginning at 24 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.
Time-lapse movie of Pk1b morphant zCREST1:membRFP transgenic embryo, beginning at 24 hpf. Dorsal view, anterior is to the left. Scale bar = 20 μm.