Abstract
To formally explore the potential therapeutic effect of histone deacetylase inhibitors (HDACIs) and DNA-methyltransferase inhibitors (DNA-MIs) on sarcomas, we treated a large sarcoma cell line panel with five different HDACIs in the absence and presence of the DNA-MI decitabine. We observed that the IC50 of each HDACI was consistent for all cell lines while decitabine as a single agent showed no growth inhibition at standard doses. Combination HDACI/DNA-MI therapy showed a preferential synergism for specific sarcoma cell lines. Subsequently we identified and validated (in vitro and in vivo) a two gene set signature (high CUGBP2; low RHOJ) that associated with the synergistic phenotype. We further uncover that the epigenetic synergism leading to specific upregulation of CDKI p21 in specific cell lines is a function of the differences in the degree of baseline chromatin modification. Finally, we show that these chromatin and gene expression patterns are similarly present in the majority of high grade primary sarcomas. Our results provide the first demonstration of a gene set that can predict responsiveness to HDACI/DNA-MI and links this responsiveness mechanistically to the baseline chromatin structure.
INTRODUCTION
Epigenetic modifying agents such as DNA-methyltransferase inhibitors (DNA-MIs) and histone deacetylase inhibitors (HDACIs) are independently FDA approved for the treatment of hematopoietic malignancies (e.g. myelodysplastic syndromes and cutaneous-T-cell lymphomas, respectively) (1). However, there is no obvious mechanistic link between epigenetic modification and either of these two tumor types that would necessarily exclude other cancer subtypes from responding based on a similar concept of epigenetic modulation, be it global or specific. Yet early clinical trial work with these agents in solid tumors has not been as successful as in hematopoietic tumors (2). To further rationally develop a therapeutic strategy for epigenetic therapy in solid tumors we focused on sarcomas. We chose sarcomas since these solid tumors share many developmental features with hematologic malignancies: (1) Both are mesodermally derived (3, 4); (2) both are believed to arise from developing progenitor cells (mesenchymal stem cells and hematopoietic stem cells, respectively) (5–9); and (3) subtypes of both harbor characteristic gene-fusions (e.g. BCR-ABL in CML (10) and COL1A1-PGFRa in dermatofibrosarcoma protuberans (4)).
As for many other solid tumors, the state of genomic methylation of sarcomas has been partially examined. Synovial sarcomas and osteosarcomas have both been reported to have global changes in DNA methylation; the former most likely due to the activity of its characteristic fusion-gene product, SYT-SSX (7, 11), and the latter as part of a complex karyotype pattern (12). Uterine leiomyosarcomas have characteristic suppression of BRCA1 via DNA promoter methylation (13). Additionally DNA promoter methylation of CDKN2A (p16), MST1, MST2, and RASSF1A in various other sarcoma subtypes have all been reported (14, 15). However, as is apparent from this brief summary, rarely has the DNA methylation pattern been consistent across sarcoma subtypes. Global histone acetylation patterns have not yet been examined.
Consistent with the notion that reversal of DNA-methylation silenced characteristic tumorsuppressor genes may result in therapeutic yield, both DNA-MIs 5-azacytidine (vidaza) and 5-aza-2′-deoxycytidine (decitabine) have been tried in pre-clinical sarcoma studies. While in all reported cases the addition of a DNA-MI was able to reverse expression of the initially DNA-methylation suppressed gene, the effect of that reversal was either not examined when studied in rhabdomyosarcomas (16), found to result in minor apoptosis when studied in osteosarcomas (17), or extremely dependent on time of exposure and choice of cell line (18) even within the same sarcoma subtype. In contrast, pre-clinical data indicates that treatment of multiple sarcoma subtypes with multiple HDACIs demonstrates significant cellular growth inhibition at (depending on HDACI) doses therapeutically active in hematologic malignancies (19–24). Combination DNA-MI/HDACI pre-clinical studies have shown marked synergism for the combination in some specific sarcoma cell lines and/or mouse models (25–27). However, there remains a significant lack of consistency of the response even within cell lines from the same subtype of sarcoma which must be overcome for this therapy to develop more effectively at the clinical level.
In this report we extend the field of epigenetic therapy to solid tumors focusing on sarcomas. We formally explore the potential therapeutic effect of HDACIs and DNA-MIs alone and in combination on a large sarcoma cell line panel and build on the observed responses to a) identify a gene signature that predicts epigenetic synergism in sarcomas and b) propose a mechanism by which this synergism is selectively mediated in specific situations.
MATERIALS AND METHODS
Cell lines
A complete description, characterization and authentication of all cell lines used (including primary source from which they were obtained; either ATCC or primary human sarcoma tissue) within this manuscript were recently reported by us (28). Briefly, MG63, U2OS, HT1080, SAS2, Sw1353, & SW982O were obtained from ATCC and authenticated by short tandem repeat analysis. MFH & LS141 from Sam Singer (MSKCC) and SKUT1, SKUT1B, LMS, and AX from Eva Hernando (NYU) as described (28). Once thawed, all cell lines are routinely tested for Mycoplasma and authenticated through conformity to their baseline analysis of cell morphology monitoring and growth curve analysis as defined by us (28) upon original acquisition.
MTS assays
Detailed information on cell lines, growth conditions, and properties have all been previously described by us (28). MTS assays: On day 0, 5000 cells in 100ul were plated per well in 96-well plate. At 24hrs, drug (or solvent as control) was added in increasing concentrations and/or varying combinations to a total of 200ul of media per well in triplicates. On day 4, 80ul of media was removed from each well, and 20ul of CellTiter 96 Aqueous One Solution Assay (Promega) were added to each well. Plate was incubated at 37 C for 1 hour and then read at 490nm on a plate reader. Cell viability was calculated by dividing treated well absorbance by control well absorbance, with control arbitrarily defined at “1” or 100%. Romidepsin, LBH589, and Belinostat were kindly provided by Owen O’Connor (NYU). Valproate and decitabine were purchased from Sigma and SAHA from Toronto Research Chemicals.
In vivo studies
1 million sarcoma cells in 250ul phosphate buffered saline were resuspended in 250ul Matrigel (BD Bioscience) and introduced subcutaneously in the flank of NOD/SCID mice (Jackson Labs). Treatments were initiated when tumors reached a palpable nodule (4-5mm in greatest dimension or approximately 20 mm2 in cross sectional area). Mice were treated on the following schedule: M, W, F via intraperitoneal injections for approximately three weeks, SAHA: 50mg/kg, valproate: 300mg/kg, Dec: 1mg/kg. Experiments were deemed concluded when the untreated mouse xenograft controls reached the largest cross-sectional diameter of 10–12mm (or a tumor cross-sectional area of 100mm2).
The mice were randomized to no treatment (control), or treatments as indicated in the results. Each experimental group consisted of five animals. The entire experiment as depicted in the manuscript was performed three times, thus 15 animals were included per arm in the final analysis depicted in Figure 2. The size of tumors were measured individually twice per week with microcalipers. Tumor size was calculated as a cross-sectional area defined as length × width. The individual relative tumor size (RTS) was calculated as the ratio of the initial tumor at which therapy was initiated to the tumor size on a given day. Statistic analyses were performed using one-way analysis of variance for comparison of tumor growth in different treatment groups.
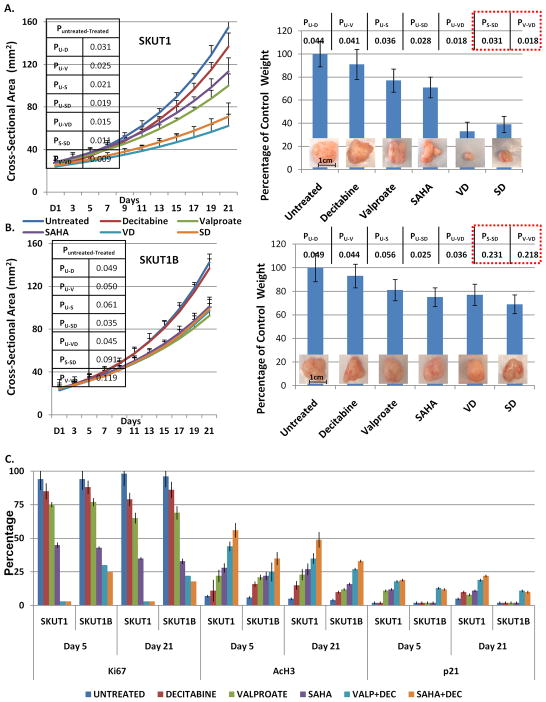
Figure 2.
(A, B) Growth curves representing changes in size of tumor volumes as a function of time and the indicated treatment in the indicated cell lines with graphical representation and actual visualization of tumor sizes upon conclusion of experiments. 15 animals were analyzed per treatment arm. Representative results are shown. C) Qualitative differences in histology and immunohistochemistry is shown. Quantitative assessment of the qualitative immunohistochemial findings (please see Materials and Methods for details), confirming that the differences observed in terms of response to combined epigenetic therapy were only statistically significant for p21 and only in the SKUT1-xenografts; thus confirming via analysis of epigenetically regulated genes the specificity of the HDACI/DNA-MI synergy for SKUT1 and not SKUT1B xenografts. High resolution IHC pictures are provided in Supplemental Figures 6–9. Statistical significance of each difference isindicated in the inserted tables. All analyses are based on day 21 xenografts.
All mice were sacrificed within 24 hours of final treatment in accordance with Columbia University Animal Welfare and IUCAC policy under IRB protocol AAAD9669 and all tumors were surgically excised. Tumors were fixed in 10% formalin, embedded in paraffin and stained with hematoxylin and eosin as previously described (28). To quantitatively assess the qualitative immunohistochemical findings, three pathologists with significant sarcoma expertise (Carlos Cordon-Cardo MD PhD, Mireia Castillo-Martin MD PhD, and Fabrizio RemottiMD, Columbia University) independently scored all a minimum of three sections from each xenograft for percentage of positively staining cells for each marker. Hematoxylin and eosin was graded as percentage cellularity per visualized field. Averages of all were scores obtained and plotted.
Immunoprecipitation-Immunoblotting (IP-IB), Immunohistochemistry (IHC)
All protein gels were performed as sequential IPs-IBs using the same antibody for both steps to increase sensitivity of detection as previously described (29). Antibodies: AV40323 (Sigma) Anti-CUGBP2; (Abcam) Mouse Anti-RHOJ Monoclonal Antibody. IHC was performed as previously described by us using the following antibodies: Ki67 (Dako; Cat # M7240), p21/waf1 (Calbiochem, Cat # OP64), AcH3 (Cell Signaling, Cat # 9677S).
qRT-PCR, Chromatin Immunoprecipitation (ChIP)
Total RNA was extracted from the indicated cell lines using an RNA extraction kit (Qiagen) according to the manufacturer’s instructions. 1 μg of RNA was transcribed into cDNA using Super-Script III First Strand Synthesis System for RT-PCR (Invitrogen). To assess the expression levels of CUGBP2, RHOJ, SLC2A13, and SMPDL3A quantitative RT-PCR reactions containing 500 ng cDNA, QuantiTect SYBR Green PCR mix (Qiagen) and corresponding primers were performed on a Stratagene MX3000P QPCR System with ROX reference dye (Stratagene). Expression levels of the abovementioned genes were normalized to expression levels of the β-actin control gene per sample. All experiments were performed twice and each time in triplicate. T-test was used to assess the significance of differences in expression between indicated treatments. Primers sequences were as follows: CUGBP2: CCTTCCACAGGAGTTTGGAGA and TCAGTAAGGTTTGCTGTCGTT (30); RHOJ: GCTACGCCAACGACGCCTTC’ and TTGAGCTCGGGGACCCATTC (31); and β-actin as previously described by us (6). ChIP was performed as previously described by us (6) using the following p21 promoter primers: 5′-ACCAAC GCAGGCGAGGGACT-3′ and 5′-GGT GTCTAGGTGCTCCAGGT-3′ (32).
Gene Expression Analysis
RNA from the indicated cell lines were hybridized to HG U133 Plus 2.0 oligonucleotide arrays per standard protocols of the Columbia Genomics Core Facility (described in detail in (33)). Class-comparison analysis using two-sided Student t-tests identified mRNAs that were differentially expressed between indicated samples (p<0.05). Cluster analysis was done with Cluster 3.0 (http://linus.nci.nih.gov/BRB-ArrayTools.html ) and displayed using TreeView program (http://rana.lbl.gov/EisenSoftware.htm). Raw data will be deposited in the public repository. Class prediction analysis was performed using BRB Array Tools as described in the supplemental.
Statistical analyses
IC50 values were calculated independently by both BioDataFit and CompuSyn software. Data is presented as mean ± SD (in vitro) or mean ± SE (in vivo), and significance was assessed with Student’s t-test. Differences in measurements were considered significant at p < 0.05.
Results
Selective Epigenetic Synergism
To explore the potential for different classes of epigenetic agents to synergize we treated a 13 sarcoma cell line panel with five HDACIs (i.e., Romidepsin, LBH589, Belinostat, SAHA, and valproate) in the presence or absence of 1uM of the DNA-MI 5-aza-2′deoxycytidine (decitabine). [Please see two recent reviews, Mai et al and Mercurio et al (34, 35), for structural and developmental history of these drugs.] Our 13 sarcoma cell lines were previously comparatively characterized and defined by us to have similar proliferative patterns (28) thus a priori discounting the possibility that differences in observed growth inhibitory effects may be attributed to differential proliferative patterns of the cell lines. All 13 sarcoma cell lines treated with the indicated HDACI exhibited little inter-cell line variation. The IC50 as calculated independently using BioDataFit and CompuSyn software were as follows: Romidepsin IC50≈ 20nm; LBH589≈50nm; Belinostat 1uM; SAHA≈5uM; valproate≈25mM using MTS as a cell growth assay and are in agreement with the IC50s of these agents for other cancer cell lines (Supplemental Table 1). These results were independently confirmed using crystal violet staining of treated cells (data not shown). Interestingly, treatment with decitabine or another DNA-DI, 5-azacytidine (vidaza) (data not shown) did not result in any detectable growth inhibition at doses as high as 10 fold higher than those necessary to ensure global DNA demethylation (Supplemental Figure 1).
Four of the 13 cell lines (MG63, MFH, SKUT1, and U2OS) showed clear synergism (compare bar pairs, Figure 1A) for all HDACIs when combined with 1uM decitabine, further referred to as Epigenetically Synergistically Responsive (ESR) cell lines; while four other cell lines did not (HT1080, SKUT1B, LMS, and LS141; Figure 1A) as observed from the fractional decrease in cell growth comparing each HDACI at a given dose in the absence or presence of decitabine. Five additional cell lines (OSA, RH30, SW872, HOS, and SKNEP) showed variable synergistic responses (data not shown). In all cases, the observed synergism was confirmed using Median Effect Equation of Chou via CompuSyn Software (36–38) (data not shown).
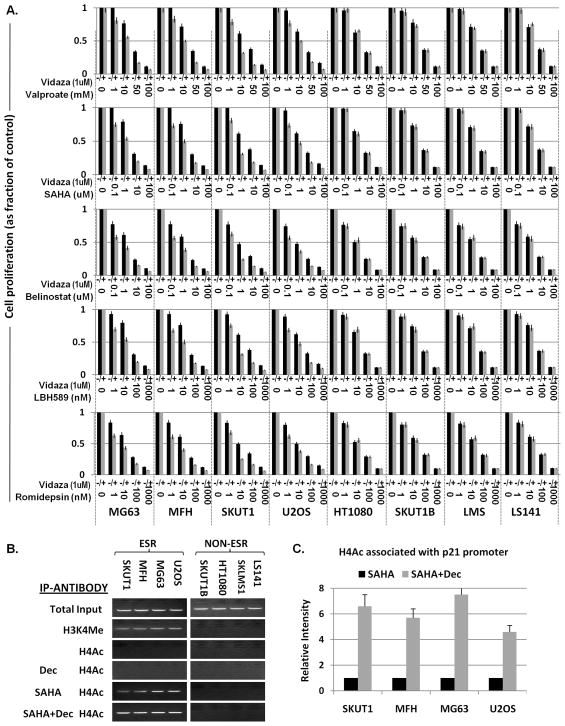
Figure 1.
(A) MTS/cell viability assays of the indicated cell lines treated with the indicated epigenetic agents. The Y-axis represents “cell proliferation” plotted as a fraction of the proliferation of the untreated cell line. The X-axis represents the individual cell lines treated with the indicated epigenetic agents at the dose ranges and combinations indicated on the graphs. All cell lines were treated for 72 hours. Each point was assayed three independent times. Average is shown and error bars represent standard deviation at each point. (B) ChIP assay using the indicated antibodies following the indicated treatment for detection of binding the p21 promoter. H3K4=Histone H4 Lysine 4 Trimethylation; H4Ac= Histone 4 Acetylation (C) Quantitative PCR comparing p21 promoter association following SAHA vs SAHA+Dec treatment from (C). Each sample and its controls were repeated three times in the respective assay. The values were averaged and graphed. Standard deviation bars are shown. D=Decitabine, V=Valproate, S=SAHA, B=Belinostat, L=LBH589, R=Romidepsin. ESR= Epigenetically Synergistically Responsive; NON-ESR= Non Epigenetically Synergistically Responsive Cell Lines.
In each case, and regardless of synergistic activity, HDACI and DNA-MI activity was independently confirmed (see Supplemental Figures 1,2 and accompanying text).The enhancement of HDACI mediated growth inhibition by the ‘cytotoxically’ inactive agent decitabine meets the formal definition of synergism (37). Finally, treating all sarcoma cell lines with the indicated HDACI in the absence and presence of decitabine showed that on average there is a statistically significant decrease in the HDACI IC50 (ranging from 20-80% depending on the HDACI) in the presence of decitabine (data not shown). This analysis provides rationale to suppose that even an unselected group of patients may benefit from such treatment.
To examine whether the decrease in cellular growth observed in the presence of epigenetic agents was cytostatic or cytotoxic, we performed crystal violet staining (data not shown), viability (trypan blue exclusion), cell cycle, and caspase activity assays on all cell lines using the five HDACIs (at a wide spectrum of doses) in the presence and absence of decitabine. Our results indicate that at the IC50 of each respective HDACI primarily corresponds to a G1-arrest, while at doses higher than the IC50 significant apoptosis is observed (Supplemental Figures 3, 4 and 5 and accompanying text).
Epigenetic synergism is associated with baseline chromatin modification
Our preliminary analysis did not reveal any differences in either baseline global histone acetylation or global DNA-methylation comparing ESR to non-ESR cell lines (Supplemental Figures 1, 2). To explore the possibility that gene specific, rather than global, epigenetic modification may account for the observed effect, we sought to address whether the ESR cell lines may have in general a more active or open chromatin; thus being more accessible to epigenetic agents. Since our data indicated that DNA-MIs were augmenting an HDACI mediated decrease in cellular growth, we focused on examining the status of p21, a well-studied cell cycle inhibitor known to be responsive to HDACIs (32), in ESR and non-ESR cell lines. We first specifically assessed if p21 associated with the presence of the classical ‘histone 3 lysine 4 trimethyl’ (H3K4me) active mark by performing chromatin immunoprecipitation (ChIP) using an anti-H3K4 antibody to precipitate nucleosomes and assessed for the presence of the p21 promoter (32). As could be seen in Figure 1B, only the p21 promoter in the ESR cell lines was marked by active H3K4me indicative of an open chromatin pattern. We also examined the repressive marks H3K9 and H3K27 (39, 40). We found mixed results in that there does not seem to be a direct relationship between either of these two repressive marks and the pattern of ESR versus non-ESR cell lines at the p21 promoter (data not shown).
Since HDACIs are able to active the p21 promoter, we further sought to determine the acetylation status of the p21 promoter. As could be seen in Figure 1B, a similar ChIP experiment using an antibody directed against acetylated Histone H4 (anti-H4Ac) indicated that at baseline the p21 promoter is not acetylated in either ESR or non–ESR cell lines (Figure 1B, fourth pair of panels from the top). Treatment of the eight cell lines with the HDACI SAHA (at the previously determined IC50 for 24 hours) and subsequent ChIP using an anti-H4Ac antibody indicated that acetylated H4 now associated with the p21 promoter, but only in the ESR cell lines (Figure 1B, fifth pair of panels from the top). Finally, treatment of all eight cell lines with SAHA and decitabine similarly resulted in association of only the ESR cell lines of p21 with acetylated histones (Figure 1B, sixth pair of panels from the top). As a control decitabine alone was not able to induce changes in the association of p21 with the acetylated Histone H4 (Figure 1B, third pair of panels from the top).
A quantitative analysis comparing SAHA treated vs. SAHA/Dec treated ESR cell lines in terms of relative amount of acetylated histones associating with p21 (Figure 1C) indicated that the combination therapy is able to dramatically increase the amount of acetylated histones associating with the p21 promoter. These observations indicate that mechanistically a baseline active or open chromatin structure at specific genes and/or their promoters facilitates epigenetic agents from activating those promoters and thus explains the preferential upregulation of specific genes in specific cell lines and the resultant phenotypic effect via treatment of epigenetic agents.
Synergistic Epigenetic Treatment is Active In Vivo
To assess if our in vitro observations translated to the in vivo setting, we chose tumorigenic cell lines that, based on our in vitro data, were either an ESR cell line (SKUT1) or non-ESR cell line (SKUT1B). Both of these are morphologically similar leiomyosarcoma cell lines (28) that also share a high degree of gene expression similarity (discussed below), but experimentally differ greatly in epigenetic responsiveness. NOD/SCID mice were subcutaneously inoculated with 1 million cells. Treatments were initiated with SAHA, valproate, or decitabine alone and SAHA or valproate in combination with decitabine when tumors reached a palpable nodule (4–5mm in greatest dimension). Mice were treated on the following schedule: M, W, F via intraperitoneal injections for 3 weeks, SAHA: 50mg/kg, valproate: 300mg/kg, Dec: 1mg/kg. Mice were observed until the untreated mouse xenograft controls reached the largest cross-sectional diameter of 10–12mm (or a tumor ‘cross-sectional area = 100mm2).
As seen in Figure 2A the combination of HDACIs with decitabine was much more effective than any of the single agent treatments and furthermore that the combination treatment was more effective than single agent treatment to a statistically significant degree (see table insert Figure 2A). Final analysis of xenografted tumors and comparative analysis by weight confirmed the results obtained from cross sectional measurements (Figure 2A right). Similar results, especially the benefit of double epigenetic agent treatment, were NOT observed for SKUT1B-derived xenografts (Figure 2B). Final pathology showed no tumor necrosis in any samples suggesting a cytostatic rather than cytotoxic effect of the treatments (hematoxylin and eosin staining, Supplemental Figure 6); in agreement with our in vitro observations at IC50 doses. Ki67, a reliable marker of sarcoma cell proliferation (see Supplemental Figure 7 and text) decreased in response to both valproate and SAHA and in both cell lines. However when SAHA or valproate was combined with decitabine, Ki67 staining cells were rarely found in SKUT1 xenografts by day five, and completely absent on day 21. In contrast, SKUT1B cells continued to have Ki67 staining positive cells even at day 21 of treatment, although those levels were significantly reduced as compared to untreated samples (Figure 2C, compare SKUT1 vs. SKUT1B Day5 and Day 21 yellow (SAHA+Dec) and light blue (Valproate+Dec) bars, and Supplemental Figure 7, green and red boxes). Interestingly, in both SAHA and valproate treated xenografts the acetylation of histone H3 was notably increased by the addition of decitabine, but more so in SKUT1 as opposed to SKUT1B xenografts (Figure 2C and Supplemental Figure 8, compare SKUT1 vs. SKUT1B Day 5 and Day 21 (yellow SAHA+Dec) and light blue (Valproate+Dec) bars.)
To further explore the mechanism by which SKUT1 cells are preferentially growth inhibited in the presence of HDACIs/DNA-MIs in our xenografts, we once again turned our attention to p21. We noted that p21 was upregulated by both HDACIs (but not by decitabine) and only in SKUT1 cells (Figure 2E and Supplemental Figure 9, purple box). Furthermore, only SKUT1 cells were able to show a synergistic response as noted by further increase in p21 in the presence of combined HDACI/DNA-MI (Figure 2E and supplemental Figure 9; compare green and red boxes).
A Two-Gene Pattern Can Cluster Epigenetic Synergistic Responders vs. Non-responders
To identify genes and/or a gene pattern that may a priori predict which sarcoma cell lines may show a synergistic response to HDACIs and DNA-MIs, we extracted total cellular RNA from the four ESR cell lines (i.e., MG63, MFH, SKUT1 & U20S) and the four non-ESR cell lines (i.e., HT1080, SKUT1B, LMS & LS141); and labeled and hybridized the RNA to Affymetrix U133.0 Plus 2.0 Arrays in conjunction with the Columbia Genomics Core. Unsupervised hierarchical clustering showed no inherent synergistic related clustering pattern of the eight cell lines (Figure 3A). We then used multiple predictive algorithms (Supplemental Table 2) to identify genes that not only strongly associate with the ESR (cell lines shown in green)/non-ESR (cell lines shown in red) phenotype but may also identify untested samples for appropriate synergism. A two gene set was identified consisting of CUGBP2 and RHOJ. [Of note using ‘Significance Analysis of Microarrays’ (SAM) comparing ESR cell lines to the non-ERS cell lines identified four genes to be significantly differentially expressed: CUGBP2, RHOJ (as identified by the predictive algorithms), and SLC2A13, and SMDDPL3A. Differences in the latter two could not be validated either by RT-PCR or immunoblot analysis (data not shown) and not further pursued.]
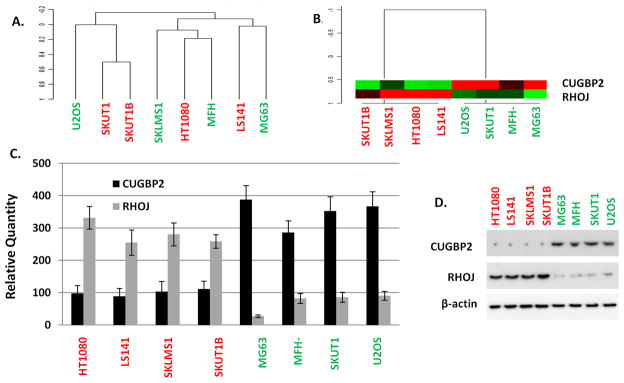
Figure 3.
(A) Unsupervised hierarchical clustering of the indicated samples using a full U133.0 Plus2 gene set. (B) Unsupervised hierarchical clustering of the indicated samples using the indicated two-gene set. (C) qRT-PCR of the indicated genes in the indicated cell lines. (D) Immunoprecipitation-Immunoblot analysis of the indicated proteins from total cellular protein extracts obtained from the indicated cell lines. Red colored cell lines = Non-ESR (Non Epigenetically Synergistically Responsive Cell Lines); Green colored cell lines = ESR (Epigenetically Synergistically Responsive Cell Lines).
Unsupervised hierarchical clustering using the two-gene predictive set was able to cluster the samples by the experimentally determined phenotype - ERS/non-ERS (Figure 3B). We went on to confirm that CUGBP2 and RHOJ are significantly differentially expressed between ESR and non-ESR cell lines using RT-PCR and immunoblot analysis (Figure 3C, D). Similar analysis of the cell lines having a mixed response (data not shown) to combined HDACI/DNA-MI therapy (i.e., SKNEP, SW872, HOS, OSA143, RH30) showed lack of differential expression of these genes between the samples and absence of the inverse patterns of CUGBP2 and RHOJ (Supplementary Figure 10A, B) that appeared to define the ESR versus the non-ESR cell lines.
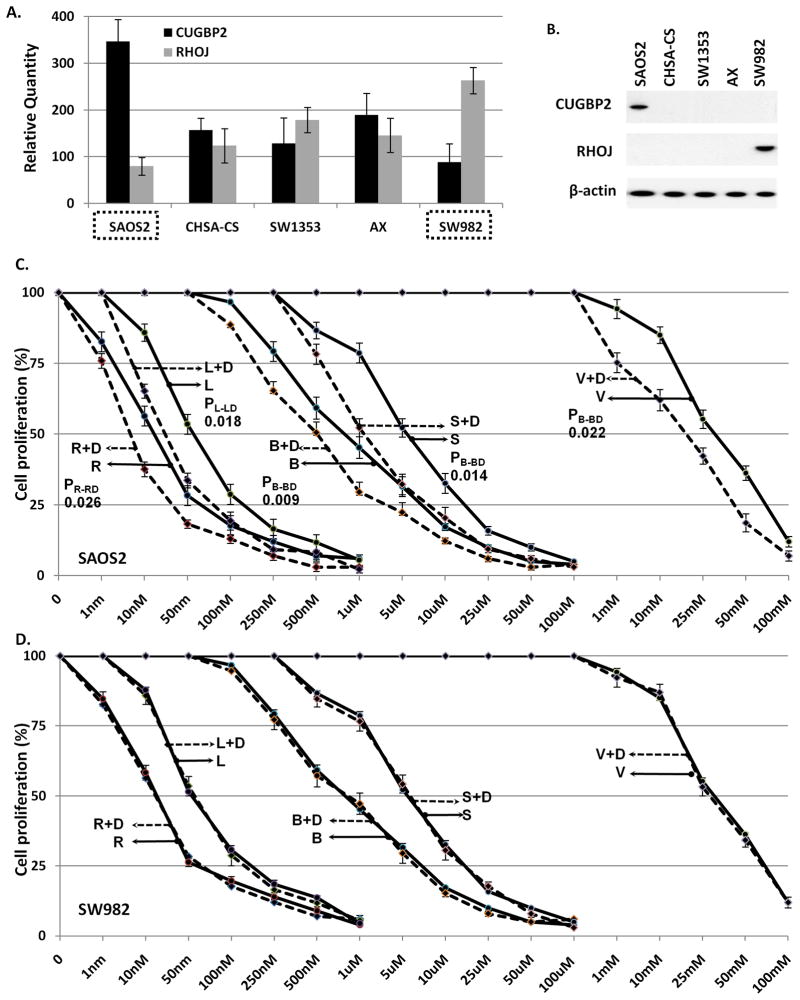
We subsequently screened an additional five sarcoma cell lines (CHSA-CS, SAOS2, SW982, SW1353, AX) from our collection (28) attempting to identify sarcoma cell lines that have inverse patterns of CUGBP2 and RHOJ. SAOS2 was identified as a cell line that had relatively high levels of CUGBP2 and low levels of RHOJ at both the RNA and protein levels suggesting that it might be a cell line that would respond to synergistic epigenetic treatment; SW982 showed the inverse pattern, suggesting a pattern similar to non-ESR cell lines (Figure 4A, B). To formally assess the responsiveness of SAOS2 and SW982 to combinations of HDACIs and DNA-MIs, we treated these two cell lines with the five HDACIs (as outlined for Figure 1 above) either alone or in combination with decitabine (at 1uM). SAOS2 showed a decrease in the HDACIs IC50 in the presence of decitabine while SW982 did not (Figure 4C, D). Taken together this data suggests that inverse levels CUGBP2 and RHOJ can correlate sarcoma cell lines that are either ESR or non-ESR.
Figure 4.
(A) qRT-PCR of the indicated genes in the indicated cell lines. (B) Immunoprecipitation-Immunoblot analysis of the indicated proteins from total cellular protein extracts obtained from the indicated cell lines. (C, D) MTS/cell viability assays of the indicated cell lines treated with the indicated epigenetic agents. The Y-axis represents “cell proliferation” plotted as a fraction of the proliferation of the untreated cell line. X-axis represents HDACI concentration.
Next we assessed the changes to CUGBP2 and RHOJ following epigenetic therapy. CUGBP2 decreased and RHOJ increased in response to combined epigenetic therapy (Supplementary Figure 10C, D); however interestingly this reversal occurred only in the original ESR cell lines. To assess if CUGBP2 and RHOJ were markers or effectors of synergistic response we downregulated CUGBP2 in SAOS2 cells and RHOJ in SW982 cells using a small interference RNA approach, in essence reversing the ratio of CUGBP2 and RHOJ levels in these cell lines. Treating each cell line with either siRNAs targeting CUGBP2 or RHOJ was only able to downregulate the intended target while not affecting the non-intended target, thus confirming specificity of the siRNAs (Supplemental Figure 11A, B). Treating the siRNA pre-treated cells with five HDACIs alone and in combination with decitabine yielded almost identical results (Supplemental Figure 11C, D) to those seen in the absence of siRNA treatment. The lack of change to epigenetic treatment in growth inhibition following the reversal of the CUGBP2 and RHOJ ratios in these cell lines suggests that these genes are markers rather than mediators of the epigenetic response.
CUGBP2/RHOJ and H3K4 patterns are recapitulated in primary high grade sarcomas
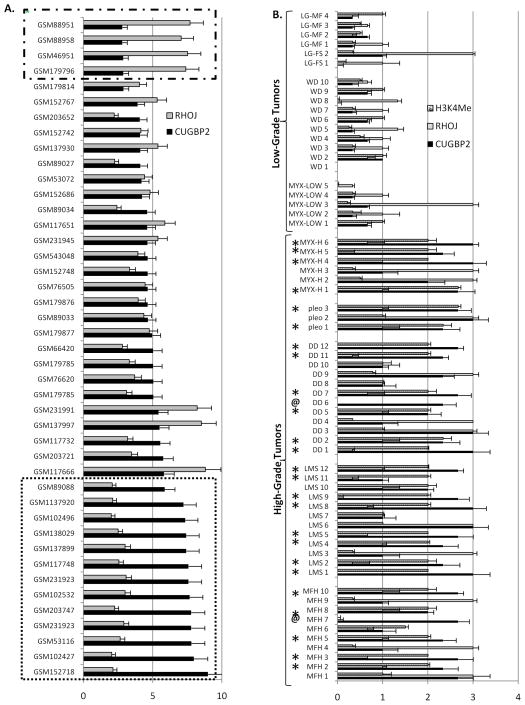
To assess whether our findings are clinically relevant we examined the expression profiles CUGBP2 and RHOJ in a large publicly available sarcoma gene expression database (International Genomics Consortium; https://expo.intgen.org/geo/home.do). As seen in Figure 5A, 13 of 44 (29.5%) primary human sarcomas (bottom dashed box) have a CUGBP2/RHOJ ratio comparable to those seen in the ESR cell lines. Furthermore, an IHC analysis of CUGBP2, RHOJ, and H3K4 levels of an extensive tissue microarray (previously described by us (6, 39)) demonstrates that a high CUGBP2/low RHOJ ratio is detected in 23 of 41 (56.5%) high grade sarcomas (Figure 5B; stars), and correlates tightly with the exception of two cases (Figure 5B; marked “@”) with detectable H3K4. This data suggests that combined HDACI/DNA-MI treatment is likely to be most effective for the more aggressive, high grade sarcomas.
Figure 5.
(A) Gene expression profiles of the indicated samples for the indicated genes of primary human sarcomas. Bottom dashed box =sarcomas with CUGBP2/RHOJ ratio profiles similar to ESR cell lines, Top dashed Box= sarcomas with CUGBP2/RHOJ ratio profiles similar to non- ESR cell lines. Error bars represent standard deviation of the expression of unique Affymetrix IDs corresponding to either CUGBP2 or RHOJ. (B) IHC analysis scored 0–3 of microtissue array. stars (*) = High CUGBP2/low RHOJ ratio and high grade sarcoma, @ = exceptions to pattern. Error bars represent standard deviation of the independent review of three pathologists (see Materials and Methods) of three core replicates.
Discussion
Our data suggests that a) HDACIs are potential potent cytostatic drugs in the treatment of sarcomas; b) a two-gene (CUGBP2, RHOJ) signature can predict the response of sarcoma cells to synergistic (i.e., HDACI/DNA-MI) epigenetic treatment; high CUGBP2/ low RHOJ = responsive; low CUGBP2/ high RHOJ = non-responsive; c) HDACI/DNA-MI mediated synergism is mediated via concurrent activation of the CDKI p21 and subsequent downregulation of CDK4 and G1-arrest; d) mechanistically, cell line specific synergism is dependent on a gene specific, not global, baseline DNA and histone epigenetic modification pattern; and e) combined HDACI/DNA-MI would be most effective for the more aggressive, high grade sarcomas.
Our conclusion that a two gene signature can predict epigenetic agent synergism is based on the following four points: 1) gene expression comparisons of cell lines that were experimentally determined to exhibit HDACI/DNA-MI synergism to those that did not identify CUGBP2 and RHOJ as two of the four differentially regulated genes and the only two genes who also display the inverse pattern at both the qRT-PCR and immunoblot levels; 2) gene expression analysis of cell lines that showed mixed responses to combination therapy did not exhibit the CUGBP2/RHOJ inverse pattern; 3) a screen of additional sarcoma cell lines identified one cell line with high CUGBP2–low RHOJ, and one cell line with low CUGBP2–high RHOJ; both cell lines responded to synergistic epigenetic treatment as predicted, thus validating our initial observations; and 4) in vivo treatment of xenografted cell lines displaying responsiveness/non-responsiveness to synergistic treatment, responded similarly in vivo. Furthermore, our data would suggest that the inverse pattern of CUGBP2/RHOJ is a predictive marker of response rather than the mediator of that response. This conclusion is based mainly on our observations that siRNA knockdown strategies either against CUGBP2 in an attempt to downregulate CUGBP2 in a cell line (SAOS2) that is initially responsive to epigenetic synergism in order to make it non-responsive; or vice versa, siRNA against RHOJ, in an attempt to make a cell line that is initially non-responsive to epigenetic treatment become sensitive did not result in the intended effect. It is of course possible that reversing the ratios is not sufficient and what is really needed is absolute levels; in other words, for example, not only does CUGBP2 need to be reduced, but RHOJ needs to be exogenously overexpressed in SAOS2 cells to make SAOS2 cells non-responsive to epigenetic synergy. Functionally, relatively little is known about these genes. RHOJ is a poorly studied RAS homologgene family member (40) and CUGBP2 is the CUG triplet repeat, RNA-binding protein 2 involved in RNA splicing (30, 41). Ingenuity pathway analysis did not identify any potential link direct or remote (data not shown). At this point the lack of functional association between these two genes and the fact that neither one alone was able to even subtly change the responsive pattern to epigenetic treatment argues against these two genes serving as mediators of the epigenetic synergism.
From a mechanistic point of view our data indicates that HDACI/DNA-MI mediated synergism is mediated via concurrent activation of the CDKI p21. Our data directly shows that p21 is upregulated via HDACIs and occurs in only those cells that have an open or active chromatin structure (as identified by the H3K4 mark). In cells not having this active mark, HDACIs appear to be incapable of upregulating p21. Upregulation of p21 appears to be more effective in decreasing CDK4 activity and likely explains the growth inhibitory effect observed in our MTS assays.Simultaneous treatment with both HDACIs/DNA-MIs increases the overall CDKI level to those needed to significantly inhibit CDK4 activity (and possibly others) and lead to G1-arrest.
Perhaps most interesting is that our observations explain the mechanism underlying the specific cell line preferential response to combined epigenetic therapy. To this point it is possible that the observed synergism of HDACIs and DNA-MIs may be explained by the independent and additive upregulation of p16 and p21. However, our observations that combined HDACI/DNA-MI treatment was specifically able to increase the amount of acetylated histones associated with the p21 promoter suggests (Figure 1) that HDACIs and DNA-MIs synergize mechanistically on the promoter of their targets (as previously reviewed by us (42) and others (43, 44)). Our observation that this only occurs in cell lines with open or active chromatin structure further leads to the mechanistic understanding that cell lines with chromatin configurations permissive or accessible to epigenetic modifying agents are the cell lines/samples that are likely to benefit from this treatment.
Finally, through an examination of primary human sarcomas we demonstrate that a high CUGBP2/low RHOJ pattern is present in the majority of high grade sarcomas and correlates strongly with H3K4 markings suggesting that the most aggressive tumors would be the ideal targets of combined HDACI/DNA-MI therapies.
In conclusion, in this report we not only describe the cytostatic effect of common epigenetic agents on sarcomas, but also identify genes and propose a mechanism of synergistic action that accounts for the preferential response of specific sarcoma cell lines. We believe that our data adds to our understanding of epigenetic therapy and would be appropriate to be used in designing clinical trials using combination epigenetic therapies as the majority of primary high grade sarcomas have this gene signature. Although there have been several reports looking at genes that might predict responsiveness to either HDACIs or DNA-MIs, to the best of our knowledge this is the first report of genes and a mechanistic understanding that may predict responsiveness to combined epigenetic therapy. Our data further suggests that scrutiny of chromatin structure may itself be used to predict epigenetic responsiveness and that perhaps chromatin configuration itself may be further modifiable to make unresponsive tumors responsive. Although our work has centered on sarcomas, there is no clear reason why these observations may be not be equally applicable to other solid tumors.
Supplementary Material
Acknowledgments
The authors would like to greatly acknowledge Carlos Cordon-Cardo MD PhD for his critical reading, review and general support of this work. We would also like to thank Mireia Castillo-Martin MD PhD and Fabrizio RemottiMD for assistance with validation of pathology review.
Financial Support:
This work was performed in the laboratory of Igor Matushansky and supported by grants to Igor Matushansy from the NIH/NCI, Columbia’s Irving Institute’s Clinical and Translational Science Award and the Gerstner Development Award.
Footnotes
Conflict of Interest: None.
References
- 1.Tuma RS. Epigenetic therapies move into new territory, but how exactly do they work? Journal of the National Cancer Institute. 2009;101:1300–1. doi: 10.1093/jnci/djp342. [DOI] [PubMed] [Google Scholar]
- 2.Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009;15:3970–7. doi: 10.1158/1078-0432.CCR-08-2786. [DOI] [PubMed] [Google Scholar]
- 3.Baron MH. Embryonic origins of mammalian hematopoiesis. Experimental hematology. 2003;31:1160–9. doi: 10.1016/j.exphem.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 4.Matushansky I, Maki RG. Mechanisms of sarcomagenesis. Hematology/oncology clinics of North America. 2005;19:427–49. v. doi: 10.1016/j.hoc.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 5.Matushansky I, Hernando E, Socci ND, Matos T, Mills J, Edgar MA, et al. A Developmental Model of Sarcomagenesis Defines a Differentiation-Based Classification for Liposarcomas. Am J Pathol. 2008 doi: 10.2353/ajpath.2008.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matushansky I, Hernando E, Socci ND, Mills JE, Matos TA, Edgar MA, et al. Derivation of sarcomas from mesenchymal stem cells via inactivation of the Wnt pathway. The Journal of clinical investigation. 2007;117:3248–57. doi: 10.1172/JCI31377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cironi L, Provero P, Riggi N, Janiszewska M, Suva D, Suva ML, et al. Epigenetic features of human mesenchymal stem cells determine their permissiveness for induction of relevant transcriptional changes by SYT-SSX1. PLoS ONE. 2009;4:e7904. doi: 10.1371/journal.pone.0007904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tirode F, Laud-Duval K, Prieur A, Delorme B, Charbord P, Delattre O. Mesenchymal stem cell features of Ewing tumors. Cancer cell. 2007;11:421–9. doi: 10.1016/j.ccr.2007.02.027. [DOI] [PubMed] [Google Scholar]
- 9.LeBien TW, Tedder TF. B lymphocytes: how they develop and function. Blood. 2008;112:1570–80. doi: 10.1182/blood-2008-02-078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar C, Purandare AV, Lee FY, Lorenzi MV. Kinase drug discovery approaches in chronic myeloproliferative disorders. Oncogene. 2009;28:2305–13. doi: 10.1038/onc.2009.107. [DOI] [PubMed] [Google Scholar]
- 11.de Bruijn DR, Allander SV, van Dijk AH, Willemse MP, Thijssen J, van Groningen JJ, et al. The synovial-sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene (de)regulation. Cancer research. 2006;66:9474–82. doi: 10.1158/0008-5472.CAN-05-3726. [DOI] [PubMed] [Google Scholar]
- 12.Sadikovic B, Yoshimoto M, Al-Romaih K, Maire G, Zielenska M, Squire JA. In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma. PLoS ONE. 2008;3:e2834. doi: 10.1371/journal.pone.0002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing D, Scangas G, Nitta M, He L, Xu X, Ioffe YJ, et al. A role for BRCA1 in uterine leiomyosarcoma. Cancer research. 2009;69:8231–5. doi: 10.1158/0008-5472.CAN-09-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, et al. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Molecular carcinogenesis. 2007;46:865–71. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- 15.Takahira T, Oda Y, Tamiya S, Yamamoto H, Kawaguchi K, Kobayashi C, et al. Alterations of the p16INK4a/p14ARF pathway in clear cell sarcoma. Cancer science. 2004;95:651–5. doi: 10.1111/j.1349-7006.2004.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch CA, Tycko B, Bestor TH, Walsh CP. Reactivation of a silenced H19 gene in human rhabdomyosarcoma by demethylation of DNA but not by histonehyperacetylation. Molecular cancer. 2002;1:2. doi: 10.1186/1476-4598-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Romaih K, Sadikovic B, Yoshimoto M, Wang Y, Zielenska M, Squire JA. Decitabine-induced demethylation of 5′ CpG island in GADD45A leads to apoptosis in osteosarcoma cells. Neoplasia (New York, NY) 2008;10:471–80. doi: 10.1593/neo.08174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Numoto K, Yoshida A, Sugihara S, Kunisada T, Morimoto Y, Yoneda Y, et al. Frequent methylation of RASSF1A in synovial sarcoma and the anti-tumor effects of 5-aza-2′-deoxycytidine against synovial sarcoma cell lines. Journal of cancer researchand clinical oncology. 2009 doi: 10.1007/s00432-009-0632-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hrzenjak A, Kremser ML, Strohmeier B, Moinfar F, Zatloukal K, Denk H. SAHA induces caspase-independent, autophagic cell death of endometrial stromal sarcoma cells by influencing the mTOR pathway. J Pathol. 2008;216:495–504. doi: 10.1002/path.2434. [DOI] [PubMed] [Google Scholar]
- 20.Kutko MC, Glick RD, Butler LM, Coffey DC, Rifkind RA, Marks PA, et al. Histone deacetylase inhibitors induce growth suppression and cell death in human rhabdomyosarcoma in vitro. Clin Cancer Res. 2003;9:5749–55. [PubMed] [Google Scholar]
- 21.Liu S, Cheng H, Kwan W, Lubieniecka JM, Nielsen TO. Histone deacetylase inhibitors induce growth arrest, apoptosis, and differentiation in clear cell sarcoma models. Molecular cancer therapeutics. 2008;7:1751–61. doi: 10.1158/1535-7163.MCT-07-0560. [DOI] [PubMed] [Google Scholar]
- 22.Sakimura R, Tanaka K, Nakatani F, Matsunobu T, Li X, Hanada M, et al. Antitumor effects of histone deacetylase inhibitor on Ewing’s family tumors. International journal of cancer. 2005;116:784–92. doi: 10.1002/ijc.21069. [DOI] [PubMed] [Google Scholar]
- 23.Sonnemann J, Dreyer L, Hartwig M, Palani CD, Hong le TT, Klier U, et al. Histone deacetylase inhibitors induce cell death and enhance the apoptosis-inducing activity of TRAIL in Ewing’s sarcoma cells. Journal of cancer research and clinical oncology. 2007;133:847–58. doi: 10.1007/s00432-007-0227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamamoto S, Tanaka K, Sakimura R, Okada T, Nakamura T, Li Y, et al. Suberoylanilide hydroxamic acid (SAHA) induces apoptosis or autophagy-associated cell death in chondrosarcoma cell lines. Anticancer research. 2008;28:1585–91. [PubMed] [Google Scholar]
- 25.Chavez-Blanco A, Perez-Plasencia C, Perez-Cardenas E, Carrasco-Legleu C, Rangel-Lopez E, Segura-Pacheco B, et al. Antineoplastic effectsof the DNA methylation inhibitor hydralazine and the histone deacetylase inhibitor valproic acid in cancer cell lines. Cancer cell international. 2006;6:2. doi: 10.1186/1475-2867-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ecke I, Petry F, Rosenberger A, Tauber S, Monkemeyer S, Hess I, et al. Antitumor effects of a combined 5-aza-2′deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer research. 2009;69:887–95. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 27.Hurtubise A, Bernstein ML, Momparler RL. Preclinical evaluation of the antineoplastic action of 5-aza-2′-deoxycytidine and different histone deacetylase inhibitors on human Ewing's sarcoma cells. Cancer cell international. 2008;8:16. doi: 10.1186/1475-2867-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mills J, Matos T, Charytonowicz E, Hricik T, Castillo-Martin M, Remotti F, et al. Characterization and comparison of the properties of sarcoma cell lines in vitro and in vivo. Hum Cell. 2009;22:85–93. doi: 10.1111/j.1749-0774.2009.00073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matushansky I, Radparvar F, Skoultchi AI. Reprogramming leukemic cells to terminal differentiation by inhibiting specific cyclin-dependent kinases in G1. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:14317–22. doi: 10.1073/pnas.250488697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D, Houchen CW, Kennedy S, Dieckgraefe BK, Anant S. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–26. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 31.de Toledo M, Senic-Matuglia F, Salamero J, Uze G, Comunale F, Fort P, et al. The GTP/GDP cycling of rho GTPase TCL is an essential regulator of the early endocytic pathway. Molecular biology of the cell. 2003;14:4846–56. doi: 10.1091/mbc.E03-04-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10014–9. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Segal NH, Pavlidis P, Antonescu CR, Maki RG, Noble WS, DeSantis D, et al. Classification and subtype prediction of adult soft tissue sarcoma by functional genomics. Am J Pathol. 2003;163:691–700. doi: 10.1016/S0002-9440(10)63696-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mai A, Altucci L. Epi-drugs to fight cancer:from chemistry to cancer treatment, the road ahead. Int J Biochem Cell Biol. 2009;41:199–213. doi: 10.1016/j.biocel.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 35.Mercurio C, Minucci S, Pelicci PG. Histone deacetylases and epigenetic therapies of hematological malignancies. Pharmacol Res. 2010;62:18–34. doi: 10.1016/j.phrs.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Chou TC. Preclinical versus clinical drug combination studies. Leuk Lymphoma. 2008;49:2059–80. doi: 10.1080/10428190802353591. [DOI] [PubMed] [Google Scholar]
- 37.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacological reviews. 2006;58:621–81. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 38.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer research. 2010;70:440–6. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 39.Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, et al. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nature medicine. 2007;13:748–53. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- 40.Vignal E, De Toledo M, Comunale F, Ladopoulou A, Gauthier-Rouviere C, Blangy A, et al. Characterization of TCL, a new GTPase of the rho family related to TC10 andCcdc42. J Biol Chem. 2000;275:36457–64. doi: 10.1074/jbc.M003487200. [DOI] [PubMed] [Google Scholar]
- 41.Dembowski JA, Grabowski PJ. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS genetics. 2009;5:e1000595. doi: 10.1371/journal.pgen.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siddiqi S, Mills J, Matushansky I. Epigenetic remodeling of chromatin architecture: exploring tumor differentiation therapies in mesenchymal stem cells and sarcomas. Current stem cell research & therapy. 2010;5:63–73. doi: 10.2174/157488810790442859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Molecular cancer therapeutics. 2009;8:1409–20. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 44.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.