Abstract
The opioid system is involved in the action of opiate drugs, opioid addiction, pain experience and analgesia. Individual differences in opioid effect may be attributed in part to genetic variations. Long-range cis regulatory elements and intronic variants are potential sources of functional diversity. Recently, we have detected association of two intronic OPRM1 variants with heroin addiction in European Americans. In the current study, we analyzed the genetic variations in the OPRM1 100 kb 5′ flanking region and intron 1 in the HapMap Caucasian population. Four major linkage disequilibrium (LD) blocks were identified, consisting of 28, 22, 15 and 42 SNPs, respectively. The locations of these blocks are (−100 – −90), (−90 – −67), (−20 - −1) and (+1 – +44) kb, respectively. The two intronic variants, indicated in our recent study, are part of a distinct haplogroup that include SNPs from intron 1, and the proximal 5′ region. The 118G (rs1799971) allele is part of a different haplogroup that includes several variants in the distal 5′ region that may have a regulatory potential. These findings were corroborated by genotyping eight SNPs in a sample of European Americans and suggest an extended OPRM1 locus with potential new regulatory regions.
Keywords: LD blocks, distal 5′ region, opioid addiction, genetic variations, haplotype
Introduction
The G protein-coupled mu opioid receptor (encoded by the OPRM1 gene) affects signal transduction pathways that mediate the effects of opioids and plays an important role in opioid reward, tolerance, analgesia and dependence. The vulnerability to develop opioid addiction is partially inherited and individual differences in opioid effects may be attributed to genetic variations. Understanding the effects of this variability is of clinical importance.
The non-synonymous variant rs1799971 (118A>G, Asn40Asp) was shown to remove a potential N-glycosylation site in the extracellular domain, to be more potent in beta-endorphin binding and receptor activity, and to reduce receptor signaling efficacy.1 Healthy subjects with the 118G allele showed an increased basal level of cortisol and greater cortisol responses to opioid receptor blockade with naloxone in a population-specific manner.2-4 Several studies reported positive association of the 118G variant with opioid dependence and other substance dependencies, in diverse populations,5-7 while other studies did not detect association.8, 9 The clinical relevance of this polymorphism for opioid analgesia and opioid adverse effects is still debatable.10-12 The 118G allele frequencies varied between populations, with high frequency in Asian populations (0.35-0.48), moderate frequency in European populations (0.1-0.17), and low frequency in African populations (<0.04).13
Another potential source of functional diversity is splice variants. At least half of the genome is expressed in alternatively spliced isoforms14 and gene functions may also be modified by SNPs in alternatively spliced exons. A number of OPRM1 alternatively spliced variants have been reported in rodents and humans.15-20 The receptors encoded by two human splice variants that retain different parts of intron 1 were shown to form heterodimers with the wild type protein.19 An alternatively spliced exon with an alternative promoter, in the 5′ upstream region (~28-30 kb), was identified in rodents and humans, along with cell-specific splice variants.15, 19, 21-23 A SNP in intron 1 showed an effect on the OPRM1 gene expression in vitro and was associated with pain sensitivity.20
The proximal promoter region does not necessarily contain all elements required for tissue-specific gene expression, and regulatory elements can be located great distances from the gene they regulate.14, 24 Only a few potential OPRM1 regulatory variants have been functionally characterized. SNP −554G>A decreased OPRM1 activity in a neuroblastoma cell line, and SNP −1320A>G showed increased OPRM1 activity.22, 25 Association studies of potential regulatory OPRM1 variants with substance dependence were limited to the proximal regulatory region. An association with substance dependence was shown with SNPs −1793T>A, −1699insT, and −2044C>A.26, 27
In a recent association study of heroin addiction we have detected association of two variants in intron 1 (rs510769 and rs3778151) in European Americans.9 Of note, the “addiction-focused” array28 used in this study does not include OPRM1 SNPs upstream of exon 1. These results are supported by two studies in which intron 1 SNPs showed association with drug dependence in European Americans,29 or with positive response to heroin after first use in Chinese.30 We did not find a similar association in an African American cohort.31 A 10K genome-wide association study of a subset of the same cohort from our laboratory identified association of a SNP rs1074287, located 11.6 kb upstream of exon 1, with heroin addiction.32
Our hypothesis is that there may be additional non-coding variants, possibly distant from OPRM1, that contribute to the individual phenotype (e.g. response to opiates, vulnerability to develop opioid addiction, and perception of pain). In addition, a combination of linked variants could contribute to the phenotypic difference in a different way than a single polymorphism.
Toward this goal, we performed an analysis of genetic variations and haplogroups in the OPRM1 100 kb 5′ flanking region, exon 1, and the 50 kb intron 1, and searched for regions in high LD with the 118G and/or the intron 1 variants. Genotype data were obtained from the International HapMap Project and from tag SNPs in a sample of European Americans.
Materials and Methods
Comparative genomic analysis
Comparative genomic analysis was obtained from the UCSC Genome Browser (http://genome.ucsc.edu/) and is based on the Vertebrate Multiz Alignment & PhastCons Conservation. Regulatory Potential data were based on the ESPERR Regulatory Potential analysis that is computed from alignments of human, chimpanzee, macaque, mouse, rat, dog, and cow. 33
HapMap data
HapMap genotype and phased haplotype data were obtained from the genome browser of The International HapMap Project Phase 2 (http://www.hapmap.org/). The sample populations used were CEU (Utah residents with Northern and Western European ancestry from the CEPH collection), YRI (Yoruba in Ibadan, Nigeria) and the combined JPN/CHB (Japanese in Tokyo and Han Chinese in Beijing). Only the founders in these sample populations were included. The phased haplotype data was generated using a maximum likelihood algorithm. The chromosomal region was set to position 154,300–154,450 kb (build 37.1).
Subjects
An exploratory representative group of 103 subjects was selected, based on their 118A>G genotype, from our European American cohort, which was collected for genetic studies with heroin addiction as described.9 To reduce the potential effect of population stratification, only subjects with a European ancestry proportion of >0.7 based on AIMs analysis were included (see below). The group can be divided as follows: 1) ”118G”/”118G” (n=5); 2) “REF”/”118G” (n=52); 3); “IVS1”/”118G” (n=17); and 4) ”REF”/“REF” (n=29). Since the 118G allele was not found to be associated with heroin addiction in this cohort, both cases (former heroin addicts in methadone maintenance treatment) and controls were included, except for subgroup 3 that included only control subjects. Subjects were recruited at the Rockefeller University Hospital, Weill Medical College of Cornell University, and the Dr. Miriam and Sheldon G. Adelson Clinic for Drug Abuse Treatment and Research in Las Vegas, NV. The Institutional Review Boards of the Rockefeller University Hospital and Cornell University approved the study for the three institutions. All subjects signed informed consent for genetic studies.
Ancestry informative markers (AIMs)
One hundred seventy-four AIMs with adequate quality were employed to calculate the proportion of European ancestry using STRUCTURE34 with the CEPH diversity panel of 1051 individuals, as a reference.9, 28, 35
Linkage disequilibrium (LD) structure, haplotypes and multi-locus genotype patterns (MLGPs)
The pattern of pair-wise LD between the three SNPs (#5, 6 and 8) in the original sample9 was measured by D’ and r2 metrics. Haplotypes were reconstructed using the accelerated expectation-maximization algorithm implemented in Haploview version 4.2.36 Multi-locus genotype patterns were generated manually.
SNP selection and genotyping
Three SNPs (rs1799971(#5), rs510769 (#6) and rs3778151 (#8)) were genotyped using the Illumina custom array.9 Additional five tag SNPs were selected for genotyping from the 100 kb 5′ flanking region (blocks 1: rs1551808 (#1), rs7758009 (#2), and rs7760028 (3#), and block 4: rs1074287 (#4)), and intron 1 (block 4: rs3778146 (#7)) (Table 1, Figure 1, and Supplement Table 1). The selection was based on information from HapMap, chromosomal location and frequency (MAF> 0.1 in CEU).
Table 1.
LD haplotype block distributionof OPRM1 SNPs in the HapMap CEU population
| # | LD Block | Position | SNPs | MAF | Referencesa | |||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Haplo- group 1 |
Haplo- group 2 |
Haplo- group 3 |
Haplo- group 4 |
|||||
| 1 | Block 1 (5′ region) |
154301883 | rs3829284 | 0.21 | ||||
| 2 | 154302640 | rs155180 | 0.28 | |||||
| 3 | 154303286 | rs116638 | 0.39 | |||||
| 4 | 154304373 | rs282095 | 0.39 | |||||
| 5 | 154304587 | rs1885629 | 0.25 | |||||
| 6 | 154304631 | rs1885628 | 0.25 | |||||
| 7 | 154304972 | rs282096 | 0.39 | |||||
| 8 | 154305678 | rs204426 | 0.39 | |||||
| 9 | 154307218 | rs605249 | 0.36 | |||||
| 10 | 154307715 | rs1708472 | 0.25 | |||||
| 11 | 154308258 | rs7745695 | 0.35 | |||||
| 12 | 154308291 | rs9397672 | 0.25 | |||||
| 13 | 154308367 | rs7741685 | 0.34 | |||||
| 14 | 154308402 | rs9397673 | 0.25 | |||||
| 15 | 154308430 | rs7745892 | 0.38 | |||||
| 16 | 154308693 | rs9397674 | 0.25 | |||||
| 17 | 154308840 | rs2169411 | 0.22 | |||||
| 18 | 154309097 | rs1033817 | 0.35 | |||||
| 19 | 154309787 | rs6557329 | 0.25 | |||||
| 20 | 154310237 | rs1033816 | 0.25 | |||||
| 21 | 154311203 | rs775800 | 0.22 | |||||
| 22 | Block 2 (5′ region) |
154312749 | rs472615 | 0.25 | ||||
| 23 | 154312816 | rs626174 | 0.25 | |||||
| 24 | 154 312831 | rs626150 | 0.25 | |||||
| 25 | 154313470 | rs612692 | 0.25 | |||||
| 26 | 154313492 | rs612665 | 0.25 | |||||
| 27 | 154313761 | rs611357 | 0.25 | |||||
| 28 | 154314057 | rs610040 | 0.25 | |||||
| 29 | 154314311 | rs598832 | 0.16 | |||||
| 30 | 154314407 | rs525479 | 0.26 | |||||
| 31 | 154314435 | rs5005184 | 0.25 | |||||
| 32 | 154315619 | rs582542 | 0.25 | |||||
| 33 | 154315643 | rs493335 | 0.25 | |||||
| 34 | 154316767 | rs9384167 | 0.23 | |||||
| 35 | 154319896 | rs613515 | 0.25 | |||||
| 36 | 154320006 | rs613030 | 0.25 | |||||
| 37 | 154320077 | rs1217420 | 0.22 | |||||
| 38 | 154320301 | rs601509 | 0.28 | |||||
| 39 | 154323894 | rs546 536 | 0.28 | |||||
| 40 | 154324902 | rs647303 | 0.24 | |||||
| 41 | 154325561 | rs488966 | 0.28 | |||||
| 42 | 154326611 | rs282108 | 0.24 | |||||
| 43 | 154329645 | rs282103 | 0.25 | |||||
| 44 | 154329725 | rs6900805 | 0.24 | |||||
| 45 | 154330276 | rs1196694 | 0.23 | |||||
| 46 | 154335481 | rs484565 | 0.20 | |||||
| 47 | 15433 6766 | rs776002 | 0.23 | |||||
| 48 | 154354079 | rs1252742 | 0.22 | |||||
| 49 | 154363627 | rs1708486 | 0.22 | |||||
| 50 | 154364271 | rs1708487 | 0.22 | |||||
| 51 | Block 3 (5′ region) |
154382310 | rs12214130 | 0.05 | ||||
| 52 | 154383069 | rs9478498 | 0.23 | |||||
| 53 | 154383251 | rs12216066 | 0.06 | |||||
| 54 | 154383485 | rs12216210 | 0.06 | |||||
| 55 | 154386982 | rs9478499 | 0.14 | |||||
| 56 | 154388025 | rs12192681 | 0.06 | |||||
| 57 | 154389936 | rs7776131 | 0.06 | |||||
| 58 | 154390181 | rs7751510 | 0.06 | |||||
| 59 | 154 390298 | rs7776341 | 0.06 | 20 | ||||
| 60 | 154390502 | rs1074287 | 0.23 | 20,32, | ||||
| 61 | 154391394 | rs6910065 | 0.06 | |||||
| 62 | 154396793 | rs6936615 | 0.15 | |||||
| 63 | 154399679 | rs12210856 | 0.06 | |||||
| 64 | 154400626 | rs12205732 | 0.05 | 26 | ||||
| 65 | 154402201 | rs6912029 | 0.05 | 26 | ||||
| 66 | Exon 1 | 154402490 | rs179997 | |||||
| 67 | Block 4 (Intron 1) |
154403712 | rs510769 | 0.23 | 9,30,4 | |||
| 68 | 154403947 | rs9322445 | 0.15 | |||||
| 69 | 154405080 | rs477292 | 0.19 | |||||
| 70 | 154405995 | rs557748 | 0.18 | |||||
| 71 | 154406133 | rs634479 | 0.18 | |||||
| 72 | 1544 08773 | rs3778145 | 0.14 | |||||
| 73 | 154409772 | rs668394 | 0.18 | |||||
| 74 | 154409860 | rs514980 | 0.18 | |||||
| 75 | 154410240 | rs511435 | 0.18 | 29,41 | ||||
| 76 | 154410479 | rs509544 | 0.18 | |||||
| 77 | 154414130 | rs607759 | 0.18 | |||||
| 78 | 154414385 | rs499796 | 0.19 | |||||
| 79 | 154416785 | rs524731 | 0.18 | 20,29,4 | ||||
| 80 | 154417218 | rs520321 | 0.18 | |||||
| 81 | 154419618 | rs3778146 | 0.14 | |||||
| 82 | 154420432 | rs3778147 | 0.14 | |||||
| 83 | 154420845 | rs3823010 | 0.14 | 29 | ||||
| 84 | 154421627 | rs9285542 | 0.14 | |||||
| 85 | 154421843 | rs7748401 | 0.05 | |||||
| 86 | 154422705 | rs3778148 | 0.14 | |||||
| 87 | 154423832 | rs3778149 | 0.14 | |||||
| 88 | 154424060 | rs7773995 | 0.14 | |||||
| 89 | 154424166 | rs7772959 | 0.14 | |||||
| 90 | 154424751 | rs9479754 | 0.14 | |||||
| 91 | 154425351 | rs3778150 | 0.14 | |||||
| 92 | 154426224 | rs9478501 | 0.14 | |||||
| 93 | 154429999 | rs6927269 | 0.14 | |||||
| 94 | 154432766 | rs10457090 | 0.05 | |||||
| 95 | 154433062 | rs1461773 | 0.14 | |||||
| 96 | 154434368 | rs9478503 | 0.15 | |||||
| 97 | 154434951 | rs1381376 | 0.15 | 20,29,30 | ||||
| 98 | 154435373 | rs3778151 | 0.17 | 30 | ||||
| 99 | 154435524 | rs3778152 | 0.04 | |||||
| 100 | 154435577 | rs3778153 | 0.15 | |||||
| 101 | 154436852 | rs9478504 | 0.15 | |||||
| 102 | 154438148 | rs17209711 | 0.15 | |||||
| 103 | 154438165 | rs17275521 | 0.15 | |||||
| 104 | 154445511 | rs3778155 | 0.14 | |||||
| 105 | 154447394 | rs3778157 | 0.15 | |||||
| 106 | 154447423 | rs3778158 | 0.15 | |||||
| 107 | 154447744 | rs3798678 | 0.15 | |||||
| 108 | 154449660 | rs563649 | 0.08 | 20,41 | ||||
SNPs in bold italics were genotyped forthis study.
References indicating SNPs that were analyzed in other studies.
Figure 1.
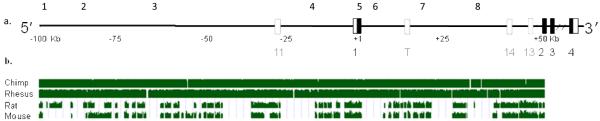
a. Schematic representation of the OPRM1 gene region and the location of the 8 SNPs genotyped: rs1551808 (1), rs7758009 (2), rs7760028 (3), rs1074287 (4), rs1799971 (118A>G, 5), rs510769 (6), rs3778146 (7), and rs3778151 (8). Black boxes indicate exons, open boxes indicate untranslated regions (UTR) and dotted boxes indicate alternatively spliced exons.
b. A customized view from the UCSC genome browser that covers 100 kb of the 5′ region, exon 1 and 50 Kb of intron 1, showing pair-wise alignment of each of four species with the human genome. The tracks from top to bottom show the positions of segments aligned with the indicated comparison species. Green bars indicate conserved regions.
DNA was extracted from blood and genotypes were determined by TaqMan® technology. The TaqMan® Pre-Designed SNP Genotyping Assay catalog numbers (Applied Biosystems (ABI), Foster City, CA) are listed in Supplement Table 1. PCR was performed in duplicate on a GeneAmp® PCR 9700 using TaqMan® universal PCR master mix with AmpErase uracil-N-glycosylate (ABI) according to the manufacturer’s protocol. Briefly, TaqMan® assay mix (40x) and universal PCR master mix (2x) were mixed and the volume was adjusted to 4 μl in a 384-well optical reaction plate; 10 ng of genomic DNA (1 μl) were added. PCR amplification lasted for 2 min at 50°C; 10 min at 95°C followed by 40 cycles of 15 sec at 92°C and 1 min at 60°C. Genotype analysis was performed on an ABI Prism® 7900 sequence detection system using SDS 2.1 software (ABI).
For verification of the TaqMan® assay, PCR amplification was performed using Platinum PCR Supermix (Invitrogen, Carlsbad, CA). Primers were designed using software Primer3.37 PCR amplification consisted of 2 min at 94°C; eight ‘touch-down’ cycles of 30 sec at 94°C, 30 sec at 63-56°C and 30 sec at 72°C; 30 cycles of 30 sec at 94°C, 30 sec at 56°C and 30 sec at 72°C; and a final step of 7 min at 72°C. PCR products were purified and sequenced on an ABI Prism 3700® capillary sequencer (ABI). Electropherograms were scored using the Sequencer 4.5 software (Gene Codes Corporation, Ann Arbor, MI).
Results
The genomic organization of the OPRM1 gene region is shown in Figure 1a. OPRM1 is mapped to chromosome 6q25.2 at position 154.4–154.7 Mb (NCBI build 37.1) and is flanked at the 5′ end by three non-functional pseudogenes. The next gene upstream to OPRM1, the regulator of G-protein signaling 17 gene (RGS17), is located at a 1 mega base distance at position 153.4. At the 3′ end, OPRM1 is overlapped by the phosphoinositide-binding protein PIP3-E gene (IPCEF1), in the opposite orientation.
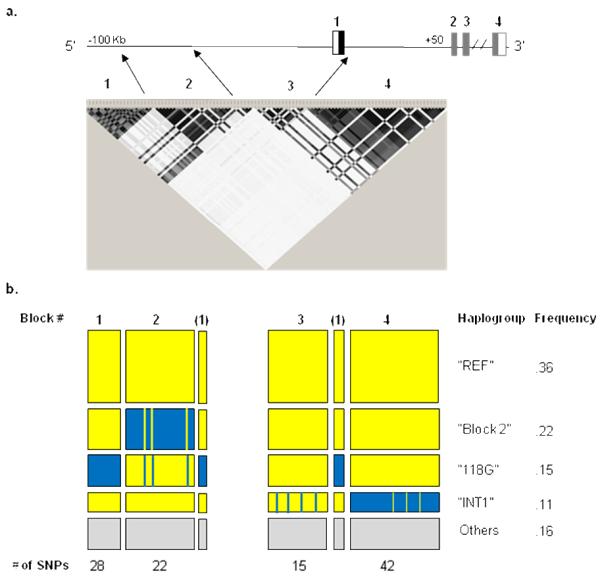
In this study, we focused on the region spanning the ~100 kb 5′ flanking region, exon 1, and the 50 kb intron 1. Sequence comparison of this region between humans, non-human primates and rodents is shown on Figure 1b. Analysis of the LD matrix of the selected region in the HapMap CEU sample revealed that 108 SNPs are located in four major blocks of high LD (Figure 2a, Table 1). The first block spans ~10 kb with 28 SNPs, the second block spans ~23 kb with 22 SNPs, the third block spans ~20 kb with 15 SNPs and the fourth block spans intron 1 (~44 kb) with 42 SNPs. SNPs analyzed in related association studies (except for 118A>G that were studied in many studies) are indicated by their references (also see Discussion).
Figure 2.
a. Plot of pair-wise linkage disequilibrium (LD) of the 150 kb OPRM1 genomic region, in the HapMap CEU sample. The plot is not drawn to scale and the arrows show the correct position of the SNPs. The color scheme indicates the magnitude of r2. Black blocks indicate r2= 1, and white blocks indicate r2= 0. b. The pattern and frequency of the major haplogroups in this population. The height of the boxes is proportional to the frequency in the population. Blue boxes indicate variant alleles, yellow boxes indicate reference alleles and gray boxes indicate all the other haplogroups.
The HapMap CEU haplotype data revealed four major haplogroups (Figure 2b): 1) the “reference” group (“REF”, ~36%) with no variants (including the reference 118A allele and the reference intron 1 sequence); 2) the block 2 group (~22%); 3) the 118G group (~15%) with additional variants in block 1; and 4) the intron 1 variants of block 4 (“INT1”, ~11%). The minor haplogroups are: block 3 (4%) and 12% of the haplotypes that could not be assigned to one of the major haplogroups (grouped as “Others” in Figure 2b). Of the 59 HapMap CEU subjects, ten (17%) were homozygous for the “REF” haplogroup, two (3%) were homozygous for the 118G haplogroup, and five (8%) were homozygous for the block 2 haplogroup. No individual was homozygous for the “INT1” haplogroup and one was heterozygous for the “118G”/“INT1” groups. All the rest were heterozygotes with different allelic combinations.
The two SNPs (rs510769 (#6), rs3778151 (#8)) identified in our recent study to be associated with heroin addiction9 are part of the 42-SNP block 4 that spans intron 1. The average MAF of the SNPs of block 4 is 0.15 in European Americans. Interestingly, four SNPs (rs9478498, rs9478499, rs1074287 (#4) and rs6936615), located at the −20 kb 5′ region (block 3 region) are in high LD with the SNPs of block 4. Four SNPs (rs7748401, rs10457090, rs3778152 and rs563649) located at intron 1 are not part of block 4 and are in high LD with the SNPs of block 3. Haplogroup “INT1” includes the reference 118A allele (Table 1). Seven SNPs (rs9384167, rs12174208, rs11966947, rs7760028 (#3), rs12527423, rs17084868, and rs17084870) are in high LD with SNPs in block 1, but are physically located in or adjacent to the region of block 2 (Table 1, Figure 2a). The region that spans block 1 (−100 kb - −90 kb) shows several regions with a regulatory potential (Supplement Figure 1).
We searched for similar haplogroups in the other HapMap populations (the combined JPT/CHB and YRI). The “INT1” haplogroup that includes the intron 1 variants of block 4 accounts for ~4% in these populations. In contrast, the “118G” haplogroup has a very different pattern. In concordance with the 118G allele frequency, the 118G haplogroup that includes the 5′ variants of block 1, is very frequent in the Asian population and can be divided into two subgroups, the major one (28%) includes the variants in block 1, similar to European Americans, and the second (14%) has other combinations (data not shown). The 118G allele was not found in the African sample but a similar haplogroup with block 1 variants (without the 118G variant allele) was represented by 7.5% of the haplotypes. Interestingly, there was a significant representation (~30%) of the block 3 haplogroup, with variants at the proximal 5′ region, in the African sample, in contrast to the low frequency in the Asian and Caucasian samples.
Three major haplogroups were identified in our previous study of European Americans (350 cases and 184 controls) based on OPRM1 exon 1 SNP rs1799971 (118A>G, #5), and intron 1 SNPs rs510769 (#6) and rs3778151 (#8).9 The major haplogroups identified are: 1) “REF” (65%, 56%); 2) “118G” (13%, 11%); and 3) “INT1” (13%, 20%) in controls and cases, respectively. The higher frequency of the “REF” group may be explained by the fact that the haplogroup of “block 2” cannot be distinguished from the reference group by the three SNPs genotyped. These results demonstrate the higher frequency of the “INT1” haplogroup in the cases that was the basis for the association of this haplogroup with heroin addiction. The allele frequencies in controls are compatible with the HapMap data from Caucasians.
To further explore the finding that the 118G haplogroup includes variant alleles in the distant 5′ region, a representative group of 103 subjects with and without the 118G allele was selected from the original cohort. This exploratory sample does not represent the general population and was selected to enable unambiguous phase determination. To reduce population admixture, only subjects with a European ancestry proportion of >0.7, based on AIMs analysis, were included. The exploratory representative sample is composed of the following subgroups (Table 2): 1) “118G”/”118G” (homozygotes for the 118G allele, n=5); 2) “REF”/”118G” (heterozygotes for 118G allele that are non-carriers of the IVS 1 variants, n=52); 3) “IVS1”/”118G” (double heterozygotes for 118A>G and the two IVS1 SNPs, n=17); and 4) “REF”/”REF” (represents of the reference genotype group, n=29, all controls).
Table 2.
Multi-locus genotype patterns (MLGPs) in a representative sample of the 118A>GGenotype subgroups
| rs1551808 | rs7758009 | rs7760028 | rs1074287 | rs1799971 | rs510769 | rs3778146 | rs3778151 | ||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Phenotype | |
| 1. “118G”/”118G“ | |||||||||
| 1 | CC | GG | CC | AA | GG | CC | TT | TT | control |
| 2 | CC | GG | CC | AA | GG | CC | TT | TT | control |
| 3 | CC | GG | CC | AA | GG | CC | TT | TT | control |
| 4 | CC | GG | CC | AA | GG | CC | TT | TT | case |
| 5 | CC | GG | CC | AA | GG | CC | TT | TT | case |
| 2. “REF”/”118G” | |||||||||
| 6 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 7 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 8 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 9 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 10 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 11 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 12 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 13 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 14 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 15 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 16 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 17 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 18 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 19 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 20 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 21 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 22 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 23 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 24 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 25 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 26 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 27 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 28 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 29 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 30 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 31 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 32 | TC | AG | TC | AA | AG | CC | TT | TT | case |
| 33 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 34 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 35 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 36 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 37 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 38 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 39 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 40 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 41 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 42 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 43 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 44 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 45 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 46 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 47 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 48 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 49 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 50 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 51 | TC | AG | TC | AA | AG | CC | TT | TT | control |
| 52 | CC | AA | TT | AA | AG | CC | TT | TT | control |
| 53 | CC | AA | TT | AA | AG | CC | TT | TT | control |
| 54 | CC | AA | TT | AA | AG | CC | TT | TT | case |
| 55 | CC | AA | TT | AA | AG | CC | TT | TT | case |
| 56 | CC | GG | CC | AA | AG | CC | TT | TT | case |
| 57 | CC | GG | CC | AA | AG | CC | TT | TT | case |
| 3. “IVS1”/”118G” | |||||||||
| 58 | TC | AG | TC | AG | AG | CT | TC | TC | control |
| 59 | TC | AG | TC | AG | AG | CT | TC | TC | control |
| 60 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 61 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 62 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 63 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 64 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 65 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 66 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 67 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 68 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 69 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 70 | TC | AG | TC | AG | AG | CT | TC | TC | case |
| 71 | CC | AG | TC | AG | AG | CT | TC | TC | case |
| 72 | CC | AG | TC | AG | AG | CT | TC | TC | case |
| 73 | TC | GG | TC | AG | AG | CT | TC | TC | case |
| 74 | CC | GG | CC | AG | AG | CT | TC | TC | case |
| 4. “REF“/”REF” | |||||||||
| 75 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 76 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 77 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 78 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 79 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 80 | CC | AA | TT | AA | AA | CC | TT | TT | control |
| 81 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 82 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 83 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 84 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 85 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 86 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 87 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 88 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 89 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 90 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 91 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 92 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 93 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 94 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 95 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 96 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 97 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 98 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 99 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 100 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 101 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 102 | TT | AA | TT | AA | AA | CC | TT | TT | control |
| 103 | TT | AA | TT | AA | AA | CC | TT | TT | control |
All subjects were genotyped for an additional five SNPs (rs1551808 (#1), rs7758009 (#2), rs7760028 (#3), rs1074287 (#4), and rs3778146 (#7)) (Table 1, Figure 1). The SNPs were selected from the 100 kb 5′ flanking region, and intron 1 based on LD information from HapMap (CEU blocks 1 and 4) and frequency in CEU (MAF> 0.1). As is shown in Table 2, the five subjects with the “118G”/”118G” pattern (subgroup 1) carry two copies of the three variant alleles (#1-3) of the SNPs from the 5′ region (C-G-C, block 1), two copies of the reference A allele of the 5′ SNP rs1074287 (#4), and two copies for the reference allele of the three SNPs from intron 1 (C-T-T, #6-8). The majority of the heterozygotes for the 118G allele (“REF”/“118G”, subgroup 2) also carry at least one variant allele of the SNPs #1-3 (TC-AG-TC), and none of them carries the variant allele of SNP rs1074287 (#4) or the three IVS1 SNPs (#6-8). The subjects in subgroup “IVS1”/”118G” (subgroup 4) all carry at least one variant allele of SNPs #1-4 (TC-AG-TC-AG). Although the phase in these subjects cannot be determined unambiguously, the genotypes most probably reflect a combination of two haplogroups: 1) SNP #4/ IVS1 SNPs #6-8 and 2) 118G/ 5′ SNPs #1-3, based on the other subgroups in which the phase can be determined with certainty. This subgroup represents a small group of the original cohort (5% in controls and 8% in cases). The majority of the “REF”/REF” subgroup (subgroup 3) does not carry a variants allele of any of the genotyped SNPs. This exploratory data corroborates the HapMap data that the 118G haplogroup includes variant alleles of SNPs in the distant 5′ region and is distinct from the “INT1” haplogroup.
Discussion
Genetic diseases can be caused by disruption of the regulatory elements and the identification of these regulatory elements is an important and challenging problem. Tissue-specific and developmental-specific expression requires sophisticated expression profiles that may involve multiple enhancer elements. Long-range cis elements and intronic variants may play a role in this regulation. For example, an evolutionarily conserved enhancer was identified 86 kb upstream of the peptidylarginine deiminases gene (PADI3) that previous studies limited to the proximal promoter failed to explain its expression pattern.38
Recently, an alternatively spliced exon with a specific promoter was identified in humans, ~28 kb upstream of exon 1,15 suggesting an extended OPRM1 gene. It is also of note that except for three pseudogenes, there is no gene located at one mega base 5′ to OPRM1.
In our hypothesis-driven association study of heroin addiction, two OPRM1 intron 1 variants (#6, #8) were found at a significantly higher frequency in subjects with heroin addiction than in healthy controls, suggesting an association with heroin addiction.9 A 10K genome-wide association study from our laboratory that used a subset of the sample population, used in the hypothesis-driven study, indicated association of a SNP rs1074287 (#4) in the 5′ flanking region (−11.6 kb from exon 1) with heroin addiction.32 Analysis of the HapMap CEU LD data in this study revealed a unique haplogroup (“INT1”) of 42 SNPs that spans intron 1 and also includes at least four variants in the 20 kb 5′ flanking region, including rs1074287. This data indicate that the finding of the two studies are compatible and refer to the same “IVS1” haplogroup. Based on this data, the intron 1 SNPs that show association with heroin addiction may be markers in high LD with regulatory variants in the proximal 5′ region. It is also possible that the SNPs in the 5′ flanking region are markers for functional SNPs in intron 1.
Several lines of evidence suggest an additional functionality to intron 1 beyond the basic splicing of the constitutive exons. Several alternatively spliced exons were described in this intron.19, 20 The finding of a high LD between SNPs spanning the entirety of intron 1 and a few SNPs in the proximal 5′ flanking region, in different populations, is intriguing. This haplogroup is represented at a relatively low frequency (4%) in Asian and African populations but at a much higher frequency (15%) in the European population, suggesting the possibility of an ancient haplotype with a specific selective advantage in Europeans. It is also possible that this population’s difference occurs by random as was recently suggested in a genome-wide study of genes with highly divergent allele frequencies between populations.39
A potential functional SNP in intron 1 (rs563649) was recently indicated in association with pain perception.20 The HapMap LD data indicate that although this SNP not part of the “INT1” haplogroup, it is in high LD with SNPs of haplogroup 3 that includes variants in the proximal 5′ region (block 3). Notably, this haplogroup is rare in European Americans, but frequent in the African sample. It is possible that this SNP is a marker of regulatory variant/s in the proximal 5′ region.
Many studies provided evidence for the importance of the OPRM1 118A>G polymorphism in addiction, pain, HPA activation, and treatment response.e.g., 2-9 These studies have shown mixed results regarding the association between the 118G variant and opioid dependence, opioid analgesia and opioid adverse effects.40 Most studies have used single SNP analyses and were generally limited to the coding region or to the proximal upstream region. Several studies employed haplotype analysis but were also limited to specific regions. Hoehe et al. performed a comprehensive haplotype analysis of OPRM1 but the study did not include SNPs from intron 1 and was limited to the proximal 5′ region (−2.4 kb). 26 Lue et al. studied five variants from the proximal 5′ flanking region (−2 kb) and two SNPs from exon 1. 27 Zhang et al. examined 13 SNPs spanning the coding sequence including several SNPs from intron 1, but none in the 5′ flanking region. 29, 30 Xuei et al. genotyped 18 SNPs including one in the proximal 5′ region (−6 kb). 41 A recent study used a haplotype-based approach to predict response to naltrexone treatment based on 10 OPRM1 tag SNPs but was limited to the proximal 5′ region (−11 kb).42 Haplotype-based analysis is generally considered more powerful than single-marker analysis and may help to determine if a specific polymorphism occurs on a common genetic background.
The mixed single SNP association results for OPRM1 SNP 118A>G may be explained by different haplotype patterns. For example, the finding that about a third of the chromosomes with the 118G allele in the HapMap Asian populations do not show high LD with block 1 SNPs suggests the existence of a second 118G haplogroup that is specific to this population. It is feasible that single SNP association studies of the 118A>G SNP in this population may result in mixed results. The data obtained in this study indicate that, in European Americans, the 118G variant allele is positioned within a specific haplogroup and is in high LD with several distant variants that may possibly have a regulatory effect. It remains to be shown whether these variants are correlated with OPRM1 expression levels. . The variant 118G allele was not found in the African HapMap population suggesting that this is a relatively new variant that arose after the out-of-Africa migration of modern humans. The high prevalence of this allele in the Asian population and the lesser prevalence in European Americans may be explained by genetic drift and/or selective advantage. Interestingly, a similar haplogroup (block 1), without the 118G allele, was found in the African HapMap sample suggesting that the change to 118G may have occurred on this ancestral haplotype.
In summary, the identification of LD blocks and haplogroups in the OPRM1 intron 1 and the 5′ flanking region may be of importance to the field of opioid response and addiction. This study suggests an extended OPRM1 locus and possible new regulatory regions. The haplotype architecture described in this study may provide direction for future genetic studies. These findings will require confirmation by functional expression studies.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Pei-Hong Shen and David Goldman from the Laboratory of Neurogenetics (NIAAA) for the AIMs analysis, to the clinical staff including Elizabeth Ducat, Brenda Ray, Dorothy Melia, and Lisa Borg, for patient recruitment and ascertainment, and toVadim Yuferov, Ann Ho, David Nielsen, Matthew Randesi and Susan Russo for their contributions.
This work was supported by the National Institutes of Health [P60-DA 05130 to M.J.K.]
Footnotes
Conflict of Interest statement. None declared
Supplementary information is available at The Journal of Human Genetics website
REFERENCES
- 1.Oertel BG, Kettner M, Scholich K, Renne C, Roskam B, Geisslinger G, et al. A common human mu-opioid receptor genetic variant diminishes the receptor signaling efficacy in brain regions processing the sensory information of pain. J Biol Chem. 2009;284:6530–6535. doi: 10.1074/jbc.M807030200. [DOI] [PubMed] [Google Scholar]
- 2.Wand GS, McCaul M, Yang X, Reynolds J, Gotjen D, Lee S, et al. The mu-opioid receptor gene polymorphism (A118G) alters HPA axis activation induced by opioid receptor blockade. Neuropsychopharmacology. 2002;26:106–114. doi: 10.1016/S0893-133X(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez-Avila CA, Covault J, Wand G, Zhang H, Gelernter J, Kranzler HR. Population-specific effects of the Asn40Asp polymorphism at the mu-opioid receptor gene (OPRM1) on HPA-axis activation. Pharmacogenet Genomics. 2007;17:1031–1038. doi: 10.1097/FPC.0b013e3282f0b99c. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez-Avila CA, Wand G, Luo X, Gelernter J, Kranzler HR. Association between the cortisol response to opioid blockade and the Asn40Asp polymorphism at the mu-opioid receptor locus (OPRM1) Am J Med Genet B Neuropsychiatr Genet. 2003;118:60–65. doi: 10.1002/ajmg.b.10054. [DOI] [PubMed] [Google Scholar]
- 5.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, et al. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kapur S, Sharad S, Singh RA, Gupta AK. A118g polymorphism in mu opioid receptor gene (oprm1): association with opiate addiction in subjects of Indian origin. J Integr Neurosci. 2007;6:511–522. doi: 10.1142/s0219635207001635. [DOI] [PubMed] [Google Scholar]
- 7.Bart G, Kreek MJ, Ott J, LaForge KS, Proudnikov D, Pollak L, et al. Increased attributable risk related to a functional mu-opioid receptor gene polymorphism in association with alcohol dependence in central Sweden. Neuropsychopharmacology. 2005;30:417–422. doi: 10.1038/sj.npp.1300598. [DOI] [PubMed] [Google Scholar]
- 8.Glatt SJ, Bousman C, Wang RS, Murthy KK, Rana BK, Lasky-Su JA, et al. Evaluation of OPRM1 variants in heroin dependence by family-based association testing and meta-analysis. Drug Alcohol Depend. 2007;90:159–165. doi: 10.1016/j.drugalcdep.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain. 2009;146:270–275. doi: 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Skorpen F, Laugsand EA, Klepstad P, Kaasa S. Variable response to opioid treatment: any genetic predictors within sight? Palliat Med. 2008;22:310–327. doi: 10.1177/0269216308089302. [DOI] [PubMed] [Google Scholar]
- 12.Kosarac B, Fox AA, Collard CD. Effect of genetic factors on opioid action. Curr Opin Anaesthesiol. 2009;22:476–482. doi: 10.1097/ACO.0b013e32832e34c9. [DOI] [PubMed] [Google Scholar]
- 13.Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- 14.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Xu M, Hurd YL, Pasternak GW, Pan YX. Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene. J Neurochem. 2009;108:962–972. doi: 10.1111/j.1471-4159.2008.05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan YX, Xu J, Mahurter L, Xu M, Gilbert AK, Pasternak GW. Identification and characterization of two new human mu opioid receptor splice variants, hMOR-1O and hMOR-1X. Biochem Biophys Res Commun. 2003;301:1057–1061. doi: 10.1016/s0006-291x(03)00089-5. [DOI] [PubMed] [Google Scholar]
- 17.Pan YX. Identification of alternatively spliced variants from opioid receptor genes. Methods Mol Med. 2003;84:65–75. doi: 10.1385/1-59259-379-8:65. [DOI] [PubMed] [Google Scholar]
- 18.Pan L, Xu J, Yu R, Xu MM, Pan YX, Pasternak GW. Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience. 2005;133:209–220. doi: 10.1016/j.neuroscience.2004.12.033. [DOI] [PubMed] [Google Scholar]
- 19.Choi HS, Kim CS, Hwang CK, Song KY, Wang W, Qiu Y, et al. The opioid ligand binding of human mu-opioid receptor is modulated by novel splice variants of the receptor. Biochem Biophys Res Commun. 2006;343:1132–1140. doi: 10.1016/j.bbrc.2006.03.084. [DOI] [PubMed] [Google Scholar]
- 20.Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mol Genet. 2009;18:1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abbadie C, Pan YX, Pasternak GW. Immunohistochemical study of the expression of exon11-containing mu opioid receptor variants in mouse brain. Neuroscience. 2004;127:419–430. doi: 10.1016/j.neuroscience.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 22.Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci U S A. 2001;98:14084–14089. doi: 10.1073/pnas.241296098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan YX. Identification and characterization of a novel promoter of the mouse mu opioid receptor gene (Oprm) that generates eight splice variants. Gene. 2002;295:97–108. doi: 10.1016/s0378-1119(02)00825-9. [DOI] [PubMed] [Google Scholar]
- 24.Kleinjan DA, Lettice LA. Long-range gene control and genetic disease. Adv Genet. 2008;61:339–388. doi: 10.1016/S0065-2660(07)00013-2. [DOI] [PubMed] [Google Scholar]
- 25.Bayerer B, Stamer U, Hoeft A, Stuber F. Genomic variations and transcriptional regulation of the human mu-opioid receptor gene. Eur J Pain. 2007;11:421–427. doi: 10.1016/j.ejpain.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Hoehe MR, Kopke K, Wendel B, Rohde K, Flachmeier C, Kidd KK, et al. Sequence variability and candidate gene analysis in complex disease: association of mu opioid receptor gene variation with substance dependence. Hum Mol Genet. 2000;9:2895–2908. doi: 10.1093/hmg/9.19.2895. [DOI] [PubMed] [Google Scholar]
- 27.Luo X, Kranzler HR, Zhao H, Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet B Neuropsychiatr Genet. 2003;120:97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- 28.Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, et al. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H, Luo X, Kranzler HR, Lappalainen J, Yang BZ, Krupitsky E, et al. Association between two mu-opioid receptor gene (OPRM1) haplotype blocks and drug or alcohol dependence. Hum Mol Genet. 2006;15:807–819. doi: 10.1093/hmg/ddl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D, Shao C, Shao M, Yan P, Wang Y, Liu Y, et al. Effect of mu-opioid receptor gene polymorphisms on heroin-induced subjective responses in a Chinese population. Biol Psychiatry. 2007;61:1244–1251. doi: 10.1016/j.biopsych.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 31.Levran O, Londono D, O’Hara K, Randesi M, Rotrosen J, Casadonte P, et al. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8:531–540. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen DA, Ji F, Yuferov V, Ho A, Chen A, Levran O, et al. Genotype patterns that contribute to increased risk for or protection from developing heroin addiction. Mol Psychiatry. 2008;13:417–428. doi: 10.1038/sj.mp.4002147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor J, Tyekucheva S, King DC, Hardison RC, Miller W, Chiaromonte F. ESPERR: learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Enoch MA, Shen PH, Xu K, Hodgkinson C, Goldman D. Using ancestry-informative markers to define populations and detect population stratification. J Psychopharmacol. 2006;20:19–26. doi: 10.1177/1359786806066041. [DOI] [PubMed] [Google Scholar]
- 36.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 37.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 38.Adoue V, Chavanas S, Coudane F, Mechin MC, Caubet C, Ying S, et al. Long-range enhancer differentially regulated by c-Jun and JunD controls peptidylarginine deiminase-3 gene in keratinocytes. J Mol Biol. 2008;384:1048–1057. doi: 10.1016/j.jmb.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 39.Coop G, Pickrell JK, Novembre J, Kudaravalli S, Li J, Absher D, et al. The role of geography in human adaptation. PLoS Genet. 2009;5:e1000500. doi: 10.1371/journal.pgen.1000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the mu-opioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83:262–268. doi: 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Xuei X, Flury-Wetherill L, Bierut L, Dick D, Nurnberger J, Jr., Foroud T, et al. The opioid system in alcohol and drug dependence: Family-based association study. Am J Med Genet B Neuropsychiatr Genet. 2007;144:877–884. doi: 10.1002/ajmg.b.30531. [DOI] [PubMed] [Google Scholar]
- 42.Oroszi G, Anton RF, O’Malley S, Swift R, Pettinati H, Couper D, et al. OPRM1 Asn40Asp predicts response to naltrexone treatment: a haplotype-based approach. Alcohol Clin Exp Res. 2009;33:383–393. doi: 10.1111/j.1530-0277.2008.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.