Abstract
Mitochondrial cytochrome c oxidase plays an essential role in aerobic cellular respiration, reducing dioxygen to water in a process coupled with the pumping of protons across the mitochondrial inner membrane. An aspartate residue, Asp-51, located near the enzyme surface, undergoes a redox-coupled x-ray structural change, which is suggestive of a role for this residue in redox-driven proton pumping. However, functional or mechanistic evidence for the involvement of this residue in proton pumping has not yet been obtained. We report that the Asp-51 → Asn mutation of the bovine enzyme abolishes its proton-pumping function without impairment of the dioxygen reduction activity. Improved x-ray structures (at 1.8/1.9-Å resolution in the fully oxidized/reduced states) show that the net positive charge created upon oxidation of the low-spin heme of the enzyme drives the active proton transport from the interior of the mitochondria to Asp-51 across the enzyme via a water channel and a hydrogen-bond network, located in tandem, and that the enzyme reduction induces proton ejection from the aspartate to the mitochondrial exterior. A peptide bond in the hydrogen-bond network critically inhibits reverse proton transfer through the network. A redox-coupled change in the capacity of the water channel, induced by the hydroxyfarnesylethyl group of the low-spin heme, suggests that the channel functions as an effective proton-collecting region. Infrared results indicate that the conformation of Asp-51 is controlled only by the oxidation state of the low-spin heme. These results indicate that the low-spin heme drives the proton-pumping process.
The O2 reduction site of mitochondrial cytochrome c oxidase is composed of a high-spin heme (heme a3) and a copper ion (CuB). The electron equivalents are supplied by cytochrome c in the outside of the mitochondrial inner membrane (the intermembrane space) via a copper site (CuA) and a low-spin heme (heme a) to the O2 reduction site (1). The protons used for water formation from O2 are transferred from the inside of the mitochondrial inner membrane (the matrix space) through two hydrogen-bond networks known as the K and D pathways (2). In addition to the proton-pumping process, the transfer of protons and electrons to the O2 reduction site results in a net positive charge movement to the intermembrane space, which provides the protonmotive force required to drive ATP synthase.
Mutations of amino acid residues within the D pathway cause a decrease in proton-pumping efficiency and a decrease in O2-reduction activity (2), observations apparently consistent with a process wherein proton transfer for proton pumping occurs via the proton pathway for water formation. On the other hand, x-ray structures of bovine heart cytochrome c oxidase in the fully oxidized and reduced states at 2.3- and 2.35-Å resolution, respectively, indicate movement of Asp-51 of subunit I [the largest subunit containing heme a and the O2-reduction site (3)], from the interior of the protein to the intermembrane surface upon reduction of the enzyme (4). In the oxidized state, Asp-51 makes contact with the matrix space via a hydrogen-bond network with a channel in which water molecules in the matrix space are accessible (H pathway) (4). These structures are strongly suggestive of a proton-pumping event occurring at Asp-51 (4). However, the proton-pumping proposal is not yet widely accepted because plant and bacterial enzymes do not have an analogous Asp-51 (bovine numbering) residue and because of the D pathway mutation results described above. Furthermore, no mechanism for driving the proton pump at Asp-51 has yet been given. Mutation of Asp-51 would provide one of the most direct methods to probe the function of Asp-51. However, site-directed mutagenesis of subunit I, a large mitochondrially encoded membrane protein, has not been attempted because of expected technical difficulties (5). We report herein investigations of the role of the H pathway by site-directed mutagenesis, x-ray crystallography, and infrared spectroscopy.
Methods
Preparation procedures of expression vectors, transfectants and antibodies specific to either bovine or human subunit I are outlined in the supporting information, which is published on the PNAS web site. Western blot analyses were carried out with standard procedures, using horseradish peroxidase-conjugated anti-rabbit IgG antibodies as the second antibody and enhanced chemiluminescence (Perkin–Elmer), which was detected as an image on x-ray film and quantified with image gauge (Fujifilm) after running on a CanoScan D2400U (Canon). The band intensities of subunit I in the Western blot pattern for bovine and human mitochondria are proportional to the amount of subunit I applied. O2-reduction and proton-pumping activities of the hybrid cytochrome c oxidase were analyzed by using mitoplasts (a mitochondrial preparation from which the outer membrane has been removed to enable direct reaction with ferrocytochrome c added) isolated from the transfected cell lines. The purification of bovine heart cytochrome c oxidase and the crystallization conditions are as described (4). Freezing the crystals soaked in buffer containing 45% ethyleneglycol at 100 K in a nitrogen stream on the goniometer is critical for improvement of the resolution of the x-ray diffraction experiment. The effect of x-ray irradiation on the absorption spectrum of the enzyme in crystals under x-ray irradiation for diffraction experiments was monitored with a custom-designed visible absorption spectrometer with a focused light beam designed to measure only the area of the enzyme crystal undergoing x-ray irradiation. Under these x-ray diffraction conditions, only a minor fraction of the heme a3 moiety of the oxidized enzyme in the crystals undergoes spectral changes indicative of the heme reduction. No significant x-ray irradiation effects are detectable in the spectrum of heme a. No x-ray effect on the absorption spectrum is detectable for the reduced enzyme. Crystals diffracting up to 1.65-Å resolution were used for the x-ray diffraction experiments. The redox-coupled infrared spectral changes were determined at 20°C with a Perkin–Elmer Spectrum 2000 Fourier transform infrared spectroscopy (FTIR) system by a conventional difference spectrum method with a single cell. Other experimental details of the above methods used in this work are given in the supporting information.
Results and Discussion
Mutagenesis of Asp-51 in Subunit I of Bovine Heart Cytochrome c Oxidase. Asp-51 of subunit I (bovine) was replaced with Asn, a nonprotonatable and isosteric amino acid, by a hybrid formation method. Expression vectors were constructed for producing, in the cytosol of HeLa cells, mitochondrially encoded subunit I of the bovine enzyme with a mitochondrial targeting signal at the N terminus and a hexahistidine tag at the C terminus, which is for quantitative determination of the expressed subunit I. The x-ray structure indicates that the C terminus of subunit I has enough space for accepting the His tag without affecting the conformation of the enzyme.
The efficiency of the bovine/human hybrid enzyme production was evaluated with antibodies strictly specific to human and bovine subunit I, as shown by the Western blot analysis of SDS-treated mitochondria from bovine heart and HeLa cells. Antibody for bovine subunit I shows a clear band for bovine subunit I but not for human subunit I (Fig. 1A, lanes 1 and 2), whereas the antibody to the human protein reacts only with the human subunit I and not with the bovine subunit (Fig. 1 A, lanes 3 and 4). Dodecyl maltoside solubilizes cytochrome c oxidase from the mitochondrial membrane without denaturing the protein and yields a 210-kDa band of cytochrome c oxidase in blue native PAGE. The bovine antibody reacts with the 210-kDa fraction of dodecyl maltoside-solubilized mitochondria isolated from a HeLa cell line transfected with the bovine wild-type gene (Fig. 1B, lane 2). The corresponding fraction taken from a cell line transfected by a vector carrying no subunit I gene (mock-transfected cell line) did not produce an analogous band (Fig. 1B, lane 1). On the other hand, the human antibody reacts with the fraction from the mock-transfected cell line (Fig. 1B, lanes 4 and 6). Thus, the results show that bovine subunit I is assembled with human subunits. The presence of a weak band in the 210-kDa fraction of the solubilized mitochondria of the cell line transfected with the bovine wild-type gene (Fig. 1B, lane 5) indicates that human subunit I is not completely replaced by bovine subunit I in the transfected cells. The residual amount of the nonhybrid enzyme was estimated quantitatively by comparing the band intensities of the 210-kDa bands from the mock-transfected and wild-type gene-transfected cells as described in Methods to be ≈20% (Fig. 1B, lanes 4 and 5). Bands shown in Fig. 1B are satisfactorily as clear as those of blue native PAGE, which provides bands much less clear than those of SDS/PAGE. The visible spectrum of dodecyl maltoside-solubilized and fully reduced mitochondria from the wild-type gene-transfected cells exhibits an α-band at 604 nm, which is characteristic of the native cytochrome c oxidase. No mitochondrial component other than cytochrome c oxidase has significant absorbance at 604 nm. Furthermore, the solubilized hybrid enzyme preparation exhibits enzyme specific activity (ferrocytochrome c oxidation rate per enzyme molecule) as high as that of the solubilized enzyme preparation from the mock-transfected cells. The spectrum and enzyme activity of the hybrid enzyme provide convincing evidence that the hybrid enzyme adopts a native conformation. Cells transfected with the Asp51Asn mutant subunit I gene exhibit production of the hybrid enzyme as efficiently as the wild-type gene-transfected cell lines given above (Fig. 1B, lanes 2 and 3). The absorption spectrum and electron transfer activity of the solubilized preparation are also essentially identical to those of the wild-type hybrid enzyme. These results suggest that the Asp51Asn mutation does not perturb the three-dimensional structure of the enzyme.
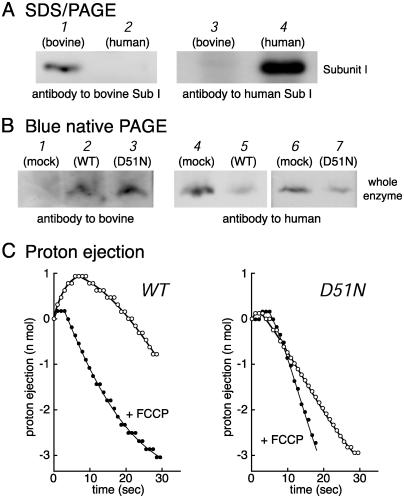
Fig. 1.
Effects of Asp-51 → Asn (Asp51Asn) mutation of subunit I on the function of cytochrome c oxidase. (A) Mitochondria from bovine heart (lanes 1 and 3) and HeLa cells (lanes 2 and 4) treated with 4% SDS/3.5 M urea and fractionated by SDS/PAGE. A sample of 70 μg of protein was loaded except for lane 1 (20 μg of protein). Antibody for bovine subunit I was used for lanes 1 and 2, and antibody for human subunit I was used for lanes 3 and 4. (B) Blue native gel electrophoresis carried out to fractionate 1.4% dodecyl maltosidesolubilized mitochondria from HeLa cells of mock-transfected (lanes 1, 4, and 6) and from HeLa cells harboring the wild-type (lanes 2 and 5) and Asp51Asn mutant (lanes 3 and 7) subunit I genes of bovine cytochrome c oxidase. In lanes 1, 2, and 3, 180, 90, and 90 μg of protein was loaded, respectively. In lanes 4–7, 70 μg of protein was loaded. The antibody specific to bovine or human subunit I was used for lanes 1–3 or for lanes 4–7, respectively. (C) Proton ejection/uptake by mitoplast samples (44.7 μg of protein) from the HeLa cells transfected with the bovine wild-type (WT) and Asp51Asn (D51N) mutant subunit I genes after addition of 8.1 nmol of ferrocytochrome c. The proton concentration changes in the presence of 10 nmol of carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) per mg of mitoplast protein are indicated by +FCCP.
Electron-transfer and proton-pumping activities of these hybrid enzymes in the mitochondria were determined by measuring the rates of ferrocytochrome c oxidation and of proton ejection by a mitoplast preparation in the presence of several reagents for blocking activities of other components in the mitochondrial inner membrane, including valinomycin and KCl for elimination of the membrane potential (supporting information). The reaction is initiated by addition of ferrocytochrome c. The ferrocytochrome c oxidation is completely inhibited by 1 mM cyanide. A mitoplast sample that includes the wild-type bovine subunit I undergoes initial acidification followed by significant alkalinization caused by O2 reduction and the proton-pumping process (Fig. 1C, WT). Similar curves are obtained for the samples from the mock-transfected cell lines (not shown). Carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) completely removes the initial acidification (Fig. 1C, WT + FCCP). The initial lag of alkalinization in the presence of FCCP indicates that the alkalinization due to O2 reduction is negligible in the initial 4 sec. Thus, the proton ejection rate can be determined from the linear phase in the first 4 sec after the initiation of the reaction. The estimated quantities of protons and electrons transferred in the initial 5 sec are 0.96 nmol of protons (Fig. 1C, WT) and 1.17 nmol of electron equivalents, respectively, giving a H+/e– ratio of 0.82. The other three measurements using the mitoplast samples, including the wild-type bovine subunit I from the same cell line, gave ratios of 0.83, 0.58, and 0.79. These values are similar to those determined for mitoplast samples prepared from a mock-transfected cell line (0.85 and 0.82) and also to the reported values for mammalian enzymes (6). These results also indicate that the native conformation of the enzyme exists in the hybrid enzymes. Mitoplast preparations including the Asp51Asn mutant bovine subunit I do not exhibit initial acidification (Fig. 1C, D51N). Acidification is also absent in the presence of FCCP (Fig. 1C, D51N + FCCP). The mutant mitoplasts exhibit cyanide-sensitive oxidation of ferrocytochrome c at an ≈70% faster rate than that of the wild type. The content of the residual human enzyme (20%, as stated above) is too low to detect proton pumping under the present experimental conditions. These results have been confirmed by data obtained from another wild-type gene-transfected cell line and three different Asp51Asn mutant gene-transfected cell lines.
The widely accepted endosymbiotic hypothesis of organelle origin implies that the original genes were massively transferred to the nucleus during evolution. However, subunit I and cytochrome b, which have 12 and 8 transmembrane α-helices, respectively, are encoded by mitochondrial DNA in all known eukaryotic organisms. The present successful import of subunit I into mitochondria is in conflict with the long-standing view that the extensive hydrophobic regions were instrumental in preventing subunit I and cytochrome b from being transferred from the mitochondrial genome during evolution (7). Establishment of the expression system of subunit I has never been attempted by the hybrid formation method used here, because attempts to transport apocytochrome b (which is smaller than subunit I) into mitochondria were unsuccessful (5).
X-Ray Structure of the H Pathway. X-ray structures of bovine cytochrome c oxidase in the fully oxidized and the reduced states at 1.8- and 1.9-Å resolution, respectively, indicate that a large conformational change of Asp-51 of subunit I occurs near the molecular surface of the intermembrane side (Fig. 2A), which includes a significant rearrangement of hydrogen-bonding interactions (Fig. 2B). The pKa of a carboxyl group is strongly influenced by its environment. For example, the pKa of acetic acid is 4.8 in water and 9.5 in methanol (8). Thus, the polar but nonaqueous environment of Asp-51 in the oxidized state, provided by the two Ser OH groups and two peptide NH groups, indicates that Asp-51 is most likely protonated. In the reduced state, the carboxyl group of Asp-51 resides at the molecular surface on the intermembrane side, in an essentially aqueous environment, indicating that the carboxyl group is most likely in the deprotonated state.
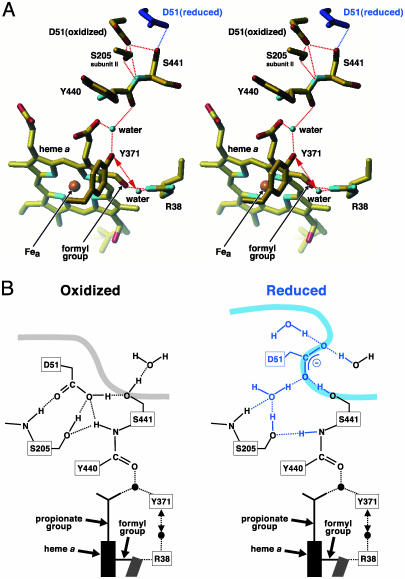
Fig. 2.
Redox-coupled conformational changes in Asp-51. (A) Stereoscopic drawing of the hydrogen-bond network in the fully oxidized and reduced (blue structure) states at 1.8- and 1.9-Å resolution, respectively, viewed from the intermembrane side. The two histidines bound to Fea (heme a iron), are not shown. (B) The hydrogen-bonding structure of Asp-51 in the oxidized (Left) and reduced (Right) states. The smooth thick curve denotes the molecular surface to which the water molecules in the intermembrane space are accessible. The conformational changes induced by reduction of the enzyme are shown by blue structures in Right. The blue (A) and black (B) balls represent the fixed water molecules. The dotted lines denote hydrogen bonds. The double-headed dotted arrows show a possible movement of the water molecule from Arg-38 to Tyr-371.
The carbonyl group of the peptide bond between Tyr-440 and Ser-441 is connected with Arg-38 by a hydrogen-bond network that includes a fixed water, Tyr-371, and a second fixed water molecule (Fig. 2). The water molecule hydrogen-bonded to Arg-38 is located ≈4 Å from Tyr-371, a distance too far to form a hydrogen bond. This water molecule could approach Tyr-371 to form a new hydrogen bond after moving away from Arg-38, an event indicated by the dotted double-headed arrows. The fixed water between Tyr-371 and the peptide carbonyl group is also hydrogen bonded to a propionate group of heme a. The structure of the hydrogen bond network from the peptide carbonyl group to Arg-38 is suggestive of much more effective proton transfer compared with the network observed in the x-ray structures at 2.3/2.35-Å resolution (4). The detailed comparison is given in the supporting information. Other improvements in the x-ray structures, compared with the previous structures (4), are also described in the supporting information.
Proton transfer through peptide bonds is possible (9). When a proton is added to a peptide carbonyl group, an imidic acid [ C(OH) N+H ] is formed. If a proton-accepting group is located near the
N+H ] is formed. If a proton-accepting group is located near the  N+H moiety, it will extract a proton to give the enol form of the peptide [ C(OH)
N+H moiety, it will extract a proton to give the enol form of the peptide [ C(OH) N ]. The much higher stability of the keto form ( CO NH ) relative to the enol form [ C(OH)
N ]. The much higher stability of the keto form ( CO NH ) relative to the enol form [ C(OH) N ] and the very small conformational change accompanied by the keto-to-enol transition are indicative of a rapid reversion to the keto form. The peptide imparts a unidirectional character to the proton transfer through the peptide that blocks reverse proton transfer from the intermembrane side.
N ] and the very small conformational change accompanied by the keto-to-enol transition are indicative of a rapid reversion to the keto form. The peptide imparts a unidirectional character to the proton transfer through the peptide that blocks reverse proton transfer from the intermembrane side.
Asp-51 in the oxidized state is hydrogen bonded to Ser-441, which is on the molecular surface, whereas in the reduced state a short hydrogen bond network including a fixed water between Asp-51 and Ser-205 connects Asp-51 with the hydrogen bond network extending to Arg-38 (Fig. 2B). Thus, Asp-51 is accessible via the hydrogen bond network to both side of the enzyme molecule in both oxidation states. The reverse proton transfer is blocked by the peptide bond. On the other hand, the previous structures at 2.3/2.35-Å resolution suggest that Asp-51 in the oxidized state is connected to the matrix space by the hydrogen-bond network and buried inside the intermembrane surface, while, upon reduction of the enzyme, it dissociates from the hydrogen-bond network and is exposed to the intermembrane space (4). This present model that the proton-pumping process is driven by the pKa change of Asp-51 and unidirectional proton transfer through the peptide bond contrasts with the previous proposal described above.
Heme a iron is bound to six nitrogens (two from histidines and four from porphyrin) in both oxidation states. In the reduced state, the two positive charges of Fe2+ are neutralized with the two negative charges of the porphyrin. However, in the oxidized state, the Fe3+ has one unneutralized positive charge. Relocation of this positive charge through the porphyrin π-electron system can promote deprotonation of any acidic group located close to the heme a. The present x-ray structures show much more clearly than the previous structures that the heme a formyl group is coplanar with the porphyrin ring and thus able to conjugate readily with the porphyrin π-electron system in both oxidation states, which is consistent with direct resonance Raman evidence for the effect of iron oxidation state on the formyl carbonyl shown by the C O stretch vibrational shift from 1610 to 1650 cm–1 upon heme a oxidation (10, 11). Thus, a change in heme a oxidation state can be expected to have a significant electrostatic influence on Arg-38, which is hydrogen bonded to the formyl group, although no significant conformational change is caused in the formyl–Arg-38 system (Fig. 2 A). The present resolution of the x-ray structures seems insufficient to detect the structural change induced by the electrostatic influence from Fea. The propionate group of heme a hydrogen bonded to the fixed water, as described above (Fig. 2), could stimulate proton transfer through the hydrogen bond network, also by the electrostatic influence upon oxidation of Fea.
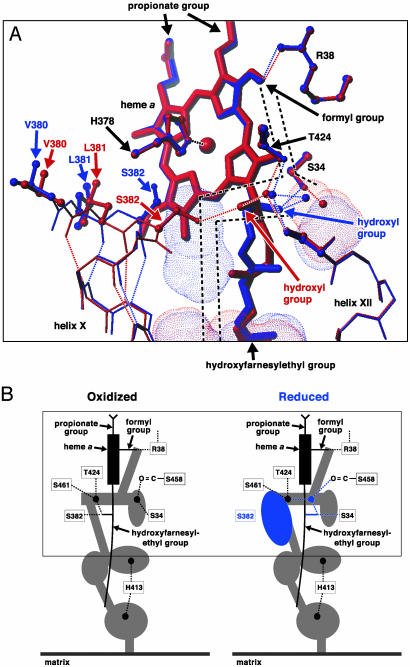
Arg-38 is hydrogen bonded to the heme a formyl group, which water molecules in the matrix space can access via a water channel. The channel near the formyl end, traced by a random walk program (4), is shown schematically by broken lines in Fig. 3A. The water-accessible space in the channel, determined by a molecular surface calculation, indicates that the channel has four cavities, each of which is large enough to contain one to three water molecules (Fig. 3B and red dotted spaces in Fig. 3A). The OH group of the hydroxyfarnesylethyl group of heme a is hydrogen bonded to Ser-382 near the water channel in the oxidized state (Fig. 3A, red structures). Upon reduction of the enzyme, the OH···Ser-382 hydrogen bond is cleaved, enabling the OH group of the hydroxyfarnesylethyl group to rotate 120° with a concomitant movement of the hydrocarbon chain near the OH group [ CH(OH) CH2 CH2 CH C(CH3) ] and enabling a 110° rotation of the OH group of Ser-382 (Fig. 3A, blue structures), coupled with a conformational change in the turn of helix X, which includes Ser-382, Leu-381, and Val-380 (Fig. 3A, red and blue amino acid structures in helix X). These conformational changes produce a new cavity between the hydroxyfarnesylethyl group and helix X (Fig. 3A, blue dotted space with no red dotted space superimposed and Fig. 3B, blue oval). The conformation change could be controlled by the oxidation state of heme a because the OH···Ser-382 hydrogen bond is located very close to heme a porphyrin π-electron system. The position and size of the four cavities observed in the oxidized state are not significantly influenced upon reduction (Fig. 3 red and blue dotted spaces). The redox-coupled conformational change indicates a water capacity change in the channel, which is likely to contribute for effective proton collection from the matrix space to Arg-38.
C(CH3) ] and enabling a 110° rotation of the OH group of Ser-382 (Fig. 3A, blue structures), coupled with a conformational change in the turn of helix X, which includes Ser-382, Leu-381, and Val-380 (Fig. 3A, red and blue amino acid structures in helix X). These conformational changes produce a new cavity between the hydroxyfarnesylethyl group and helix X (Fig. 3A, blue dotted space with no red dotted space superimposed and Fig. 3B, blue oval). The conformation change could be controlled by the oxidation state of heme a because the OH···Ser-382 hydrogen bond is located very close to heme a porphyrin π-electron system. The position and size of the four cavities observed in the oxidized state are not significantly influenced upon reduction (Fig. 3 red and blue dotted spaces). The redox-coupled conformational change indicates a water capacity change in the channel, which is likely to contribute for effective proton collection from the matrix space to Arg-38.
Fig. 3.
X-ray structure of the water channel in the H pathway. (A) Redox-coupled conformational change of the water channel. The upper part of the channel is shown. Red and blue denote structures in the oxidized and reduced states, respectively. Red and blue dotted surfaces indicate cavities detectable in the oxidized and reduced states, respectively. The broken lines schematically indicate water paths connecting these cavities. The dotted lines show hydrogen bonds. The small balls mark the positions of fixed water molecules. His-61 bound to Fea from the opposite side of the heme plane is not shown. (B) Schematic representation of the redox-coupled conformational changes in the water channel. The area given in A is marked by a square. The water channel is represented by the gray and blue area. The black and blue balls denote fixed water molecules. The structures detectable only in the reduced state are in blue.
FTIR Analysis of the Redox-Coupled Conformational Changes. FTIR was applied to identify the redox-active metal site (or sites) controlling the conformation of Asp-51. In the difference FTIR spectrum of the fully oxidized versus the fully reduced enzyme in H2O, Asp-51 gives a peak at 1,738 cm–1 and a trough at 1,585 cm–1, which are assignable to C O stretch modes of COOH and COO–, respectively (12). These results confirm previously reported results (supporting information). Redox difference spectra in the presence of cyanide and CO also give essentially identical bands at 1,738 and 1,585 cm–1. Cyanide stabilizes heme a3 in the oxidized state. Thus, the enzyme reduced in the presence of cyanide has heme a, CuA, and CuB in the reduced state and heme a3 in the cyanide-bound oxidized state. As a result, the difference spectrum between the cyanide-bound oxidized enzyme and the cyanide-bound reduced enzyme yields the sum of redox difference spectra induced by heme a, CuA, and CuB. On the other hand, CO stabilizes CuB and heme a3 in the reduced state to give the sum of redox difference spectra induced by heme a and CuA. Therefore, these results indicate that the protonation state of Asp-51 is controlled by heme a and/or CuA.
In the reductive titration of the bovine enzyme in the presence of cyanide, the decreases in intensity of the two bands are proportional to the electron equivalents added. Three electron equivalents are required for complete elimination of the two bands in the difference spectrum. The results indicate that the infrared spectral changes are caused by a single electron equivalent. The reductive titration of the cyanide-bound enzyme, monitored by the oxidation states of the redox-active metal sites, indicates that in each overall oxidation state, the reducing equivalents are evenly distributed among the three metal sites, CuA, heme a, and CuB (13). Thus, if two or three electrons were required for induction of the infrared spectral changes, the spectral change would not appear below one or two electron equivalents, respectively. Therefore, COOH of Asp-51 dissociates upon reduction of only one metal site, either heme a or CuA. The detailed infrared results are given in the supporting information.
It has been reported that only one proton equivalent per fully reduced enzyme is pumped upon complete oxidation of the fully reduced enzyme with O2 (14, 15). (Both the experimental results given in figure 2 of ref. 14 and figure 2 of ref. 15 clearly show that 1.2–1.3 protons per enzyme molecule are released upon O2 oxidation of the fully reduced enzyme, although the authors of both papers do not conclude “one proton per enzyme.”) The electron equivalents from cytochrome c are transferred from CuA to heme a and subsequently, from heme a to the O2 reduction site (16). In the reaction of the fully reduced enzyme with O2, the reduced heme a is oxidized by the O2 reduction site and then the oxidized heme a is reduced by CuA, before the O2 reduction site receives the electron equivalent. The process includes a single reduction step of heme a, but no reduction step of CuA. Thus, heme a can be assigned as the metal site controlling the conformation of Asp-51. This conclusion that proton release occurs upon heme a reduction is consistent with the reported experimental results indicating that proton pumping occurs in the reductive phase (15, 17).
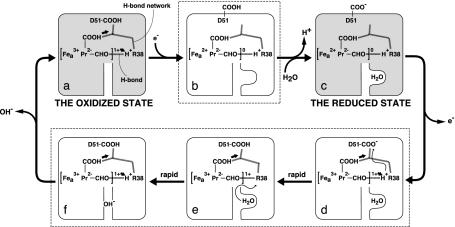
The Proton-Pumping Mechanism. The overall mechanism based on the present results is summarized as follows: while heme a is in the oxidized state (Fig. 4a), Arg-38 remains predominantly protonated even under the positive charge influence from heme a, because water molecules in the matrix space are accessible to Arg-38 via the water channel. Asp-51 is buried inside the protein and is protonated (Fig. 4a). Upon heme a reduction, which removes the net positive charge in heme a, Asp-51 is exposed to the intermembrane space and the capacity of the water channel increases (Fig. 4b) so that water molecules are taken up from the matrix space, while the proton on Asp-51 is released to the intermembrane space (Fig. 4 b and c). Upon heme a oxidation, Asp-51 moves back to the interior of the protein and the appearance of the net positive charge on heme a decreases the affinity of the formyl oxygen for the proton shared with Arg-38. This decreased affinity promotes proton transfer from Arg-38 toward Asp-51 (Fig. 4 d and e). The propionate group hydrogen-bonded to the fixed water could also accelerate proton transfer along the hydrogen-bond network. The resulting deprotonated Arg-38 extracts protons from water molecules in the water channel (Fig. 4e) before the water channel capacity decreases to expel OH– (Fig. 4 f and a). The ability of the channel to rapidly undergo a reduction in water capacity and release OH– immediately after water dissociation is expected to prevent reverse proton transfer (Fig. 4 e and f).
Fig. 4.
Proposed proton-pumping mechanism. The iron, porphyrin, and formyl side group of heme a are shown by Fea, Pr, and CHO, respectively. The COOH on Pr denotes one of the propionate groups of heme a. The brackets ([]1+ and[]0) indicate the net charge of the six-coordinated heme a. The diagrams shadowed and in the dotted squares show the structures in the stable and intermediate states, respectively. The thick arrows in a and d–f and the thin arrows in d and e indicate the electrostatic influence of the net positive charge of heme a and the proton transfers upon heme a oxidation, respectively. The dotted lines in the diagrams denote the hydrogen bond network connecting Arg-38 with Asp-51, including the peptide bond that blocks the reverse proton transfer from the intermembrane side.
Involvements of heme a formyl group and water molecules surrounding heme a in proton pumping have been suggested on the basis of resonance Raman results (10, 11). The critical role of heme a in proton pumping has been proposed also on the basis of extensive investigation of redox-coupled proton ejection and uptake by the enzyme (18). Asp-51, one of the key residues of the H pathway, is conserved only in the animal enzymes as described above. However, x-ray structures of the bovine and bacterial enzymes suggest the existence of a proton pump system driven by the low-spin heme apart from the O2 reduction system in all cytochrome c oxidases (4, 19–21). Detailed discussions on the evolutionary aspects of the H pathways are presented in the supporting information. A previously reported view, proton pumping at or near the O2 reduction site, which is in contrast to the proton pumping in the H pathway, is evaluated in the supporting information, in light of the present experimental results.
Supplementary Material
Acknowledgments
We thank Dr. Takashi Ogura and Dr. Takashi Sugimura for helpful suggestions, Dr. Makoto Suematsu for helpful discussions, and Dr. Takashi Osumi, Dr. Takashi Sugimura, Dr. Kenji Watanabe, Dr. Winslow S. Caughey, and Dr. Mark Roach for critical reading of the manuscript. S.Y. is a Senior Visiting Scientist in RIKEN Harima Institute. This work was supported in part by Grants-in-Aid for Scientific Research on Priority Areas (H.S. and K.S.-I.) and for 21st Century Center of Excellence Program from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (T.T. and S.Y.) and by a research grant from the Japan Biological Informatics Consortium entrusted from the New Energy and Industrial Technology Organization (to T.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: heme a3, a high-spin heme A of cytochrome c oxidase; heme a, a low-spin heme A of cytochrome c oxidase; CuB, a copper ion located near heme a3; CuA, a copper site that receives electrons from ferrocytochrome c; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; FTIR, Fourier transform infrared spectroscopy.
Data deposition: The atomic parameters of cytochrome c oxidase have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1V54 and 1V55).
References
- 1.Ferguson-Miller, S. & Babcock, G. T. (1996) Chem. Rev. 96, 2889–2907. [DOI] [PubMed] [Google Scholar]
- 2.Gennis, R. B. (1998) Biochim. Biophys. Acta 1365, 241–248. [DOI] [PubMed] [Google Scholar]
- 3.Tsukihara, T., Aoyama, H., Yamashita, E., Tomizaki, T., Yamaguchi, H., Shinzawa-Itoh, K., Nakashima, R., Yaono, R. & Yoshikawa, S. (1996) Science 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- 4.Yoshikawa, S., Shinzawa-Itoh, K., Nakashima, R., Yaono, R., Yamashita, E., Inoue, N., Yao, M., Fei, M. J., Libeu, C. P., Mizushima, T., et al. (1998) Science 280, 1723–1729. [DOI] [PubMed] [Google Scholar]
- 5.Claros, M. G., Perea, J., Shu, Y., Samatey, F. A., Popot, J.-L. & Jacq, C. (1995) Eur. J. Biochem. 228, 762–771. [PubMed] [Google Scholar]
- 6.Wikström, M. K. F. (1977) Nature 266, 271–273. [DOI] [PubMed] [Google Scholar]
- 7.de Grey, A. D. N. J. (2000) Trends Biotechnol. 18, 394–399. [DOI] [PubMed] [Google Scholar]
- 8.Isaacs, N. S. (1995) Physical Organic Chemistry (Longman, Essex, U.K.), 2nd Ed., pp. 235–286.
- 9.Perrin, C. L. (1989) Acc. Chem. Res. 22, 268–275. [Google Scholar]
- 10.Babcock, G. T. & Callahan, P. M. (1983) Biochemistry 22, 2314–2319. [DOI] [PubMed] [Google Scholar]
- 11.Sassaroli, M., Ching, Y.-C., Dasgupta, S. & Rousseau, D. L. (1989) Biochemistry 28, 3128–3132. [DOI] [PubMed] [Google Scholar]
- 12.Venyyaminov, S. Y. & Kalmin, N. N. (1990) Biopolymers 30, 1243–1257. [DOI] [PubMed] [Google Scholar]
- 13.Yoshikawa, S., Mochizuki, M., Zhao, X.-J. & Caughey, W. S. (1995) J. Biol. Chem. 270, 4270–4279. [DOI] [PubMed] [Google Scholar]
- 14.Oliveberg, M., Hallen, S. & Nilson, T. (1991) Biochemistr y 30, 436–440. [DOI] [PubMed] [Google Scholar]
- 15.Verkhousky, M. I., Jasatis, A., Verkhovskaya, M. L., Morgan, J. E. & Wikstrom, M. (1999) Nature 400, 480–483. [DOI] [PubMed] [Google Scholar]
- 16.Hill, B. C. (1994) J. Biol. Chem. 269, 2419–2425. [PubMed] [Google Scholar]
- 17.Ruitemberg, M., Kannt, A., Bamberg, E., Fendler, K. & Michel, H. (2002) Nature 417, 99–102. [DOI] [PubMed] [Google Scholar]
- 18.Capitanio, N., Capitanio, G., Boffoli, D. & Papa, S. (2000) Biochemistry 39, 15454–15461. [DOI] [PubMed] [Google Scholar]
- 19.Soulimane, T. Buse, G., Bourenkov, G. P., Bartunik, H. D., Huber, R. & Than, M. E. (2000) EMBO J. 19, 1766–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ostermeier, C., Harrenga, A., Ermler, U. & Michel, H. (1997) Proc. Natl. Acad. Sci. USA 94, 10547–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svensson-Ek, M., Abramson, J., Larsson, G., Törnroth, S., Brzezinski, P. & Iwata, S. (2002) J. Mol. Biol. 321, 329–339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.