Summary
Notch proteins are transmembrane receptors that normally adopt a resting state poised to undergo activating proteolysis upon ligand engagement. Receptor quiescence is maintained by three LIN12/Notch repeats (LNRs), which wrap around a heterodimerization domain (HD) divided by furin cleavage at site S1 during maturation. Ligand binding initiates signaling by inducing sensitivity of the HD to proteolysis at the regulated S2 cleavage site. Here, we used hydrogen exchange mass spectrometry to examine the solution dynamics of the Notch1 negative regulatory region in autoinhibited states before and after S1 cleavage, in a proteolytically sensitive “on” state, and in a complex with an inhibitory antibody. Conversion to the “on” state leads to accelerated deuteration in the S2 region and in nearby secondary structural elements within the HD. In contrast, complexation with the inhibitory antibody retards deuteration around the S2 site. Together, these studies reveal how S2 site exposure is promoted by receptor activation and suppressed by inhibitory antibodies.
Notch signaling is an ancient cell-cell communication system that regulates embryonic and fetal development as well as adult tissue homeostasis (Bray, 2006; Kopan and Ilagan, 2009). The four human Notch receptors are large, single-pass transmembrane proteins that share a similar modular organization, with a series of EGF repeats that bind ligands, a negative regulatory region (NRR), and an intracellular effector domain that follows a single transmembrane segment (Figure 1A). The receptors are normally synthesized as precursor proteins, which are typically cleaved during transport to the cell surface (at a site called S1) by a furin-like protease (Logeat et al., 1998).
Figure 1.
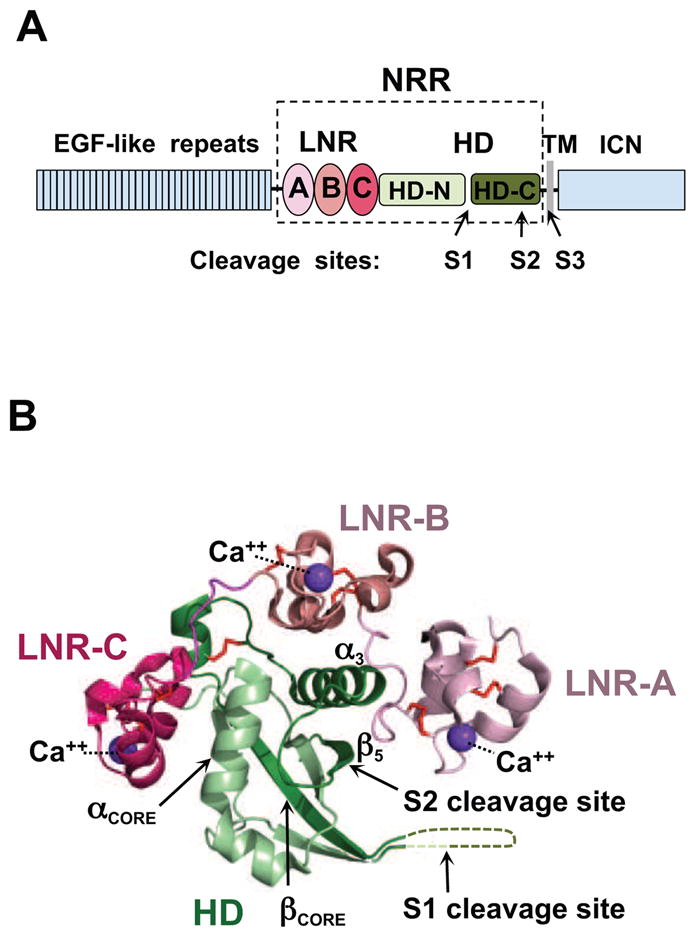
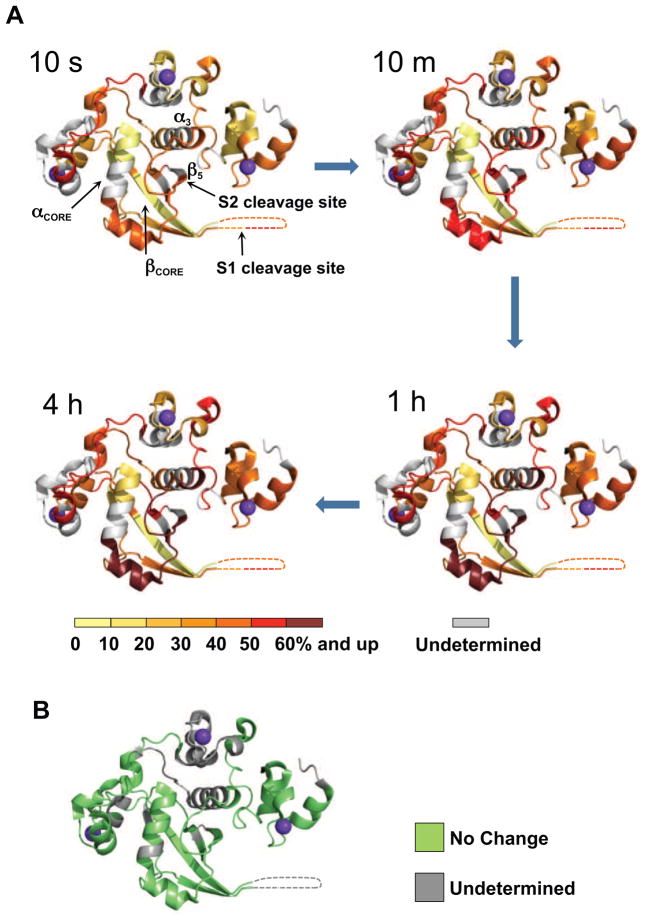
Domain organization and overview of the Notch1 NRR structure. (A) Domain organization of Notch1. The extracellular portion of the receptor consists of 36 EGF-like repeats responsible for ligand binding (blue), and the negative regulatory region (NRR, boxed) that maintains proteolytic resistance in the absence of ligands. The NRR encompasses three LIN12/Notch repeats (LNR-A, LNR-B, and LNR-C, colored in different shades of red), and the heterodimerization domain (HD, green), divided at S1 by a furin-like protease during maturation. Ligand binding to the extracellular portion of Notch triggers metalloprotease cleavage at site S2. The resulting truncated transmembrane subunit of the receptor is a substrate for cleavage at S3 by gamma-secretase, which releases the intracellular part of Notch (ICN, blue) from the membrane. (B) Ribbon diagram of the Notch1 NRR in its autoinhibited conformation (PDB ID code 3IO8). The three LNR modules are shown in different shades of pink, and the HD domain is in green. Disulfide bonds are orange, and the three calcium ions coordinated by the LNR modules are purple. Key secondary structural elements and the S1 and S2 cleavage sites are also indicated. See also Figure S1.
Notch receptors are poised to undergo activating proteolysis upon binding to transmembrane ligands on neighboring cells. The activation switch of the receptor lies within the NRR (Kopan et al., 1996; Sanchez-Irizarry et al., 2004), which includes a series of three Lin12/Notch repeats (LNRs) and a heterodimerization domain (HD) that becomes divided upon cleavage at S1 (Logeat et al., 1998). The NRR maintains proteolytic resistance in the absence of ligands by burying a proteolytic site called S2, which is situated near the C-terminal end of the HD domain (Gordon et al., 2009a; Gordon et al., 2007; Gordon et al., 2009b) (Figure 1B). In a process that remains poorly understood, ligand binding then renders S2 sensitive to cleavage by ADAM-family metalloproteases (Brou et al., 2000; Mumm et al., 2000). The truncated receptor thus generated is subsequently processed by the intramembrane protease called gamma secretase to liberate the Notch intracellular domain (ICN), which travels to the nucleus and assembles into a transcriptional activation complex to regulate target gene transcription (see Kopan and Ilagan, 2009 for a review).
Proper regulation of Notch activity is crucial, as mutations that increase or decrease Notch signal strength can produce developmental defects or diseases such as cancer. The frequent occurrence of activating point mutations within the Notch1 NRR in T cell lymphoblastic leukemia/lymphoma, for example, further highlights the consequences of even modestly destabilizing this regulatory switch to tip the balance in favor of proteolytic sensitivity (Malecki et al., 2006; Weng et al., 2004).
Here, we sought to explore the fundamental issue of protease sensitivity by using hydrogen exchange mass spectrometry (HX MS) (Wales and Engen, 2006) to examine the conformation and dynamics of various states of the Notch1 NRR, focusing on the kinetic accessibility of the S2 site. The data show that the hydrophobic core of the Notch1 HD domain exchanges more slowly than the LNR repeats, and that slow exchange of the HD core is unaffected by S1 cleavage. Interactions between the LNR repeats and the HD shield residues in this interdomain interface from exchange, and relaxation of the long-range interface between the LNRs and the HD allows more deuterium to exchange into the region of the S2 site. Conversely, an anti-Notch1 inhibitory antibody, which contacts a discontinuous epitope encompassing residues from the first LNR domain and the loop preceding the S2 site, stabilizes the S2 site and leads to a reduced amount of deuteration. Together, these studies provide new insights into the mechanism underlying Notch autoinhibition, receptor activation, and the basis for allosteric modulation of Notch signals by inhibitory antibodies.
RESULTS
Dynamics of the wild-type NRR from human Notch1
We first investigated the dynamics of the intact Notch1 NRR in its autoinhibited conformation by HX MS. This approach, which reports on the combined effects of hydrogen bonding and solvent accessibility of the backbone amides of the protein, is particularly informative when a conformational state that is not amenable to crystallography is compared to a high-resolution structure of the protein in a different conformational state. Intact protein is labeled with deuterium for various amounts of time under physiological conditions. After quenching, the protein is digested into small peptide fragments and the amount of deuterium in each peptide measured with mass spectrometry (Supplementary Figure S1). With such data, it is possible to assign dynamic properties to specific structural elements of the protein because regions that are more exposed and dynamic undergo rapid deuteration whereas regions that are more stable and less dynamic are deuterated more slowly (Wales and Engen, 2006). Importantly, it is also then possible to identify regions of conformational change when data for two conformational states are compared. The peptides analyzed in comparative analysis studies are listed in Supplementary Table S1 and mapped onto the protein sequence in Supplementary Figure S2.
Analysis of the deuteration of the wild-type Notch1 NRR in its unprocessed form (prior to S1 cleavage) shows that the hydrophobic interior of the HD domain is most resistant to deuteration (Figure 2A and Supplementary Movie S1). The most highly protected secondary structural elements in the protein include the C-terminal part of the core helix (αcore), and the central beta strand (βcore) of the HD domain immediately preceding the S1 cleavage loop. Both of these regions exchanged fewer than 15% of their backbone amide protons even after four hours of deuterium labeling. The resistance of these secondary structural elements to deuteration is indicative of their relative stability in the autoinhibited state. These secondary structural elements coincide with the regions of the protein frequently mutated in human T cell lymphoblastic leukemia/lymphoma (Weng et al., 2004), further underscoring their importance in maintaining the structural integrity of the NRR.
Figure 2.
HX MS of the Notch1 NRR in the “off” state. (A) Extent of deuteration of peptides from the Notch1 NRR. The relative percent deuteration is colored based on a sliding scale from yellow (<10% incorporation) to deep red (>60% incorporation). Portions of the protein for which it was not possible to acquire exchange information are colored gray. (B) HX MS results comparing exchange of precursor and S1 cleaved forms of the Notch1 NRR. There were no detectable differences in the rates of peptide deuteration between the two proteins (see Supplemental Figure 3 for exchange curves). An absolute difference in mass of less than 0.5 Da is reported as “no change”. See also Figures S2 and S3, Table S1 and Movie S1.
By comparison, the three LNR modules and the N-terminal end of the first helix of the HD domain, which together comprise the periphery of the structure, exhibit much more rapid deuteration than the hydrophobic core of the HD domain. The dynamics of helix three (α3) and strand five, which is the β-strand containing the S2 cleavage site (Figure 1B), are more challenging to assess, because the major peptide reporting on these regions also includes the loop that connects them. The hydrogen exchange data show that this peptide (encompassing the S2 site) exhibits partial protection from exchange at the initial time point, and incorporates additional deuterium over time. This observation suggests that the masking of the S2 site is not absolute in the autoinhibited state, and that long-range engagement of the LNRs with the regions surrounding the S2 site is more dynamic than the protein interior, which incorporates little additional deuterium over time. Finally, peptides mapping to the loop linking LNR-B and LNR-C (BC loop) and the unstructured linker spanning the S1 site (S1 loop) show rapid deuteration (Supplementary Figure S3) that remains constant throughout the time course of the experiment, implying a high degree of solvent exposure and a lack of stable hydrogen bonds. These results are in agreement with previous NMR and X-ray studies (PDB 3ETO and 3I08) pointing to the intrinsic flexibility of both the BC and S1 loops.
Comparison of the dynamics of the Notch1 NRR with and without furin cleavage
One of the leading models for ligand-induced activation of Notch receptors postulates that ligand engagement of S1-cleaved receptors triggers subunit dissociation, thereby exposing the S2 site to metalloproteases (Nichols et al., 2007). However, the hydrophobic interior of the Notch1 NRR, which includes the αcore and βcore regions straddling the S1 cleavage site, is not rapidly deuterated, indicating that it is protected from exchange and substantially less dynamic than the region surrounding the S2 site. Opening of the protein to expose the S2 site would therefore appear to be kinetically favored over subunit dissociation.
To determine directly whether or not S1 cleavage changes the conformation of the hydrophobic core or otherwise alters the dynamics of the Notch1 NRR, we investigated the effect of furin cleavage on the hydrogen exchange behavior of the protein (for experimental data, see Supplementary Figure S3A). The data show that the deuterium exchange properties of the intact and furin-cleaved, heterodimeric forms of the Notch1 NRR are virtually indistinguishable (Figure 2B). Most importantly, the exchange patterns of the αcore, βcore, and S2-site regions remain unaltered, indicating that the hydrophobic core is still more highly protected than the S2 site even after cleavage at S1.
EDTA treatment of the Notch1 NRR as a model for an activated conformation
Like the type-A modules found in proteins of the low-density lipoprotein receptor family (Blacklow and Kim, 1996; Fass et al., 1997), the LNR modules of the Notch NRR rely on three disulfide bonds and calcium ion coordination by a conserved acidic motif to maintain their structural integrity (Aster et al., 1999; Vardar et al., 2003). This observation suggests that chelation of the bound calcium ions will detach the LNR modules from the HD domain and release the autoinhibition imposed by the long-range interface between these domains.
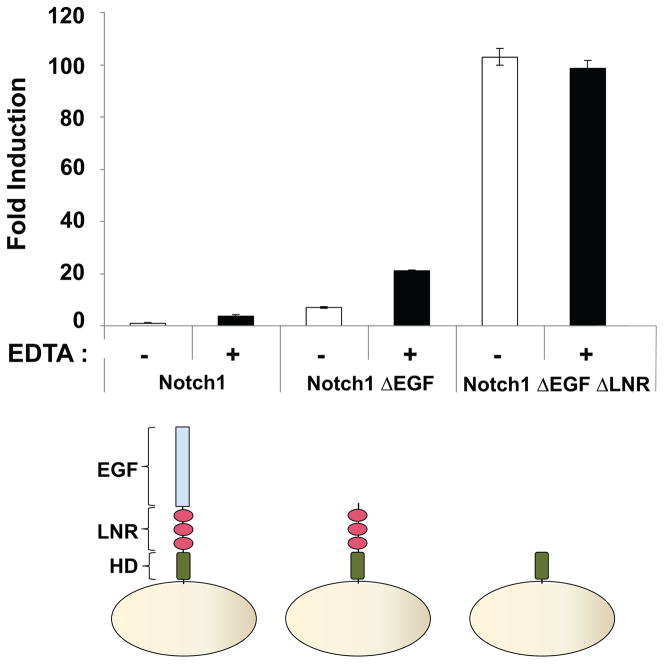
Consistent with this idea, previous studies have established that EDTA treatment activates Notch signaling in cells, and promotes shedding of the Notch ectodomain (Krejci and Bray, 2007; Rand et al., 2000). EDTA treatment of truncated receptors lacking the EGF repeats but retaining the NRR also promotes ectodomain shedding (Rand et al., 2000). To determine directly whether or not the activating effect of chelator treatment is intrinsic to the NRR, we tested whether EDTA treatment results in the activation of a truncated receptor in which the NRR has been retained but all of the EGF repeats have been deleted. The results (Figure 3) show that treatment of either full-length Notch1 or the truncated receptor with EDTA induces a five-fold increase in luciferase reporter gene transcription. Moreover, further truncation of the receptor to remove both the EGF and LNR modules creates a constitutively active protein that is unaffected by EDTA treatment. Together, these studies confirm that the induction of signaling by EDTA is intrinsic to the NRR of human Notch1.
Figure 3.
EDTA treatment induces Notch1 proteolytic sensitivity. U2OS cells transiently transfected with the indicated Notch1 variants (wt, ΔEGF, or ΔEGFΔLNR) were treated with buffer containing either CaCl2 (2.5 mM) or EDTA (5 mM). Firefly luciferase reporter gene activity was measured relative to renilla luciferase as a control. All measurements were done in triplicate. Error bars represent standard deviation.
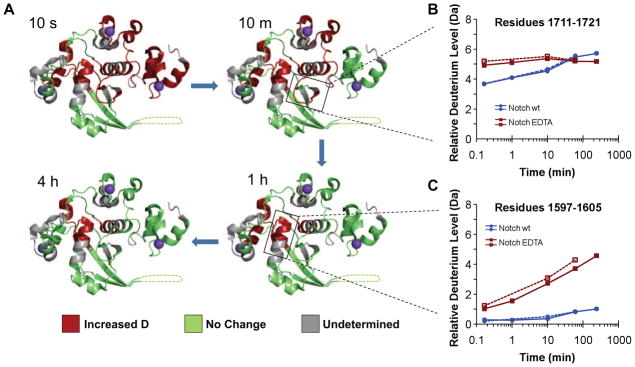
In the leading model for Notch activation, detachment of the LNR modules from the HD domain is proposed to occur in response to ligand-induced mechanical force (e.g. refs. (Kopan and Ilagan, 2009; Nichols et al., 2007)). Here, we used a chemical approach to model the activated state of the Notch1 NRR by using EDTA to disengage the LNR modules from the HD domain. Thus, we monitored the Notch1 NRR by HX MS after chelator treatment to probe the dynamics of the protein in this form of the “open” state, comparing the pattern of exchange to that of the native, autoinhibited protein (for data, see Supplementary Figure S3B). When the exchange properties of the LNR modules in their calcium-loaded and calcium-free state were compared, more rapid deuteration was observed for all three LNR repeats in their calcium-free state after EDTA treatment (Figure 4 and Supplementary Movie S2). This observation likely reflects the relaxation of structure resulting from removal of the bound calcium ion from each repeat (Figure 4A). More surprisingly, HD regions at the interdomain interface with the LNR modules, including peptides spanning the metalloprotease-cleavage site S2 (residues 1711-1721; Figure 4B) and a portion of the helix that precedes it (residues 1694-1702), also exhibited more rapid deuteration after EDTA treatment, as did the C-terminal end of αcore (Figure 4C). When the S1-cleaved form of the Notch1 NRR was also probed by HX-MS, the differences in exchange patterns between native and EDTA-treated states were largely preserved (Figure 5; for data, see Supplementary Figure S3C). Together, these studies reveal that hydrogen exchange at the site of metalloprotease cleavage is inhibited due to the stabilizing influence of the long-range interface between the LNR modules and the HD domain, explaining how the long-range interface disfavors access of metalloproteases to S2 in the autoinhibited state. In addition, the data also show that calcium coordination by the LNR repeats plays an essential role in stabilizing the interface responsible for maintaining autoinhibition.
Figure 4.
Comparison of HX MS patterns for the Notch1 NRR in “off” and “on” states. (A) Time course of exchange, comparing deuteration of the Notch1 NRR in closed and open (EDTA-treated) states. The ribbon diagram of the Notch1 NRR is colored according to the effect of EDTA treatment. Regions exhibiting faster deuteration in the presence of EDTA (with an absolute difference in mass of at least 0.5 Da at some point during the exchange experiment) are colored red, and regions with no detectable change in the deuteration rate (an absolute difference in mass of less than 0.5 Da at some point during the exchange experiment) are green. Gray regions correspond to segments for which no peptide mapping data are available. (B,C) Semi-logarithmic plots showing the relative deuterium level of two different regions (see Supplemental Figure 3B for all other peptides) of the Notch1 NRR in closed and open states. Data acquired from two independent experiments are shown to highlight the reproducibility of the measurements. Panel B encompasses the site of S2 cleavage and panel C corresponds to a section of the core helix in the hydrophobic core of the HD domain. See also Figures S2 and S3, Table S1, and Movie S2.
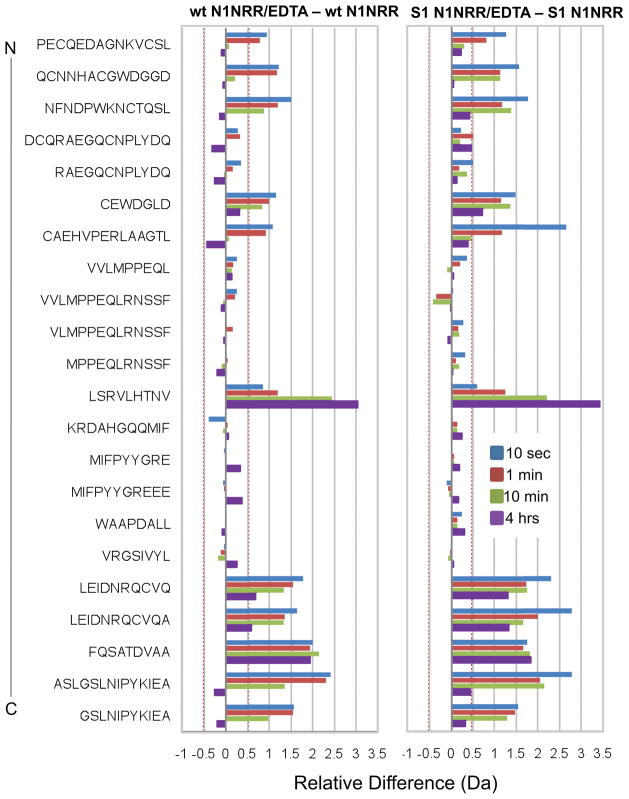
Figure 5.
HX-MS relative difference plots for unprocessed N1-NRR (left panel) and S1 cleaved N1-NRR (right panel), comparing closed and EDTA-treated states. The differences in deuterium incorporation between the EDTA treated protein and the native protein are plotted for deuterium exchange time points of 10 seconds (blue bars); 1 minute (red bars); 10 minutes (green bars) and 4 hours (purple bars). The data shown are for peptides common to both experiments, ordered from N- to C- terminus (top to bottom). See also Figures S2 and S3, and Table S1.
Molecular Basis for Anti-Notch1 NRR Antibody - Mediated Inhibition
Allosteric antibodies that inhibit Notch signaling by binding to the NRR have now been raised against Notch1 (Aste-Amezaga et al., 2010; Wu et al., 2010), Notch2 (Wu et al., 2010), and Notch3 (Li et al., 2008). The epitopes for the anti-Notch3 antibody and for one of the anti-Notch1 antibodies have been mapped, and the X-ray structure of one of the anti-Notch1 antibodies has been solved in complex with the Notch1 NRR. In both of these cases, the contact interface between the antibody and the NRR includes residues from both the LNR and HD domains, suggesting that the antibodies clamp the receptor in the off state by bridging the two domains.
Here, we examined the interaction between the human Notch1 NRR and a different anti-NRR antagonistic antibody, WC629, which has been shown to specifically target the NRR of Notch1 and no other Notch receptors (Aste-Amezaga et al., 2010). We identified the binding epitope for the WC629 antibody using a combination of HX MS and site-directed mutagenesis.
We first compared the exchange behavior of the Notch1 NRR in the presence and absence of antibody WC629 (for data, see Supplementary Figure S3D). Binding of WC629 decreased deuterium incorporation into NRR peptides derived from the loop connecting LNR-B to LNR-C (BC loop), a region within the HD domain immediately preceding and including the S2 site (S2 loop), and a region from LNR-A (Figure 6A).
Figure 6.
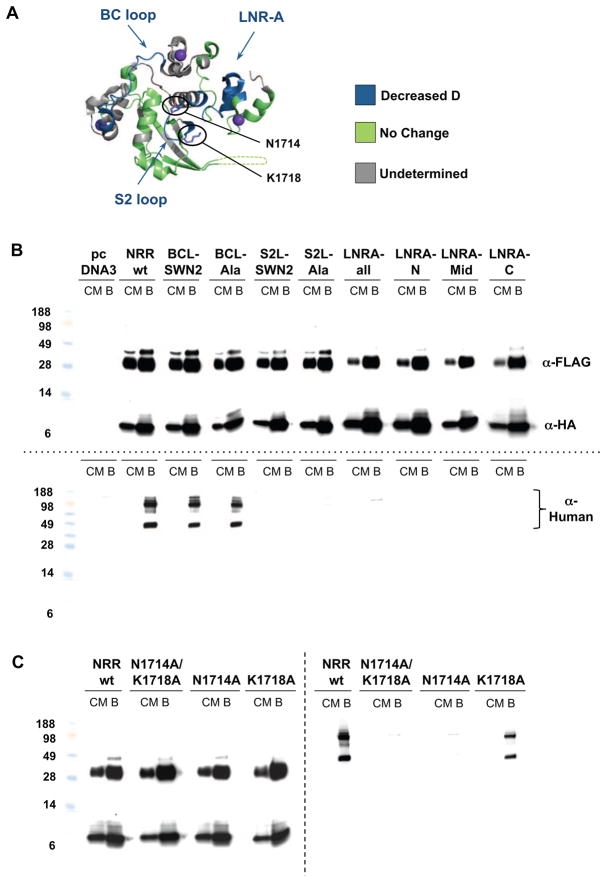
The WC629 inhibitory antibody bridges the LNR and HD domains and retards deuteration of the S2 region. (A) Ribbon diagram of the Notch1 NRR, colored according to the effect of antibody binding. Regions exhibiting slower deuteration in the presence of the antibody (with an absolute difference in mass of at least 0.5 Da) are colored blue, and regions with no detectable change in the deuteration rate (an absolute difference in mass of less than 0.5 Da) are green. Gray regions correspond to segments for which no HX MS data were obtained. (B) Epitope fine mapping. The indicated wild-type and mutated Notch1 NRR minireceptors, which are divided at S1 into two subunits during maturation, were tagged with FLAG and HA epitopes at their N- and C-termini, respectively, and complexes were precipitated with an anti-HA antibody. The top panel shows co-precipitation of the FLAG- and HA-tagged subunits for all mutants tested, verifying S1 cleavage and stable subunit association. The bottom panel shows co-immunoprecipitation of the WC629 inhibitory antibody with the minireceptor. The bound WC629 antibody was detected with a goat anti-human-HRP antibody conjugate. BCL: BC-loop; S2L: S2 loop; SWN2: Notch2 sequence swap; CM: conditioned media; B: beads. The sequences of the mutations tested are listed in Supplementary Figure 5. (C) Point mutagenesis of N1714 and K1718 of the S2 loop. Left panel shows co-precipitation of the FLAG- and HA-tagged subunits and right panel shows co-immunoprecipitation of the WC629 inhibitory antibody with the minireceptor. Co-precipitation experiments were carried out as above in panel (B). See also Figures S2, S3 and S4, and Table S1..
To determine how residues from these three regions contribute to antibody binding, we analyzed the recovery of complexes between WC629 and either wild-type or mutated Notch1-NRR “minireceptors” in immunoprecipitation assays. The minireceptors contain the three LNR modules and the complete HD domain, with an added N-terminal FLAG tag and a C-terminal HA-epitope tag to facilitate immunoprecipitation (Sanchez-Irizarry et al., 2004). The dual tags also enable assessment of S1 cleavage and subunit association of the minireceptors as surrogate markers of protein structural integrity. The wild-type mini-receptor and all of the mutants tested (Supplementary Figure S5) undergo S1 processing and the subunits remain associated after processing, as judged by recovery of the FLAG-tagged subunit following imunoprecipitation (IP) with anti-HA. To examine WC629 binding, we attempted to immunoprecipitate complexes between the different minireceptor proteins and WC629 with anti-HA. The IP data revealed that substitution of the BC loop with homologous residues of Notch2 or with alanine at six positions did not detectably interfere with WC629 binding. In contrast, substitution of the native sequences of the S2 loop or the LNR-A region with the Notch2 sequences or with alanine at exposed positions abrogated WC629 binding (Figure 6B).
To identify key residues that contributed energetically to WC629 binding, we focused on the two residues of the S2 loop that are most solvent exposed: N1714 and K1718. We made three additional alanine-substituted minireceptors: N1714A/K1718A, N1714A alone and K1718A. IP studies showed that neither N1714A/K1718A nor N1714A immunoprecipitated WC629, whereas K1718A did, but with qualitatively less recovery than the wild-type Notch1 NRR (Figure 6C). Based on these studies, we conclude that N1714 of the S2 loop is a key energetic contributor to WC629 binding by the Notch1 NRR.
Discussion
A central key to understanding the mechanism of Notch activation lies in elucidating how the NRR of Notch receptors controls access of metalloprotesases to the S2 site in response to ligand binding. Here, we have applied HX MS to study the intrinsic dynamics and conformational changes of the human Notch1 NRR in five distinct biochemical states to gain insight into Notch activation and its inhibition by allosteric antibodies that bind to the NRR.
As the reference state for these studies, we chose to use the uncleaved precursor form of the Notch1 NRR, folded into its autoinhibited, metalloprotease-resistant conformation. The most highly protected region of the Notch1 NRR is the hydrophobic interior of the HD domain, with the beta strand immediately following the S1-cleavage site exhibiting the greatest degree of protection against exchange. When deuterium exchange of the precursor protein is compared with those of the S1-cleaved molecule, there are no detectable differences, indicating no significant conformational change upon S1 cleavage. This result is consistent with previous evidence that the S1 cleavage site of Notch1 lies within a long, unstructured loop (Gordon et al., 2009b).
To model the Notch “on” state of the receptor, we released autoinhibition by incubation of the Notch1 NRR with the divalent metal ion chelator EDTA. Because the LNR modules rely on calcium coordination for their structural integrity, this treatment relaxes the structural integrity of the LNR modules (Aster et al., 1999; Vardar et al., 2003), disengages the LNR domain from the HD, and promotes proteolytic activation of Notch in cells (Figure 3 and (Rand et al., 2000)). Whether the “endstate” of the HD domain after EDTA-induced activation faithfully mimics the “endstate” of the HD after physiologic activation of Notch receptors is, of course, an open question. If ligand-induced signaling relies on mechanical force to peel the LNR repeats away from the HD domain, as many researchers have proposed (see (Kopan and Ilagan, 2009) for a review), it seems reasonable to posit that the use of EDTA to induce detachment of the LNRs from the HD chemically (as opposed to mechanically) should lead to a similar endpoint in terms of the structure and dynamics of the HD domain.
Although the peptide coverage of the LNRs is incomplete, comparison of the exchange into the autoinhibited and EDTA-treated NRRs suggests that the degree of protection of the LNR modules follows the order LNR-A < LNR-B < LNR-C. The kinetics of exchange of the region flanking the S2 site parallels the exchange behavior of the LNR-B repeat, and chelator treatment appears to promote relaxation of helix three, which immediately precedes and packs against the terminal beta strand that houses the S2 site. This finding suggests that detaching the LNR-B repeat from the HD domain is needed to relax the structural elements of the HD domain that encompass the S2 site to allow metalloprotease access, and provides an explanation for the observation that both the LNR-A and LNR-B repeats must be deleted to result in substantial ligand-independent signaling activity of the Notch1 NRR (Gordon et al., 2007).
The hydrophobic interior of the HD domain, including the core beta hairpin that straddles the S1-cleavage site, is the region of the protein most highly protected against exchange. The core beta hairpin does not exhibit greater exchange after EDTA treatment, a finding that contrasts with the increased rate of exchange that occurs at the S2 site in response to EDTA treatment. These data argue against current models for activation invoking ligand-induced dissociation of S1-cleaved Notch receptors prior to metalloprotease cleavage, because i) the core of the HD domain is intrinsicially more highly protected against exchange than the S2 site and ii) exchange of this core is not enhanced under conditions that promote metalloprotease cleavage.
It is important to emphasize that the studies reported here do not distinguish between different models for ligand-induced activation (e.g. mechanical force, allosteric switch, etc.). Although EDTA treatment relaxes the structure of the LNR modules, making it possible to probe the intrinsic dynamics of the HD in its “unmasked” state for comparison to the autoinhibited (masked) state, it is a non-physiologic activating stimulus, and thus our experiments do not address whether or not the LNR domains are peeled off mechanically from the site during physiologic activation by ligands. (It is, however, worth noting that ligands might apply enough force to peel the LNR modules away from the HD without actually dissociating the subunits before S2 cleavage takes place -- for example, if the kinetics of metalloprotease access to and cleavage of the HD domain at S2 occurs more rapidly than does the increase in force required to dissociate the subunits).
The work reported here also lays the groundwork for future experiments to study the activation dynamics in pathogenic Notch signaling, which may exhibit subtle differences from physiologic or EDTA-induced signaling. All known activating point mutations of Notch1 found in T cell acute lymphoblastic leukemia/lymphoma (T-ALL) lead to increased ligand-independent signaling. The most common mutations associated with TALL lie within the hydrophobic core of the HD and typically cause thermodynamic destabilization of the NRR (Malecki et al., 2006; Weng et al., 2004). It would be informative to determine whether these mutations increase the exchange rate of the hydrophobic core of the HD to the point at which the activation mechanism becomes dissociation driven, as anticipated for at least three of the T-ALL associated mutations that promote subunit dissociation in vitro (Malecki et al., 2006).
These studies also examined how the binding of a potent allosteric inhibitory antibody affects the exchange kinetics of Notch1 NRR to shed light on the mechanism of antibody inhibition. Mapping of the epitope by combining hydrogen exchange data with mutational studies clearly identified residues in LNR-A and in the loop preceding the S2-cleavage site as key contributors to the energetics of antibody binding. The exchange studies also established that a peptide including the S2 site is more protected against exchange upon antibody treatment, providing direct evidence that kinetic access to the metalloprotease site is reduced when antibody is bound.
Over the past several years, Notch receptors have emerged as attractive therapeutic targets in cancer as well as in other diseases (Miele et al., 2006; Rizzo et al., 2008). The HX MS approach developed here can be used to evaluate candidate molecules that emerge from screens selecting for compounds designed to inhibit Notch signaling either by stabilizing the NRR in its autoinhibited conformation or by directly masking the S2 site. Such compounds would be invaluable not only as therapeutic agents but also as experimental tools to study the consequences of signaling by individual Notch receptors.
Experimental Procedures
Protein Expression and Purification
The human Notch1 NRR (residues E1446 to Q1733; Genbank ID 148833507) was subcloned into the pET15b vector containing an N-terminal hexahistidine tag followed by a TEV cleavage site. Thus, the NRR contains an additional glycine at its N-terminus after release of the histidine tag by TEV cleavage. The Notch1 NRR-WT protein was prepared essentially as previously described (Gordon et al., 2009b). In brief, the protein was produced recombinantly in RosettaII(DE3)pLysS bacteria and recovered from the insoluble fraction using 5M urea. The protein was affinity-purified on a nickel column, eluted with imidazole and treated with TEV protease to remove the His6 tag. The protein was refolded in vitro by dialysis in a redox buffer containing 5 mM cysteine and 1 mM cystine. Refolding progress was monitored by HPLC. The refolded protein was further purified by anion exchange chromatography. To prepare the S1-cleaved form of the Notch1 NRR, purified Notch1 NRR-WT (0.5 mg) was subjected to in vitro S1 cleavage by treating with recombinant furin (40 units; New England Biolabs) overnight in a buffer containing 10mM Tris pH 8, 10 mM NaCl and 10 mM CaCl2 (Gordon et al., 2009b). Cleavage was determined to have gone to completion when the Notch1 NRR-WT precursor band on SDS-PAGE was no longer detectable. The furin-cleaved NRR was purified by size exclusion chromatography in 25 mM Tris buffer pH 8, containing 150 mM NaCl and 10 mM CaCl2. To prepare Notch1 NRR-EDTA, purified Notch1 NRR-WT (5 mg/mL) was treated with 20 mM EDTA for 2 h at room temperature. The EDTA-treated NRR was directly used in the HX MS experiment without further purification.
Hydrogen Exchange - Mass Spectrometry Experiments
Experiments were performed as reported previously (Iacob et al., 2009), with the following details tailored specifically to the Notch system. A stock solution (75 μM) of Notch1 NRR-WT, Notch1 NRR-S1 or Notch1 NRR-EDTA (75 pmol) was prepared in 25 mM Tris buffer pH 8, 150 mM NaCl and 10 mM CaCl2, H2O. For the HX MS experiment monitoring exchange of Notch1 NRR-WT in complex with WC629, we first incubated 75 pmol of Notch1 NRR-WT with 84 pmol of WC629 for 3 hours in the same buffer. Deuterium exchange was initiated by diluting the stock solution 15-fold with 25mM Tris (p2H 8), 150 mM NaCl and 10 mM CaCl2, in 2H2O at 21 °C. At each deuterium exchange time point (10s, 1m, 10m, 1h and 4h) labeling was quenched by adding an equal volume of quench buffer (200 mM citrate buffer, 0.5 M TCEP, 4M guanidine HCl, H2O, pH 2.3) and dropping the temperature rapidly to 0 °C. After 10 minutes in the quench buffer at 0 °C, a step that was requried to reduce the disulphide bonds and allow for efficient pepsin digestion, each sample was injected into a custom Waters nanoACQUITY platform equipped with an 2.1 mm × 50 mm immobilized pepsin column for digestion and UPLC system for separation of resulting peptic peptides (system described in detail in (Wales et al., 2008)). The peptides were separated with a gradient of 8–40% acetonitrile in 6 minutes (both mobile phases contained 0.05% formic acid, pH 2.6) and directed into a Waters QT of Premier mass spectrometer with conventional electrospray ionization source. The mass spectrometer settings were: ESI+ mode; capillary 3500 V; cone 35 V; desolvation and source temperatures were 175 °C and 80 °C, respectively; nitrogen desolvation gas flow of 600 L/hr; mass acquisition range of 50–2000 m/z; scan rate of 0.2 scans/second; instrument always collecting data in MSE mode. Continuous lock mass correction was accomplished with infusion of a peptide standard every 30 seconds for a mass accuracy of 3–5 ppm. The data were analyzed with HX-Express (Weis et al., 2006) and other custom macros. Each experiment was performed in duplicate. Only peptides detected in both the tests and reference conditions were used in comparative analyses (Supplementary Figure 2). The threshold for detecting differences between samples was set well outside the error of making each measurement, as determined by duplicate measurements (see replicate curves in Figure 4B,C and Supplemental Figure 3B) and prior analyses of many proteins with this experimental setup. An absolute difference in mass of 0.5 Da was used as a threshold for the comparisons. Any point in the deuterium exchange curves with the difference in mass of less than 0.5 Da was reported as “no change”. Throughout the manuscript, data that are plotted on the same graph were obtained using identical experimental conditions, thus negating the need for back-exchange correction for comparison purposes (Wales and Engen, 2006). Much more caution must be used comparing data not on the same graph because in order to compare such data on an absolute deuterium basis, a back-exchange correction which takes into account experimental variation from day-to-day must be applied. As we wished to determine the relative difference between protein states (for example as shown in Figure 4, and Supplemental Figure S3), no such back-exchange correction was made. All mass spectra indicated that all peptic peptides underwent exchange with EX2 kinetics.
Reporter Assays
Triplicate cultures were plated at 1 × 105 U2OS cells per well of a six-well dish in cell culture medium. After 24h, cells in each well were cotransfected with 750 ng of TP1-luciferase reporter plasmid, 15 ng of an internal control Renilla luciferase reporter plasmid (pRL-TK), and with 60 ng of various human Notch1 constructs in pcDNA3 (Invitrogen) or with vector alone using Lipofectamine 2000 (Invitrogen). After 24h, the transfected cells were treated with either 2.5 mM CaCl2 or 5 mM EDTA for 10m, and then resupplied with cell culture medium for an additional 6h. Cells were then harvested, and firefly and renilla luciferase activities were quantitated in cell lysates by using a dual luciferase assay (Promega).
Immunoprecipitation
Using Lipofectamine 2000, 293T cells in a 10-cm dish were transiently transfected with 3 μg of empty pcDNA3 plasmid or with pcDNA3 plasmids encoding various human Notch1 NRR minireceptors. To enable detection, these minireceptors were fused to FLAG and HA tags at the N- and C-termini, respectively (Sanchez-Irizarry et al., 2004). As a result of furin cleavage during expression, the resulting polypeptides are secreted as heterodimers, with the N-terminal subunits containing a FLAG tag, and the C-terminal subunits bearing an HA tag. 48h after transfection, conditioned media were collected and mixed with the antibody WC629 for 3h. The mixtures were incubated with anti-HA beads (Covance) overnight at 4 °C. After washing the beads three times, the bound proteins on the beads were analyzed by SDS-PAGE and Western blot. The upper part of the Western blots, which contained the FLAG-tagged portion of the NRR, was stained with rabbit anti-FLAG antibody and then goat anti-rabbit antibody conjugated to horseradish peroxidase (HRP). The lower part of the blots, which contained the HA-tagged portion of the NRR, was stained with mouse anti-HA and goat anti-mouse-HRP antibodies. Western blots to detect WC629 binding were stained using goat anti-human-HRP and WestPico chemiluminescent substrate (Pierce-Thermo).
Supplementary Material
Acknowledgments
We are grateful to Wendy Gordon for helpful discussions. This work is supported by NIH (National Institutes of Health) grants R01 CA092433 (S.C.B.), P01 119070 (S.C.B., J.C.A.), R01 070590 (J.R.E.) and R01 086507 (J.R.E.), and the Leukemia and Lymphoma Society (S.C.B., J.C.A.). K.T. is supported by the Irvington Institute Postdoctoral Fellowship program of the Cancer Research Institute.
Footnotes
Author Contributions
S.C.B., J.R.E., T.E.W., and K.T. designed the study, with input from J.C.A. K.T. and T.E.W. performed all HX MS experiments. K.T. purified all proteins, performed reporter assays, and immunoprecipitation studies. M.A.A. provided antibody. S.C.B., J.R.E., T.E.W., and K.T. analyzed data. K.T. and S.C.B. wrote the manuscript with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aste-Amezaga M, Zhang N, Lineberger JE, Arnold BA, Toner TJ, Gu M, Huang L, Vitelli S, Vo KT, Haytko P, et al. Characterization of Notch1 antibodies that inhibit signaling of both normal and mutated Notch1 receptors. PLoS One. 2010;5:e9094. doi: 10.1371/journal.pone.0009094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster JC, Simms WB, Zavala-Ruiz Z, Patriub V, North CL, Blacklow SC. The folding and structural integrity of the first LIN-12 module of human Notch1 are calcium-dependent. Biochemistry. 1999;38:4736–4742. doi: 10.1021/bi982713o. [DOI] [PubMed] [Google Scholar]
- Blacklow SC, Kim PS. Protein folding and calcium binding defects arising from familial hypercholesterolemia mutations of the LDL receptor. Nat Struct Biol. 1996;3:758–762. doi: 10.1038/nsb0996-758. [DOI] [PubMed] [Google Scholar]
- Bray SJ. Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- Fass D, Blacklow S, Kim PS, Berger JM. Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module. Nature. 1997;388:691–693. doi: 10.1038/41798. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Roy M, Vardar-Ulu D, Garfinkel M, Mansour MR, Aster JC, Blacklow SC. Structure of the Notch1-negative regulatory region: implications for normal activation and pathogenic signaling in T-ALL. Blood. 2009a;113:4381–4390. doi: 10.1182/blood-2008-08-174748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, Histen G, Sanchez-Irizarry C, Aster JC, Blacklow SC. Structural basis for autoinhibition of Notch. Nat Struct Mol Biol. 2007;14:295–300. doi: 10.1038/nsmb1227. [DOI] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, L’Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, Blacklow SC. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009b;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacob RE, Pene-Dumitrescu T, Zhang J, Gray NS, Smithgall TE, Engen JR. Conformational disturbance in Abl kinase upon mutation and deregulation. Proc Natl Acad Sci U S A. 2009;106:1386–1391. doi: 10.1073/pnas.0811912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci U S A. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Li Y, Wu W, Gordon WR, Chang DW, Lu M, Scoggin S, Fu T, Vien L, Histen G, et al. Modulation of Notch signaling by antibodies specific for the extracellular negative regulatory region of NOTCH3. J Biol Chem. 2008;283:8046–8054. doi: 10.1074/jbc.M800170200. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki MJ, Sanchez-Irizarry C, Mitchell JL, Histen G, Xu ML, Aster JC, Blacklow SC. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L, Golde T, Osborne B. Notch signaling in cancer. Curr Mol Med. 2006;6:905–918. doi: 10.2174/156652406779010830. [DOI] [PubMed] [Google Scholar]
- Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- Nichols JT, Miyamoto A, Olsen SL, D’Souza B, Yao C, Weinmaster G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J Cell Biol. 2007;176:445–458. doi: 10.1083/jcb.200609014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand MD, Grimm LM, Artavanis-Tsakonas S, Patriub V, Blacklow SC, Sklar J, Aster JC. Calcium depletion dissociates and activates heterodimeric notch receptors. Mol Cell Biol. 2000;20:1825–1835. doi: 10.1128/mcb.20.5.1825-1835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo P, Osipo C, Foreman K, Golde T, Osborne B, Miele L. Rational targeting of Notch signaling in cancer. Oncogene. 2008;27:5124–5131. doi: 10.1038/onc.2008.226. [DOI] [PubMed] [Google Scholar]
- Sanchez-Irizarry C, Carpenter AC, Weng AP, Pear WS, Aster JC, Blacklow SC. Notch subunit heterodimerization and prevention of ligand-independent proteolytic activation depend, respectively, on a novel domain and the LNR repeats. Mol Cell Biol. 2004;24:9265–9273. doi: 10.1128/MCB.24.21.9265-9273.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardar D, North CL, Sanchez-Irizarry C, Aster JC, Blacklow SC. Nuclear magnetic resonance structure of a prototype Lin12-Notch repeat module from human Notch1. Biochemistry. 2003;42:7061–7067. doi: 10.1021/bi034156y. [DOI] [PubMed] [Google Scholar]
- Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, Finkle D, Venook R, Wu X, Ridgway J, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464:1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.