Abstract
The cystic fibrosis transmembrane conductance regulator (CFTR) is a unique ion channel in that its gating is coupled to an intrinsic enzymatic activity (ATP hydrolysis). This enzymatic activity derives from the evolutionary origin of CFTR as an ATP-binding cassette transporter. CFTR gating is distinct from that of a typical ligand-gated channel because its ligand (ATP) is usually consumed during the gating cycle. However, recent findings indicate that CFTR gating exhibits allosteric properties that are common to conventional ligand-gated channels (e.g. unliganded openings and constitutive mutations). Here, we provide a unified view of CFTR gating that combines the allosterism of a ligand-gated channel with its unique enzymatic activity.
Keywords: ABC Transporter, Allosteric Regulation, Chloride Channels, Cystic Fibrosis, Ion Channels
CFTR and ABC Transporters
CFTR2 is an essential anion channel whose dysregulation causes multiple disorders (1–3). It is the only known ion channel that links an enzymatic activity (ATP hydrolysis) to opening and closing of the pore (channel gating). CFTR owes this property to its origin as an ABC transporter, many others of which are active transport ATPases or pumps (4–6). The fact that CFTR consumes its ligand (ATP) by hydrolysis during the gating cycle makes it a unique ion channel. However, recent findings indicate that CFTR gating by ATP also exhibits features of an allosteric activation mechanism that are characteristic of more conventional ligand-gated channels that reversibly bind their ligands. Our main goals in writing this minireview are (i) to compare those features of CFTR gating that are unique to its origin as an ABC transporter with those that are shared with other ligand-gated channels, (ii) to propose a conceptual model of CFTR gating that merges the allosterism of a ligand-gated channel with its enzymatic activity, and (iii) to illustrate the importance of considering allosteric principles when interpreting CFTR data.
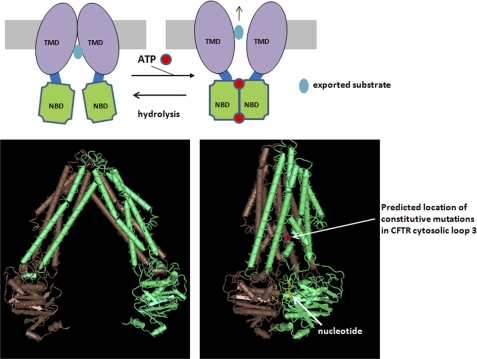
Fig. 1 illustrates the basic operating principles of an ABC transporter. The crystal structures of the bacterial MsbA exporter are shown for reference (7). The core structural components include two TMDs that form the permeation pathway and two NBDs that mediate the ATP hydrolysis that fuels active substrate transport. Two ATP molecules bind in pockets at the interface of an NBD dimer; each binding pocket is lined with residues from both NBDs (i.e. Walker A and B sequences from one NBD and an ABC signature sequence from the opposite NBD). ATP binding to both sites creates or stabilizes the NBD dimer with an associated rearrangement of the translocation pathway by a proposed tweezer-like mechanism (7, 8). For an exporter, this involves a shift from an inward-facing conformation with high substrate affinity to an outward conformation with low substrate affinity. Long cytosolic loops connect the NBDs to the TMDs and mediate the coupling between ATP binding and structural rearrangements of the TMDs. The transporter is reset to the inward-facing conformation following ATP hydrolysis at one or both sites. Like this generic ABC transporter, CFTR also possesses two TMDs and two NBDs (1, 9), binds two ATP molecules at the interface of an apparent NBD dimer (10, 11), and exhibits ATPase activity (12), albeit predominately at one site (13, 14). The difference for CFTR is that this enzymatic activity is coupled to a cycle of channel opening and closing rather than to substrate transport through the translocation pathway.
FIGURE 1.
Transport mechanism of an ABC exporter. Upper, schematic view. Lower, crystal structures of the bacterial exporter MsbA in the nucleotide-free state (left; Protein Data Bank code 3B5W) and the AMP-PNP-bound state (right; code 3B60). Structures were downloaded from www.ncbi.nlm.nih.gov/structure (see also Ref. 7).
Accepted View of the CFTR Gating Mechanism
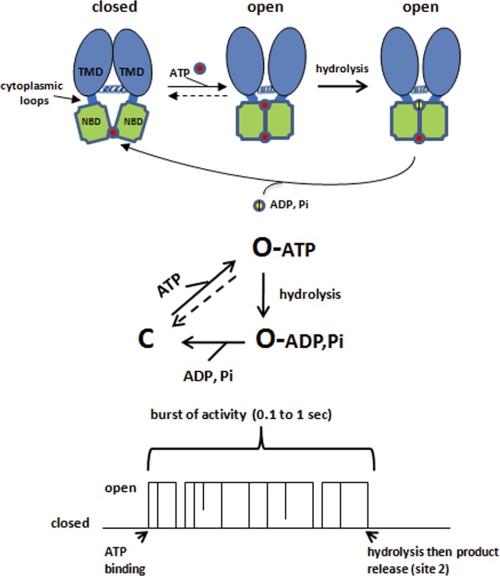
A simple scheme that illustrates the well accepted features of CFTR gating is shown in Fig. 2 (see also Refs. 15–17). Channel opening is normally associated with ATP binding at both composite sites, which promotes or stabilizes an NBD1-NBD2 dimer that can be detected functionally (10) or biochemically by cysteine cross-linking (11). What makes CFTR different from typical ligand-gated channels is the exceptionally slow rate of ATP unbinding (koff < 0.2 s−1), which is due presumably to the tightness of the NBD1-NBD2 dimer. This is where the enzymatic activity of CFTR comes in; ATP hydrolysis and subsequent product release destabilize the dimer and increase the probability of channel closure.
FIGURE 2.
Scheme illustrating well accepted features of CFTR gating. Upper, the model assumes ATP turnover at site 2 only and highly phosphorylated channels. Middle, corresponding gating scheme. Lower, ATP binding promotes open channel bursts that are terminated by hydrolysis and product release. The essential role of magnesium ions as cofactors for ATP binding and hydrolysis is omitted for simplicity. Note that ATP binding promotes a burst of channel openings that are punctuated by short closings (see text and Footnote 3).
The link between ATP hydrolysis and channel closing has been deduced from two sorts of experiments: (i) adding poorly hydrolyzable ATP analogs to channels that first have been opened by ATP, which greatly prolongs channel openings (the former are ineffective for opening the channel per se) (18, 19), and (ii) mutagenesis of key residues for ATP hydrolysis at composite site 2 (e.g. Lys-1250 in NBD2) (10, 20), which also stabilizes channel openings (mean open times of >10 s).3 The strong effect of site 2 mutations on channel closing argues that this is the important site for promoting channel closure. This conclusion is supported by biochemical evidence that site 1 binds ATP very tightly and has a very low hydrolytic rate, whereas hydrolysis at site 2 occurs more rapidly (13, 14). The overall ATP turnover rate of purified CFTR reconstituted in liposomes occurs within the same time domain as channel gating (i.e. approximately one ATP molecule hydrolyzed per s versus gating cycles of similar duration) (12, 16). Presumably, this rate reflects primarily the turnover rate at site 2.
Channel closing is enhanced by the dissociation of the hydrolysis products (ADP and phosphate or Pi) from site 2 rather than hydrolysis per se. The best evidence for this point is the large stabilizing effect of phosphate analogs (e.g. orthovanadate) on open channel bursts, which presumably form stable complexes with ADP following Pi release (21). In sum, the best available data indicate that most channel openings occur when ATP has bound to both sites. The great majority of closings follow ATP hydrolysis and product release at site 2 (estimated to be >95% by Csanády et al. (16)).
An important feature of CFTR gating that is not addressed in Fig. 2 is the phosphorylation dependence of channel activity. The main physiologic stimulus of CFTR activity is phosphorylation of multiple sites within its large cytosolic R domain by PKA (23–25). How CFTR channel activity is optimized by phosphorylation of the R domain, which is located between NBD1 and TMD2, is still unclear. NBD1-NBD2 dimerization, as monitored by cysteine cross-linking, is enhanced by PKA phosphorylation (11, 26). Conceivably, the unphosphorylated R domain limits channel activity by precluding the NBD dimer, but this cannot be the only mechanism (discussed below).
Ligand-gated Channels and Allosteric Activation Mechanisms
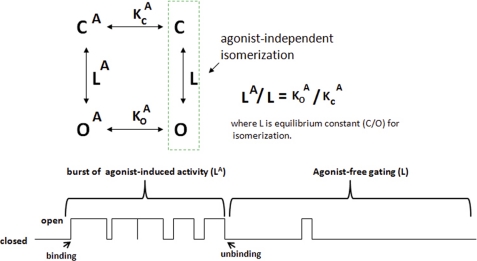
Other ligand-gated channels do not consume their ligands to promote channel closing. Unbinding occurs sufficiently fast to permit reversible gating under equilibrium conditions. The gating of a typical ligand-gated channel (e.g. a nicotinic acetylcholine receptor (27, 28)) obeys allosteric principles that date back 40–50 years to the prescient writings of J.-P. Changeux, S. J. Edelstein, and colleagues (e.g. the classic MWC model) (29–31). The relevant concepts were originally developed for multimeric proteins, but it is now clear that monomeric proteins exhibit similar allosteric properties (32, 33). The first principle of an allosteric activation scheme for a typical ligand-gated channel is the concept that the open pore conformation is accessible in the absence of the ligand although usually with low single channel open probability (Po) (27–31, 34). Ligand binding shifts the equilibrium between closed and open states to favor the latter. This shift in equilibrium occurs in part because ligand binding stabilizes the open state, a process termed conformational selection (34, 35). Fig. 3 illustrates this type of activation scheme for a hypothetical channel with one agonist (A)-binding site. The closed-to-open transitions of this channel are described as isomerizations with equilibrium constants L (number of closed channels (C)/number of open channels (O); terminology from Ref. 31). Closed-to-open isomerizations can occur in the absence of agonist (termed unliganded openings) (27) but with lower Po values for the unliganded channel than for the agonist-bound channel (L > LA).
FIGURE 3.
Simplest allosteric activation scheme for a conventional ligand-gated channel. This hypothetical channel binds one agonist molecule (A). L represents isomerization equilibrium constants (L = C/O, and LA = CA/OA). K represents equilibrium constants for agonist binding to the closed and open states (e.g. O/OA = KOA/[A]). For this reversible system, the equilibrium constants for agonist binding (K) to the open and closed states must vary in proportion to the asymmetry in isomerization equilibrium constants (where L > LA).
Such cyclic allosteric schemes, in which all open states are connected to corresponding closed states through isomerization reactions, have provided good approximations of the behaviors of numerous proteins and ligand-gated channels (27–38). Of course, this macroscopic view is oversimplified in that the liganded and unliganded active conformers (or open channels) cannot be structurally identical. Ligand binding must have some effect on protein structure, but in those few cases where detailed structural information is available, the primary effect is local or small-scale, i.e. in the vicinity of the binding site (39). Conversely, the conformational changes that underlie protein activation are sufficiently large-scale to be possible even without ligand binding. More nuanced views of agonist activation that combine an initial conformational selection step followed by secondary structural changes that are induced by ligand binding (a secondary “induced fit” step) have appeared in the recent literature motivated largely by the results of detailed NMR structural studies (e.g. of the PBX1 homeodomain DNA-binding protein) (35, 40). For a ligand-gated channel, this more nuanced view would predict that the open state conformations in the presence and absence of agonist binding are similar but not identical. To what extent such structural differences influence the properties of the pore (conductance, selectivity, blocker sensitivity) is largely unexplored and may vary between channel types.
The important aspect of the type of ligand activation mechanism illustrated in Fig. 3 is that it predicts features of channel regulation that are not obvious for sequential, strict coupling mechanisms (see Refs. 31, 32, 34, and 41 for comparisons of cyclic and sequential activation schemes). The first point is that a cyclic allosteric scheme that permits unliganded gating predicts that the open state of a ligand-gated channel must have a higher agonist affinity than the closed state (27, 32, 38). The disparate affinities of the open and closed state conformations in this reversible system are easily derived mathematically (Fig. 3) (32, 41). The consequences of these disparate affinities are significant. Notably, any factor that promotes ligand-free isomerization (decreases L) and thereby biases the equilibrium toward the open state concomitantly increases agonist sensitivity. This is the classic reciprocity principle that holds for all cyclic allosteric activation schemes (27–32, 37, 38). The functional consequence of this reciprocity is a high degree of synergy between regulatory inputs, i.e. varying one input (e.g. phosphorylation) affects the sensitivity to all other inputs (e.g. agonist sensitivity) (see Ref. 37 and below for examples pertinent to CFTR). Another feature of such an activation scheme is the possibility of mutations that increase Po in the absence of ligand (termed constitutive mutations in Ref. 31). A number of such mutations have been produced for the well studied neurotransmitter-gated channels (e.g. the acetylcholine (27) and GABAA (38) receptors). These constitutive mutations often localize to the interfaces between channel subunits and/or to the symmetry axis that links the ligand-binding site to the channel gate (27, 31, 32, 38) apparently because mutations here most strongly impact the free energy difference (ΔG) between the unliganded open and closed states. By definition, constitutive mutations decrease the isomerization constant L; thus, they also should increase agonist sensitivity in accord with the reciprocity principle above (42). This prediction has been confirmed for a number of constitutive mutations in typical ligand-gated channels (27, 38).
CFTR Gating and Allosterism
Does CFTR gating by ATP exhibit allosteric properties that are shared with other ligand-gated channels? Aleksandrov, Riordan, and co-workers (43–46) argued for an allosteric mechanism based in part on a thermodynamic analysis of CFTR gating in synthetic lipid bilayers. Their conclusion that CFTR gating is a thermodynamically reversible process and their use of analytic approaches that depend on this assumption have been challenged (47, 48). However, the notion that CFTR gating shares allosteric features with typical ligand-gated channels is supported by three lines of evidence: (i) ATP-free CFTR openings occur (17, 49–51); (ii) constitutive mutations that enhance unliganded CFTR channel activity have been produced (17, 51); and (iii) some of these mutations have pleiotropic effects on CFTR channel regulation by ATP and by PKA, as predicted by the reciprocity principle above (27–32, 51).
Regarding the first line of evidence, several groups reported wild-type CFTR openings in excised patches in an ATP-free bath that also included an ATP-scavenging enzyme to eliminate contaminating ATP (with estimated Po values in the absence of ATP ranging from 0.0003 (51) to ∼0.004 (49)). Hwang and co-workers (50) also noted that channel openings could be detected for a CF mutant form of CFTR that is completely unresponsive to ATP (G551D mutation in NBD1 signature sequence). Also, Riordan and co-workers (52) and our group (51, 53) observed that CFTR channels that lack one of the two NBDs that are essential for ATP-dependent gating open spontaneously at low frequency in excised patches or lipid bilayers. In all of these cases, the unitary currents and blocker sensitivities of the observed unliganded CFTR openings were similar if not identical to those that are characteristic of openings in the presence of ATP. Thus, it seems likely that the same (or similar) open state can be achieved with or without ATP binding (or NBD dimerization).
Concerning the second line of evidence, mutations in the cytosolic loops (51) and in NBD2 (17) that increase ATP-independent channel activity have recently been reported. These mutants behave like constitutive mutants in the nomenclature of Changeux and Edelstein (31), although the ATP-independent activities of the cytosolic loop mutants remain sensitive to PKA phosphorylation of the R domain (see below) (51). The constitutive mutations in cytosolic loops 1 and 3 that we reported localize near the presumed symmetry axis of the channel when mapped onto the available crystal structures of related ABC transporters (see Fig. 1 for presumed location of residue 978 in CFTR loop 3, a hot spot for constitutive mutations, when mapped onto the MsbA structure). This location is consistent with the prediction of Changeux and Edelstein (31, 32) for constitutive mutations in allosteric proteins and with the locations of like mutations in ligand-gated channels such as the acetylcholine and GABAA receptors (27, 38). The conclusion that the cytosolic loop mutations enhance unliganded CFTR Po is further supported by their abilities to enhance the otherwise low activities of CFTR-G551D and CFTRΔ1198 (NBD2 deletion construct) channels when introduced into these ATP-insensitive constructs (51). Some of the constitutive loop mutations had large effects on the energetics of unliganded gating (3–5-fold decrease in ΔG between ATP-free open and closed states), which implies that the conformation of the cytosolic loops substantially retards spontaneous channel openings. In previous work (51), we modeled the loops as a compression spring that resists unliganded channel opening. ATP binding and NBD dimerization would compress this normally stiff spring to increase Po (see also Ref. 47). Certain loop mutations are imagined to reduce the stiffness of this spring (e.g. by disrupting loop-loop interactions) and thereby increase Po in the absence of ATP. This heuristic model may be applicable to the conformational switching of ABC transporters in general given that large rearrangements of the cytosolic loops occur during the switch from the inward-facing to outward-facing conformations of bacterial ABC transporters (7, 54).
Importantly, the constitutive loop mutations strongly increased both the ATP and PKA sensitivities of CFTR gating in addition to enhancing unliganded Po (decreasing L) (51). Such “multiple phenotypes” of the constitutive mutants are expected for an activation scheme in which channel gating, ligand binding, and other regulatory inputs (i.e. phosphorylation) are reciprocally coupled (37, 38, 42). Of course, the reciprocity between CFTR channel gating and ATP binding is more complicated than that for a typical ligand-gated channel given that most open channel bursts terminate following ATP hydrolysis and product release rather than ligand unbinding. Still, as we argue below, reciprocity between CFTR gating and nucleotide occupancy can be expected for an activation mechanism that permits unliganded gating.
Expanded CFTR Gating Schemes That Incorporate Allosteric Principles Common to Ligand-gated Channels
The aforementioned findings indicate that CFTR channel gating shares allosteric properties commonly ascribed to conventional ligand-gated channels, notably (i) unliganded openings, (ii) constitutive mutations that promote unliganded activity, and (iii) pleiotropic effects of these mutations on CFTR regulation by ATP and PKA. On the other hand, CFTR is a special case in that it binds its agonist so tightly that ATP hydrolysis normally is required to inactivate the channel on a physiologically relevant (subsecond) time scale (as argued by Csanády, Gadsby, and co-workers (16, 47, 48). Given this relationship between channel activity and ATP hydrolysis, wild-type CFTR gating is a slow process overall, with kinetics that are orders of magnitude less than for typical ligand-gated channels (see Ref. 55). The length of the gating cycle (open to closed to open) is determined at least in part by the hydrolytic rate at site 2, but post-hydrolytic steps such as ATP/ADP exchange may also slow the opening rate and limit CFTR channel activity at maximal ATP concentrations. (In this regard, wild-type Po is 0.3–0.5 at maximal ATP and PKA activities (24).)
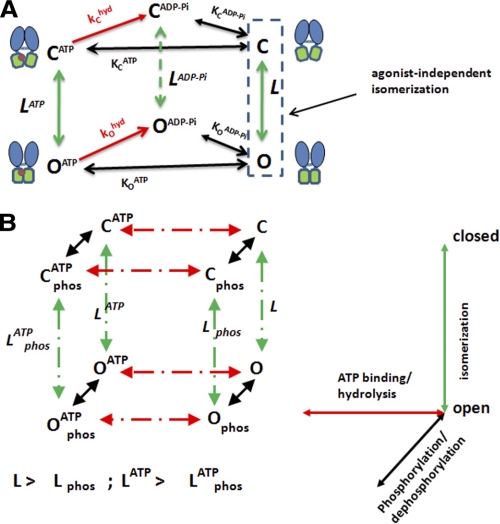
How do we reconcile these recently described features of CFTR gating with its hydrolytic activity? Fig. 4A shows a gating scheme for which the allosterism of a ligand-gated channel is merged with the enzymatic activity of CFTR. We propose a cyclic scheme in which all open states are connected to corresponding closed states because such schemes describe well the gating of typical ligand-gated channels (28, 37, 38), and they predict reciprocal effects of constitutive mutations on ligand sensitivity (as observed for certain cytosolic loop mutations in CFTR) (51). Each vertical closed-to-open transition is quantified by an isomerization equilibrium constant (L for unliganded openings). Fig. 4A assumes that the channels are highly phosphorylated (see below for consideration of phosphorylation as a regulatory input). Fig. 4A also includes irreversible transitions that correspond to ATP hydrolysis in red; thus, this is a non-equilibrium gating scheme unlike those for conventional ligand-gated channels.4
FIGURE 4.
Expanded CFTR gating schemes that incorporate allosteric principles. A, simplest cyclic scheme that combines unliganded gating and closed-open isomerizations with irreversible hydrolytic (hyd) steps (trigonal prism scheme). Channels are assumed to be highly phosphorylated. ATP turnover is assumed to occur at one site only (site 2). Note that the schematic representations of the closed and open NBD dimer structures are shown only to indicate that these structures have a higher probability to be associated with the open and closed channel conformations, respectively (see text and Footnote 4). B, phosphorylation (phos) is introduced as a distinct regulatory input (cubic scheme). ATP turnover again is assumed to occur at one site only. Predicted asymmetries in isomerization equilibrium constants are indicated. See text and supplement data for details.
Fig. 4A predicts asymmetries in the nucleotide occupancies of the closed and open channels, as for other cyclic allosteric activation mechanisms (see supplement data). In particular, Fig. 4A predicts that the open channel should have a higher affinity both for ATP and for the hydrolysis products, ADP and Pi. This could explain the higher ATP sensitivities and the slower deactivation rates upon ATP removal observed for constitutive loop mutants (i.e. mutants that bias the equilibrium toward the open channel) (51). Given that the macroscopic deactivation rate of wild-type CFTR following ATP removal is determined by the rate of ATP hydrolysis and subsequent product release from site 2 (13, 14, 16, 21), the slower deactivation of these constitutive mutants (51) could be due to a slower release of ADP and/or Pi after hydrolysis. Conceptually, this can be explained by recognizing that pore opening and NBD1-NBD2 interactions are reciprocally coupled such that mutations that affect one (gating) must also bias the equilibrium toward the other (NBD dimerization).
Fig. 4B is a related scheme that includes PKA phosphorylation as a distinct regulatory input in parallel with ATP binding/hydrolysis. Treating phosphorylation as a separate input is based on two considerations: (i) phosphorylation is a reversible process like ligand binding and unbinding/hydrolysis, reversible in this case by the competing actions of kinases and phosphatases (see also Ref. 33); and (ii) recent evidence indicates that R domain phosphorylation regulates CFTR gating independently of ATP binding and NBD dimerization (50, 51, 53). The strongest evidence for the latter is the observation that CFTR channels that lack NBD2 but possess one of the constitutive loop mutations (e.g. CFTR-K978C/Δ1198) are strongly stimulated by PKA phosphorylation of the R domain even though their activity is otherwise independent of ATP binding (or, obviously, of NBD1-NBD2 dimerization) (51). This finding does not rule out other roles of the R domain in regulating CFTR gating such as effects on NBD dimerization. Of course, the reported enhancement of NBD dimerization by PKA (11, 26) may be a secondary consequence of a primary effect of phosphorylation on channel opening that is mediated by R domain interactions elsewhere within the polypeptide. As noted above, reciprocity between pore opening and NBD1-NBD2 interactions is expected for a cyclic allosteric gating mechanism.
Implications for CFTR Gating
Earlier, we noted that cyclic allosteric activation mechanisms have features that are not predicted by sequential, strict coupling models. These features include the disparate ligand affinities of the closed and open conformations and the consequent reciprocity between gating, ligand occupancy, and other regulatory inputs. The “other regulatory inputs” can come in a variety of forms ranging from phosphorylation (as for CFTR) to voltage (as for BK-type potassium channels gated by both calcium and voltage) (37). The important functional consequence of this reciprocity is strong synergy between regulatory inputs. This synergy is illustrated by the common finding that varying one regulatory input to an ion channel (e.g. calcium activity for a BK channel) changes the sensitivity of that channel to all other inputs (e.g. its voltage dependence). This probably explains the pleiotropic effects of constitutive loop mutations on the sensitivity of CFTR gating to ATP and PKA (51). It may also explain an earlier report that PKA phosphorylation enhances the ATP sensitivity of CFTR gating (58). Fig. 4B predicts that the converse also should be true, i.e. factors or mutations that affect ATP binding or NBD dimerization should also affect the phosphorylation state of the channel (i.e. by reciprocal coupling between pore opening and R domain conformation). The latter prediction has not been directly tested to our knowledge, but it is interesting to note that Hwang and co-workers (59) reported 10 years ago that the most common CF mutation (ΔF508 in NBD1) markedly decreases the PKA sensitivity of CFTR activation. It is now clear that the ΔF508 mutation substantially reduces CFTR Po at maximally activating ATP and PKA concentrations probably by disrupting the structural link between NBD1 and the cytosolic loops (i.e. disrupts gating) (60–62). Thus, the low PKA sensitivity observed by Wang et al. (59) could be due to reciprocal coupling between gating (channel opening/closing) and R domain phosphorylation (i.e. the kinase and phosphatase accessibilities of the PKA sites within this domain). Of course, Fig. 4B is an oversimplification in that it ignores the multiplicity of the phosphorylation sites within the R domain. A more accurate scheme would include the phosphorylation of each site as a distinct regulatory input. All of these inputs would be reciprocally coupled to each other and to ATP binding and hydrolysis, leading to synergy at multiple levels (including synergistic interactions between the different phosphorylation sites).
Unfortunately, the reciprocity between regulatory inputs that leads to this synergy also complicates the design and interpretation of experiments. For example, if mutating a given residue reduces the effects of a modulator (e.g. drug) on CFTR activity, does that mean that this residue is near a binding site for that modulator? Absolutely not, as has been well recognized by pharmacologists for many years, mutating this residue could indirectly affect modulator efficacy by “reciprocally” influencing other regulatory inputs (e.g. ATP binding/hydrolysis) or ligand-free isomerization (L) (see also Ref. 41). Two other examples were described above: the uncertainties in interpreting the effects of PKA phosphorylation on NBD dimerization (11, 26) and of the ΔF508 mutation on the PKA sensitivity of activation (59). Deducing whether these are primary or secondary effects is impossible without other data.
On a more positive note, the fact that unliganded CFTR gating can occur and be modulated by mutations (17, 51) or by compounds (53) improves the prospect of CF therapies that circumvent defects in ATP binding or NBD dimerization. The rescue of CFTR-G551D function by constitutive mutations in the cytosolic loops supports this concept (51). Some of the small molecule activators of mutant CFTR channels such as G551D that have been discovered recently (63, 64) may target the cytosolic loops or TMDs along the channel symmetry axis to promote unliganded activity. This area can be explored further as a complement to other therapeutic approaches.
Acknowledgments
We thank Drs. David Dawson, T.-C. Hwang, David Gadsby, and Laszlo Csanády for helpful comments.
This minireview will be reprinted in the 2011 Minireview Compendium, which will be available in January, 2012.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data.
Although the terms “channel openings” and “open channel bursts” are often used interchangeably, the latter term is correct. ATP binding promotes a burst of channel openings that are punctuated by short closings (e.g. see Ref. 22). Hydrolysis and subsequent product release terminate these bursts (Fig. 2).
Regarding Fig. 4A, it is important to note that we do not consider the open pore conformation at the TMDs (whatever that looks like structurally) to be strictly demanded by a tight NBD1-NBD2 dimer, nor do we consider the closed pore conformation to be demanded by the absence of the NBD1-NBD2 dimer. In this view, the pore can open occasionally even if the NBDs have not dimerized, and it can close even in the presence of a tight NBD dimer. Support for the former are the ATP-free openings of wild-type CFTR and channels lacking NBD2 (51–53). Evidence for the latter is the bursting behavior of ATP-activated channels for which open channel bursts are punctuated by short closings (22, 56, 57). Some of these intraburst closings may be caused by voltage-dependent pore block by bulky anions or by a charged component of the CFTR protein itself (56), but brief intraburst closings are evident at all voltages especially in single channel records that are not heavily filtered (e.g. see Fig. 3 in Ref. 57). We suggest that some intraburst closings reflect open-to-closed isomerizations of the ATP-bound channel (LATP transition in Fig. 4A), i.e. fast intraburst gating versus the slow gating controlled by ATP binding and hydrolysis. These considerations are consistent with (indeed expected for) an allosteric gating scheme in which the links between ATP binding/NBD dimerization and channel opening/closing are probabilistic. The logical extension of these considerations is that occasionally a closed channel can hydrolyze ATP, i.e. when the pore has closed in the presence of the NBD dimer that is necessary for ATP hydrolysis, and occasionally, an open channel can bind ATP when that channel first opened spontaneously prior to ATP binding.
- CFTR
- cystic fibrosis transmembrane conductance regulator
- ABC
- ATP-binding cassette
- TMD
- transmembrane domain
- NBD
- nucleotide-binding domain
- CF
- cystic fibrosis
- AMP-PNP
- adenosine 5′-(β,γ-imidotriphosphate).
REFERENCES
- 1. Riordan J. R., Rommens J. M., Kerem B., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Plavsic N., Chou J. L. (1989) Science 245, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 2. Gabriel S. E., Brigman K. N., Koller B. H., Boucher R. C., Stutts M. J. (1994) Science 266, 107–109 [DOI] [PubMed] [Google Scholar]
- 3. De Braekeleer M., Férec C. (1996) Mol. Hum. Reprod. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 4. Locher K. P., Lee A. T., Rees D. C. (2002) Science 296, 1091–1098 [DOI] [PubMed] [Google Scholar]
- 5. Dawson R. J., Locher K. P. (2006) Nature 443, 180–185 [DOI] [PubMed] [Google Scholar]
- 6. Aller S. G., Yu J., Ward A., Weng Y., Chittaboina S., Zhuo R., Harrell P. M., Trinh Y. T., Zhang Q., Urbatsch I. L., Chang G. (2009) Science 323, 1718–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Proc. Natl. Acad. Sci. U. S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Locher K. P. (2009) Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sheppard D. N., Welsh M. J. (1999) Physiol. Rev. 79, S23–S45 [DOI] [PubMed] [Google Scholar]
- 10. Vergani P., Lockless S. W., Nairn A. C., Gadsby D. C. (2005) Nature 433, 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mense M., Vergani P., White D. M., Altberg G., Nairn A. C., Gadsby D. C. (2006) EMBO J. 25, 4728–4739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li C., Ramjeesingh M., Wang W., Garami E., Hewryk M., Lee D., Rommens J. M., Galley K., Bear C. E. (1996) J. Biol. Chem. 271, 28463–28468 [DOI] [PubMed] [Google Scholar]
- 13. Aleksandrov L., Aleksandrov A. A., Chang X. B., Riordan J. R. (2002) J. Biol. Chem. 277, 15419–15425 [DOI] [PubMed] [Google Scholar]
- 14. Basso C., Vergani P., Nairn A. C., Gadsby D. C. (2003) J. Gen. Physiol. 122, 333–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hwang T. C., Sheppard D. N. (2009) J. Physiol. 587, 2151–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Csanády L., Vergani P., Gadsby D. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Szollosi A., Vergani P., Csanády L. (2010) J. Gen. Physiol. 136, 407–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hwang T. C., Nagel G., Nairn A. C., Gadsby D. C. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 4698–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vergani P., Nairn A. C., Gadsby D. C. (2003) J. Gen. Physiol. 121, 17–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gunderson K. L., Kopito R. R. (1995) Cell 82, 231–239 [DOI] [PubMed] [Google Scholar]
- 21. Baukrowitz T., Hwang T. C., Nairn A. C., Gadsby D. C. (1994) Neuron 12, 473–482 [DOI] [PubMed] [Google Scholar]
- 22. Carson M. R., Travis S. M., Welsh M. J. (1995) J. Biol. Chem. 270, 1711–1717 [DOI] [PubMed] [Google Scholar]
- 23. Cheng S. H., Rich D. P., Marshall J., Gregory R. J., Welsh M. J., Smith A. E. (1991) Cell 66, 1066–1072 [DOI] [PubMed] [Google Scholar]
- 24. Mathews C. J., Tabcharani J. A., Chang X. B., Jensen T. J., Riordan J. R., Hanrahan J. W. (1998) J. Physiol. 508, 365–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilkinson D. J., Strong T. V., Mansoura M. K., Wood D. L., Smith S. S., Collins F. S., Dawson D. C. (1997) Am. J. Physiol. 273, L127–L133 [DOI] [PubMed] [Google Scholar]
- 26. He L., Aleksandrov A. A., Serohijos A. W., Hegedus T., Aleksandrov L. A., Cui L., Dokholyan N. V., Riordan J. R. (2008) J. Biol. Chem. 283, 26383–26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Purohit P., Auerbach A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cadugan D. J., Auerbach A. (2010) Biophys. J. 99, 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Monod J., Wyman J., Changeux J. P. (1965) J. Mol. Biol. 12, 88–118 [DOI] [PubMed] [Google Scholar]
- 30. Edelstein S. J. (1975) Annu. Rev. Biochem. 44, 209–232 [DOI] [PubMed] [Google Scholar]
- 31. Changeux J. P., Edelstein S. J. (1998) Neuron 21, 959–980 [DOI] [PubMed] [Google Scholar]
- 32. Changeux J. P., Edelstein S. J. (2005) Science 308, 1424–1428 [DOI] [PubMed] [Google Scholar]
- 33. Volkman B. F., Lipson D., Wemmer D. E., Kern D. (2001) Science 291, 2429–2433 [DOI] [PubMed] [Google Scholar]
- 34. Tsai C. J., Ma B., Sham Y. Y., Kumar S., Nussinov R. (2001) Proteins 44, 418–427 [DOI] [PubMed] [Google Scholar]
- 35. Boehr D. D., Nussinov R., Wright P. E. (2009) Nat. Chem. Biol. 5, 789–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardino A. K., Kern D. (2007) Methods Enzymol. 423, 149–165 [DOI] [PubMed] [Google Scholar]
- 37. Cui J., Aldrich R. W. (2000) Biochemistry 39, 15612–15619 [DOI] [PubMed] [Google Scholar]
- 38. Chang Y., Weiss D. S. (1999) Biophys. J. 77, 2542–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luque I., Leavitt S. A., Freire E. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 235–256 [DOI] [PubMed] [Google Scholar]
- 40. Farber P. J., Mittermaier A. (2011) J. Mol. Biol. 405, 819–830 [DOI] [PubMed] [Google Scholar]
- 41. Colquhoun D. (1998) Br. J. Pharmacol. 125, 924–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galzi J. L., Edelstein S. J., Changeux J. P. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1853–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aleksandrov A. A., Riordan J. R. (1998) FEBS Lett. 431, 97–101 [DOI] [PubMed] [Google Scholar]
- 44. Aleksandrov A. A., Chang X., Aleksandrov L., Riordan J. R. (2000) J. Physiol. 528, 259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aleksandrov A. A., Aleksandrov L. A., Riordan J. R. (2007) Pflugers Arch. 453, 693–702 [DOI] [PubMed] [Google Scholar]
- 46. Aleksandrov A. A., Cui L., Riordan J. R. (2009) J. Physiol. 587, 2875–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Csanády L., Nairn A. C., Gadsby D. C. (2006) J. Gen. Physiol. 128, 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Csanády L. (2009) J. Gen. Physiol. 134, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hennager D. J., Ikuma M., Hoshi T., Welsh M. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 3594–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bompadre S. G., Sohma Y., Li M., Hwang T. C. (2007) J. Gen. Physiol. 129, 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang W., Wu J., Bernard K., Li G., Wang G., Bevensee M. O., Kirk K. L. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3888–3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cui L., Aleksandrov L., Chang X. B., Hou Y. X., He L., Hegedus T., Gentzsch M., Aleksandrov A., Balch W. E., Riordan J. R. (2007) J. Mol. Biol. 365, 981–994 [DOI] [PubMed] [Google Scholar]
- 53. Wang W., Bernard K., Li G., Kirk K. L. (2007) J. Biol. Chem. 282, 4533–4544 [DOI] [PubMed] [Google Scholar]
- 54. Gerber S., Comellas-Bigler M., Goetz B. A., Locher K. P. (2008) Science 321, 246–250 [DOI] [PubMed] [Google Scholar]
- 55. Chen T. Y., Hwang T. C. (2008) Physiol. Rev. 88, 351–387 [DOI] [PubMed] [Google Scholar]
- 56. Zhou Z., Hu S., Hwang T. C. (2001) J. Physiol. 532, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cai Z., Scott-Ward T. S., Sheppard D. N. (2003) J. Gen. Physiol. 122, 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Szellas T., Nagel G. (2003) FEBS Lett. 535, 141–146 [DOI] [PubMed] [Google Scholar]
- 59. Wang F., Zeltwanger S., Hu S., Hwang T. C. (2000) J. Physiol. 524, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang W., Li G., Clancy J. P., Kirk K. L. (2005) J. Biol. Chem. 280, 23622–23630 [DOI] [PubMed] [Google Scholar]
- 61. Miki H., Zhou Z., Li M., Hwang T. C., Bompadre S. G. (2010) J. Biol. Chem. 285, 19967–19975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Serohijos A. W., Hegedus T., Aleksandrov A. A., He L., Cui L., Dokholyan N. V., Riordan J. R. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3256–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van Goor F., Hadida S., Grootenhuis P. D., Burton B., Cao D., Neuberger T., Turnbull A., Singh A., Joubran J., Hazlewood A., Zhou J., McCartney J., Arumugam V., Decker C., Yang J., Young C., Olson E. R., Wine J. J., Frizzell R. A., Ashlock M., Negulescu P. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18825–18830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pedemonte N., Sonawane N. D., Taddei A., Hu J., Zegarra-Moran O., Suen Y. F., Robins L. I., Dicus C. W., Willenbring D., Nantz M. H., Kurth M. J., Galietta L. J., Verkman A. S. (2005) Mol. Pharmacol. 67, 1797–1807 [DOI] [PubMed] [Google Scholar]