Abstract
The Gram-negative myxobacterium Sorangium cellulosum So ce56 bears the largest bacterial genome published so far, coding for nearly 10,000 genes. Careful analysis of this genome data revealed that part of the genes coding for the very well conserved biosynthesis of lipopolysaccharides (LPS) are missing in this microbe. Biochemical analysis gave no evidence for the presence of LPS in the membranes of So ce56. By analyzing the lipid composition of its outer membrane sphingolipids were identified as the major lipid class, together with ornithine-containing lipids (OL) and ether lipids. A detailed analysis of these lipids resulted in the identification of more than 50 structural variants within these three classes, which possessed several interesting properties regarding to LPS replacement, mediators in myxobacterial differentiation, as well as potential bioactive properties. The sphingolipids with the basic structure C9-methyl-C20-sphingosine possessed as an unusual trait C9-methylation, which is common to fungi but highly uncommon to bacteria. Such sphingolipids have not been found in bacteria before, and they may have a function in myxobacterial development. The OL, also identified in myxobacteria for the first time, contained acyloxyacyl groups, which are also characteristic for LPS and might replace those in certain functions. Finally, the ether lipids may serve as biomarkers in myxobacterial development.
Keywords: Bacteria, Carbohydrate Structure, Endotoxin, Lipid Structure, Lipopolysaccharide (LPS), Membrane Lipids, Myxobacterium, Sphingosine
Introduction
Sorangium cellulosum So ce56 (So ce56) belongs to the quite heterogeneous group of myxobacteria, which represent Gram-negative soil δ-proteobacteria that show a complex live cycle involving a cooperative and cellular morphogenesis (1). They have the ability to form fruiting bodies under nutrition starvation and move in swarms. Gliding movement on surfaces and the differentiation processes require complex cell-to-cell signaling and reminds the process of differentiation in eukaryotic Dictyostelium. In the last decades a number of pharmaceutical metabolites (2, 3) such as epothilone, a potent antitumor compound (4) have been isolated from this group of bacteria. Members of the genus Sorangium produce 46% of new bioactive secondary metabolites isolated from myxobacteria as yet (2).
So ce56, as a member of the suborder Sorangiineae, exhibits the largest bacterial genome entirely published so far. The recent genome sequencing revealed a huge chromosome of 13,033,779 base pairs, coding for 9,367 proteins over all (5).
Gram-negative bacteria possess characteristic cell envelopes comprising an inner (IM)2 and an outer membrane (OM) and specific cell surface carbohydrates (6). One class of prominent Gram-negative surface carbohydrates are the lipopolysaccharides (LPS, endotoxin), which represent the main components of the outer leaflet of the OM. LPS have complex structures, usually consisting of three main components that are built by specific cellular machineries (7), i.e. the O-antigen that extends into the extracellular medium, the core region, and the lipid A that anchors the molecule in the OM and, in toxic LPS, represents the endotoxic moiety of this molecule. The complex bacterial glycostructures present at the cell surface are important for interactions with their animate and inanimate environment. LPS play in addition an essential structural role as key constituents of the OM. In human, animal, and plant pathogenic bacteria LPS are often recognized by the particular hosts and provoke defense reactions. Only few Gram-negative bacteria are known that lack LPS, initially discovered in Sphingomonas (8–14).
In this study the uncommon OM of So ce56 is reported, which lacks LPS and therefore is a novel representative of the small group of Gram-negative bacteria without LPS. Instead, the OM contained a mixture of sphingo-, ornithine-containing, and ether lipids. The OM lipid composition of So ce56 was elucidated.
EXPERIMENTAL PROCEDURES
Bacterial Strain and Growth
So ce56 was grown in 70 liters of the synthetic medium SG (15) in a 100-liter bioreactor. After 240 h of growth at 32 °C, a pH of 7.2 and a pO2 maintained at 40% by regulation of the stirrer velocity, cells were separated from the culture broth by centrifugation (CEPA Z61). Clarification of the supernatant was performed by filtration (Cuno, 60 SP/60 CP), and concentration to 1,373 g was done by ultrafiltration (Hydrosart, molecular cutoff of 30 kDa, Fa. Sartorius). This vesicle-containing retentate was used for further analytical work.
Extraction and Isolation
Outer membrane vesicles (OMVs) were isolated according to a modified method by Wai et al. (16). The culture supernatant (200 ml) was centrifuged at 6000 × g for 1 h and subsequently filtered through a 0.2-μm filter to remove residual cells. OMVs were obtained by ultracentrifugation of the supernatant at 100,000 × g for 4 h.
Lipid extraction was performed according to Bligh and Dyer (17). Purification and isolation of the lipids was done by subjecting the extract to a silica gel column (7 × 1 cm, Silica Gel 60, Merck, 230–400 mesh) and stepwise elution with 6 volumes of 60 ml of chloroform (fraction 1), then 2.5% methanol/chloroform (v/v; fraction 2, SL), 5% methanol/chloroform (v/v; fraction 3, GGEL), 20% methanol/chloroform (v/v; fraction 4, GSL), 30% methanol/chloroform (v/v; fraction 5, OL, PEAGEL), and finally 40% methanol/chloroform (v/v; fraction 6, AEPnSL, PEASL).
Fractions 2, 3, 5 were further purified on a silica gel column (5 × 0.5 cm, Silica Gel 60, Merck, 230–400 mesh) to yield the final fractions indicated in Fig. 1 (SL, GGEL, OL, and PEAGEL). The lipids were eluted as described above. The elution was monitored by TLC, and appropriate fractions were combined.
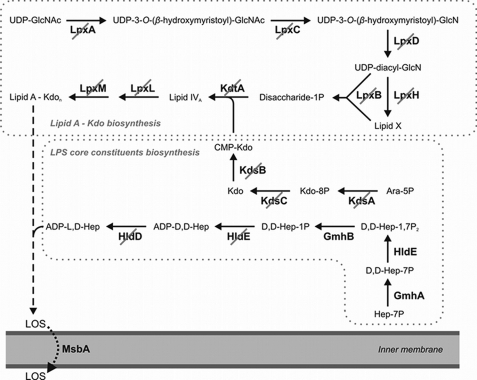
FIGURE 1.
Absence of conserved genes involved in LPS biosynthesis. The scheme depicts the biosynthetic route leading to the lipooligosaccharide (LOS), a premature form of LPS that is fundamentally conserved among Gram-negative bacteria (7). Enzyme-catalyzed reactions for which no genes were found in the genome of S. cellulosum So ce56 are crossed out in the scheme. A further characteristic component of LPS core oligosaccharides is heptose, which is however less prevalently represented in Gram-negative bacteria than lipid A and Kdo. Whereas there was some similarity to genes coding for the initial reaction steps starting with the isomerization of sedoheptulose-7-phosphate, no genes were found for the ultimate two reactions, indicating that So ce56 provided no adenosine-5′-diphospho-l-glycero-d-manno-heptose (ADP-l, d-Hep) as precursor for the LPS core. In a striking contrast to the absence of well-known genes assumed to be essential for the LOS biosynthesis, the chromosome carried several isogenes coding for the LOS exporter MsbA (supplemental Table S1).
Hot Phenol-Water Extraction
The hot phenol-water extraction of LPS was performed according to Westphal et al. (18).
Transmission Electron Microscopy
TEM was performed according to Engels et al. (19).
ESI-FT-ICR-MS
High resolution electrospray ionization fourier-transform ion cyclotron resonance mass spectrometry (ESI-FT-ICR-MS) was performed on a 7 Tesla APEX Qe instrument (Bruker Daltonics). For the negative-ion mode, the samples were dissolved in a water/2-propanol/triethylamine/acetic acid or trifluoroacetic acid (TFA) mixture (50:50:0.006:0.002, v/v/v/v). The samples were sprayed at a flow rate of 2 μl min−1. Capillary entrance voltage was set to 3.8 kV and dry gas temperature to 150 °C. Mass spectra were recorded in broad band mode, and mass scale was calibrated externally with glycolipids of known structure. The spectra were charge deconvoluted, and mass numbers given refer to the monoisotopic mass of the neutral molecule. For MS/MS lipid species were isolated in the ICR-cell and than fragmented by infrared multiphoton dissociation. For this, the unfocused beam of a 35 watt, 10.6 μm CO2 laser (Synrad) is directed through the center of the trap. Duration of laser irradiation (100–300 ms) and laser power (∼50%) were adapted to generate optimal fragmentation (20). Representative mass spectra are shown as supplemental Figs. S5–S11.
Sugar, Fatty Acid, Ether Lipid, and Sphingosine Analysis
Sugars and fatty acids were analyzed after acidic methanolysis (2 m methanolic HCl for 16 h at 85 °C) and peracetylation by GC-MS. The sphingosine was hydrogenated before methanolysis and peracetylation. The sphingolipid was dissolved in chloroform and 2 mg of platinum oxide was added. The hydrogenation was performed by bubbling with H2. The structures of ether lipids were additionally confirmed by trimethylsilyl derivatization with N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA).
Double Bond Localization
The positions of double bonds could be localized by derivatization with pyrrolidide and using linoleic acid as reference (21). The resulting products were analyzed by GC-MS.
Absolute Configuration
The determination of the absolute configuration of Glc and ornithine was performed by acidic butanolysis with (S)-2-butanol as described (22). The acetylated butyl derivatives were measured by GC-MS, and the configuration was determined by using d-Glc and l-ornithine as references.
The absolute configuration of OH-fatty acids was analyzed by GC-MS after l-phenylethylamide derivatization (23). The 2-hydroxy fatty acids were hydrogenated with platinum oxide and H2 prior to that. Representative chromatograms are shown as supplemental Figs. S1–S4.
GC-MS
Gas chromatography-mass spectrometry analysis was performed on an Agilent 5975 inert XL MSD instrument equipped with a HP-5MS capillary column (30 m × 0.25 mm, film thickness 0.25 μm) and applying a temperature gradient of 150 (kept for 3 min) to 320 °C at 5 °C/min.
Thin-Layer Chromatography
TLC was performed on high-performance TLC plates (Silica Gel 60, 10 × 10 cm, 0.2 mm thick; Merck, Darmstadt). The solvent system was chloroform/methanol/water (20:7:1, v/v/v). Lipids were visualized by dipping the TLC plate into one of the following solution and subsequently developing at 120 °C. Orcin stain solution (1 mg of orcin in 10% H2SO4 in ethanol), molybdenum stain solution (1 m H2SO4, 40 mm ammoniumheptamolybdat ((NH4)6Mo7O24·4H2O), 3 mm Cer(IV)-sulfate (Ce(SO4)2·4H2O)), and ninhydrin stain solution (0.2% ninhydrin in acetone).
NMR Spectroscopy
Deuterated solvents were purchased from Deutero GmbH (Kastellaun, Germany). NMR spectroscopic measurements were performed at 300 K on a Bruker AvanceIII 700 MHz (equipped with an inverse 5 mm quadruple-resonance Z-grad cryoprobe) and on a Bruker AvanceII 360 MHz NMR spectrometer (equipped with an inverse 5 mm multi-resonance Z-grad probe). Tetramethylsilane was used as internal standard for 1H and 13C NMR spectra, 85% of phosphoric acid was used as an external standard for 31P NMR spectra. All data were acquired and processed by using Bruker TOPSPIN V 2.1. 1H NMR assignments were confirmed by two-dimensional 1H,1H-COSY and -TOCSY experiments, 13C NMR assignments were indicated by two-dimensional 1H,13C-HSQC, based on the 1H NMR assignments. Inter-residue connectivities and further evidence for 13C assignment were obtained from two-dimensional 1H,13C-HMBC. Connectivities of phosphate groups were assigned by two-dimensional 1H,31P-HMQC. Full assignments of 1H-, 13C-, and 31P-NMR resonances for isolated compounds are reported in supplemental Tables S2–S6.
Genome Analysis
The following approach was carried out to identify putative LPS genes. DNA sequence data of well-known reference genes that code for important and often essential key steps of the LPS biosynthesis in Escherichia coli (7) was taken from reviewed UniProtKB/Swiss-Prot entries (24) to search the predicted coding-sequences (CDS) within the genomes of Myxococcus xanthus DK 1622 and S. cellulosum So ce56 as available from the NCBI genome project database by means of the Blastp algorithm (25). With the exception of a few members of abundantly occurring protein families like certain glycosyltransferases and epimerases, where a cutoff for relevant similarities was set at an E-value of 10−25, blast hits were considered when alignments had E-values falling below 10−5.
RESULTS
Absence of LPS in S. cellulosum So ce56
When the genome of So ce56 was analyzed for genes of cell surface glycostructures, no key gene for O-antigen biosynthesis was found and a complete absence of known and usually conserved genes coding for enzymes involved in the lipid A and core biosynthesis of LPS was detected (Fig. 1). In contrast to the universal absence of LPS biosynthesis genes, there were ten paralogous genes coding for MsbA-like LOS exporters (supplemental Table S1). This indicated a highly unusual composition of the outer leaflet of the OM, leading to the analysis of its composition.
To screen the envelope of So ce56 for the presence of LPS, hot phenol-water extraction (18) was applied. Neither the water nor the phenol phase contained any characteristic LPS components such as 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo). The other characteristic LPS components, namely 3-hydroxy fatty acids were found only in minor amounts, which could be assigned to another lipid class later.
Ultrastructure of S. cellulosum So ce56
Because no evidence for the presence of LPS could be found in So ce56 we analyzed the fine structure of the cell envelope. Transmission electron microscopy clearly revealed the typical Gram-negative structure comprising an IM and an OM (Fig. 2A). As indicated in Fig. 2B, So ce56 was able to produce OMVs, which are characteristic for Gram-negative bacteria. OMVs usually originate from the OM and consist of multiple types of lipids (26). A previous study of the ultrastructure of other Sorangium strains is in accordance with our findings (27).
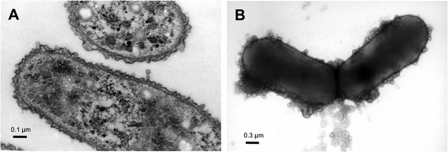
FIGURE 2.
Transmission electron micrographs of vegetative S. cellulosum So ce56 cells grown in liquid medium. A, ultra thin section of vegetative cells clearly shows the presence of two membranes enveloping the bacterial cell. B, negative contrast image of entire So ce56 cells. The cells are surrounded by globular structures representing OMVs.
Extraction and Purification of Lipids
The isolation of the IM and OM gradient ultracentrifugation did not result in a sufficient separation of both membranes, probably due to the lacking LPS in the OM and a similar lipid composition of both membranes. For this reason OMVs were harvested as a reliable source for outer membrane lipids (26). From 400 ml of concentrated culture supernatant 179.8 mg OMV (dry weight) could be obtained. The extraction of this material yielded in 51 mg of crude lipid extract, obtained by the extraction according to Bligh and Dyer (17). An aliquot (20 μg) of this extract was analyzed by thin-layer chromatography (TLC) (Fig. 3) and visualized by different staining methods. Seven different spots could be detected by developing the TLC with Mostain solution (Fig. 3, lane B). Based on the analytical data described below, spots were marked as sphingolipids (SL), glycerol ether lipids (GGEL), glycosphingolipids (GSL), phosphoethanolaminyl glycerol ether lipids (PEAGEL), ornithine-containing lipids (OL), aminoethyl phosphonyl sphingolipids (AEPnSL), and phosphoethanolaminyl sphingolipids (PEASL) according to their lipid classes. The orcin staining, characteristic for sugars, showed two different spots on the TLC plate (Fig. 3, lane C). The ninhydrin-staining resulted in three positive spots (Fig. 3, lane A). The crude lipid extract was applied to further isolation and purification steps by silica gel column chromatography, which delivered the separated fractions SL, GGEL, GSL, and the mixed fractions PEAGEL+OL as well as AEPnSL+PEASL for chemical and structural studies. Their structures were elucidated as described in the following. The chemical characteristics of the different lipids are presented in Table 1 and Fig. 4.
FIGURE 3.
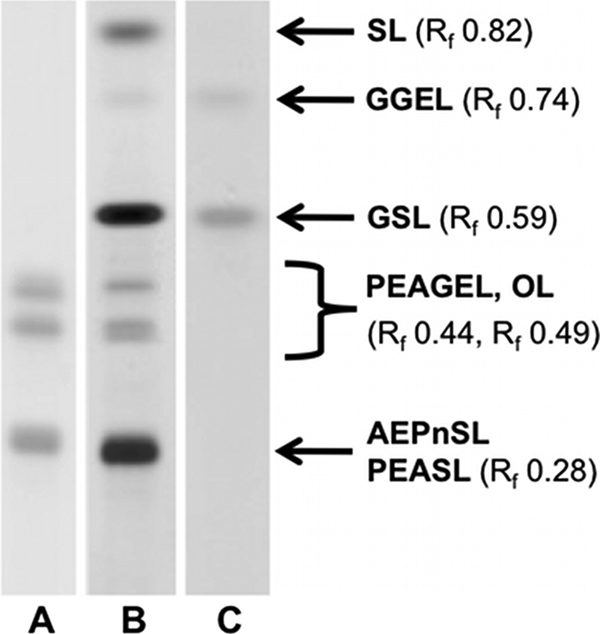
HPTLC analysis of crude lipid extract from outer membrane vesicles of S. cellulosum So ce56. Lipids were stained with A: ninhydrin, B: Mostain, C: Orcin. The bands are marked as sphingolipids (SL), glycerol ether lipids (GGEL), glycosphingolipids (GSL), phosphoethanolaminyl glycerol ether lipids (PEAGEL)), ornithine-containing lipids (OL), aminoethyl phosphonyl sphingolipids (AEPnSL), and phosphoethanolaminyl sphingolipids (PEASL) for the different lipid classes. The retention factors (Rf) are given in brackets.
TABLE 1.
Composition of the S. cellulosum So ce56 outer membrane
The list of lipids identified by high resolution ESI-FT-ICR mass spectrometry is shown.
| Lipid class | Abbr.a | Exact mass (measured) | Exact mass (calculated) | Mass accuracy |
|---|---|---|---|---|
| Sphingolipids | SL1a | 589.5064 | 589.5071 | 1.1026 |
| SL1b | 603.5217 | 603.5227 | 1.6569 | |
| SL1c | 617.5371 | 617.5384 | 2.0242 | |
| SL2a | 591.5220 | 591.5227 | 1.1834 | |
| SL2b | 605.5375 | 605.5384 | 1.4037 | |
| SL2c | 619.5530 | 619.5540 | 1.6141 | |
| GSL1a | 751.5590 | 751.5598 | 1.0645 | |
| GSL1b | 765.5743 | 765.5755 | 1.5021 | |
| GSL1c | 779.5901 | 779.5911 | 1.2827 | |
| GSL2a | 753.5746 | 753.5755 | 1.1280 | |
| GSL2b | 767.5902 | 767.5911 | 1.1725 | |
| GSL2c | 781.6074 | 781.6068 | 0.8316 | |
| AEPnSL1a | 696.5203 | 696.5206 | 0.4307 | |
| AEPnSL1b | 710.5351 | 710.5363 | 1.6185 | |
| AEPnSL1c | 724.5507 | 724.5519 | 1.6562 | |
| AEPnSL2a | 698.5357 | 698.5363 | 0.7874 | |
| AEPnSL2b | 712.5515 | 712.5519 | 0.5614 | |
| AEPnSL2c | 726.5676 | 726.5676 | 0.0688 | |
| PEASL1a | 712.5149 | 712.5155 | 0.8421 | |
| PEASL1b | 726.5305 | 726.5312 | 0.8947 | |
| PEASL1c | 740.5456 | 740.5468 | 1.6204 | |
| PEASL2a | 714.5314 | 714.5312 | 0.3499 | |
| PEASL2b | 728.5467 | 728.5468 | 0.1373 | |
| PEASL2c | 742.5623 | 742.5625 | 0.2020 | |
| Ornithine-containing lipids | OL1c | 622.5302 | 622.5284 | 2.8914 |
| OL1b, OL2a | 624.5460 | 624.5441 | 3.1223 | |
| OL2c | 636.5461 | 636.5441 | 3.2205 | |
| OL2b, OL1d | 638.5621 | 638.5597 | 3.7585 | |
| OL1f | 648.5461 | 648.5441 | 3.1609 | |
| OL2d | 652.5766 | 652.5754 | 1.9155 | |
| OL2f | 662.5617 | 662.5597 | 3.0186 | |
| OL1a | 610.5299 | 610.5284 | 2.4569 | |
| OL2e | 666.5930 | 666.5910 | 3.0003 | |
| Glycerol ether lipids | PEAGELba | 621.5117 | 621.5098 | 3.0571 |
| PEAGELbb | 635.5273 | 635.5255 | 2.8323 | |
| PEAGELdd | 659.5268 | 659.5255 | 1.9711 | |
| PEAGEL3d | 673.5064 | 673.5046 | 2.6726 | |
| PEAGEL1b | 649.5067 | 649.5046 | 3.2332 | |
| PEAGEL3b, PEAGEL1d | 661.5062 | 661.5046 | 2.4187 | |
| PEAGEL3c, PEAGEL2d | 675.5223 | 675.5202 | 3.1087 | |
| GGELba | 660.5556 | 660.5539 | 2.5736 | |
| GGELbb | 674.5715 | 674.5695 | 3.0019 | |
| GGEL1a | 674.5352 | 674.5331 | 3.1133 | |
| GGEL3a | 686.5323 | 686.5331 | 1.1653 | |
| GGEL2b, GGEL1c | 702.5643 | 702.5644 | 0.1423 | |
| GGEL3b, GGEL1d | 700.5493 | 700.5487 | 0.8565 | |
| GGELbd | 686.5714 | 686.5697 | 2.4761 | |
| GGEL1b, GGEL2a | 688.5484 | 688.5488 | 0.5809 | |
| GGELdd | 698.5703 | 698.5695 | 1.1452 |
a Abbreviations can be correlated with the corresponding structures in Fig. 4.
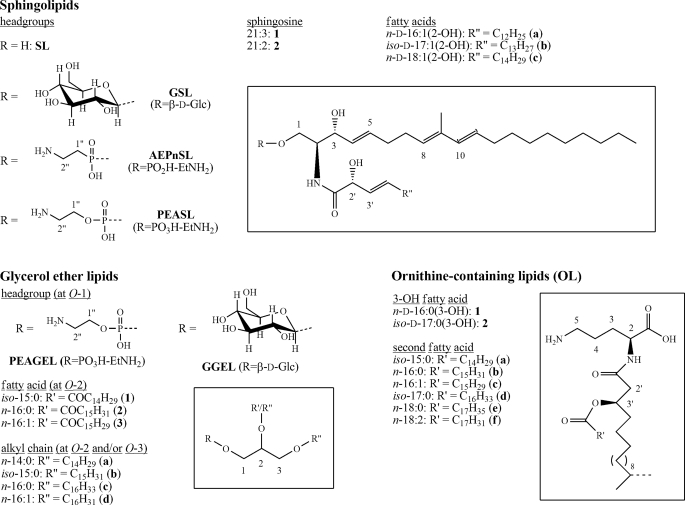
FIGURE 4.
Summary of all lipid structures of the S. cellulosum So ce56 outer membrane. The basic structures of the three observed lipid classes are presented in boxes. Specified structures can be received by combination of abbreviations (e.g. GSL1a: (2S,2′R,3R,4E,8E,10E)-1-O-β-d-glucopyranosyl-N-2-(2′-hydroxy-3′-hexadecenoyl)-9-methyl-4,8,10-icosatrien-1,3-diol; GGEL2b:1-O-β-d-glucopyranosyl-2-O-hexadecanoyl-3-O-iso-pentadecyl-sn-glycerol).
Structures of Sphingolipids
The crude extract of So ce56 comprised four different types of sphingolipids, all of which possessed a C21-sphingosine (proven by GC-MS analysis) as the basic structure. As indicated in Fig. 4 1-O-unsubstituted SL TLC: Rf 0.82 (Fig. 3), GSL TLC: Rf 0.59 (Fig. 3), AEPnSL and PEASL were identified. The two latter types were isolated as single fraction (TLC: Rf 0.28 (Fig. 3)), in which phosphonosphingolipids were the major compounds (89% AEPnSL, 11% PEASL).
1H NMR and MS/MS analysis (for detailed assignment of 1H- and 13C-NMR shifts see the supporting information (SL: supplemental Table S2; GSL: supplemental Table S4; AEPnSL+PEASL: supplemental Table S6) and for MS/MS (supplemental Fig. S8)) of these three TLC fractions showed a high structural similarity of the isolated sphingolipid types, only differing in signals of the respective headgroup and a changed NMR shift for protons 1a-H, 1b-H, 2-H, and 3-H (Fig. 5). In case of PEASL, only the headgroup protons (1″-H and 2″-H from the ethanolamine residue) were distinguishable from the signals resulting from AEPnSL. Possible minimal changes in NMR shifts for other protons could not be excluded, but were not evaluable (only for 3-H a separated signal was visible). Furthermore, NMR analysis showed that all sphingolipid types occurred in two different variants of the C21-sphingosine, namely as 9-methyl-4,8,10-icosatrien-1,3-diol (21:3) and as 9-methyl-4,8-icosadien-1,3-diol (21:2; see marked signals in Fig. 5). The double bonds in the sphingosine part at position C-4 and C-10 (in case of 21:3) were determined to be trans-type by the respective coupling constants (J4,5 15.0 or 15.4 Hz and J10,11 15.4 Hz). The double bond at C-8 was identified as trans-type based on the chemical shift (δC 16.0/16.1(in 21:2), δC 12.7/12.8 (in 21:3)) of the methyl group at C-9. These findings are consistent with the published data of Zhang et al. for 21:2-type SL and GSL (28). In all cases 21:3 was the predominant variant (SL: 75%, GSL: 90%, AEPnSL+PEASL: 83%; ratios were determined by integrating the respective methyl groups at C-9). The absolute stereochemistry of position C-2 and C-3 of the sphingosine was presumed by comparision of observed NMR shifts and coupling constants with those published earlier. A coupling constant for J2,3 of ∼7 Hz clearly identifies an erythro-sphingosine, while a coupling constant between 3.5 and 4.5 Hz indicates the threo-form (29). The determined coupling constants for J2,3 were ∼6 Hz for SL, 7.0 Hz for GSL, and 7.5 Hz for AEPnSL+PEASL. This is in agreement with the fact that only d-erythro-sphingosine is known to occur in nature (30). Thus, we proposed for the determined sphingosine variants the absolute stereochemistry d-erythro (2S,3R), which allowed the two variants of C21-sphingosine to be described more precisely as (2S,3R,4E,8E,10E)-9-methyl-4,8,10-icosatrien-1,3-diol (21:3) and (2S,3R,4E,8E)-9-methyl-4,8-icosadien-1,3-diol (21:2), corresponding to earlier reports (28, 31, 32).
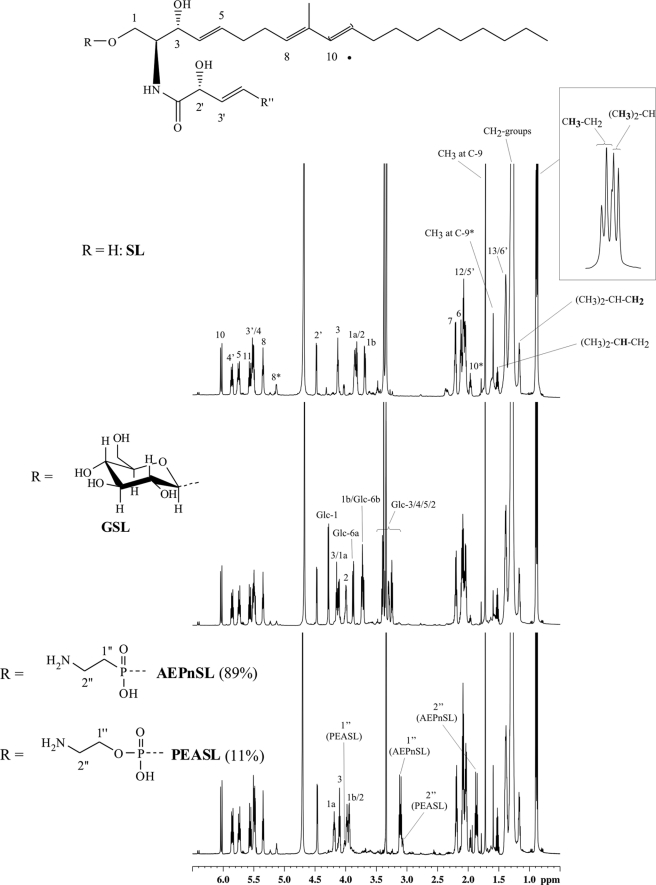
FIGURE 5.
Comparative 1H NMR analysis of sphingolipid-containing TLC fractions showing high structural similarity. Top: SL, center: GSL, bottom: AEPnSL+PEASL. A complete assignment of protons is given for SL, for other compounds only shifted protons 1-H, 2-H, and 3-H as well as protons from the head groups are indicated. Signals marked with (*) belong to the 21:2-type sphingosine. Enlargement of the region for methyl groups in SL shows the presence of two different types (n-form (triplet) and iso-form (doublet)) as it is also the case for both other fractions.
The glycosyl moiety in GSL was identified as Glc by GC-MS analysis. The determination of the absolute configuration was performed by acidic butanolysis and revealed the d-configuration. NMR analysis confirmed this result and showed a β-linkage (J1,2 7.9 Hz) to the sphingosine moiety. This result was also in accordance with reported structures (28, 31).
In each TLC fraction three fatty acids were detected by GC-MS. These fatty acids were identified as 2-hydroxy-3-hexadecenoic (16:1(2-OH); 20%), 2-hydroxy-3-heptadecenoic (17:1(2-OH); 47%), and 2-hydroxy-3-octadecenoic (18:1(3-OH); 33%) acid, respectively. In 16:1(2-OH) and 18:1(2-OH) the n-form and in 17:1(2-OH) the iso-form occurred almost exclusively. The respective opposite forms could not be detected in significant amounts (< 0.5%). NMR analysis revealed that all 2-hydroxy fatty acids had the double bond at position C-3′, which were of the trans-type due to their coupling constant (J3′,4′ 15.4 Hz). The phenylethylamide derivative showed that all 2-hydroxy fatty acids have the d-configuration.
In accordance with the results of GC-MS and NMR analysis, ESI-FT-ICR-MS analysis of the three sphingolipid-containing TLC fractions pointed to the presence of 24 different sphingolipids. The measured and calculated exact masses of all lipids are shown in Table 1. All sphingolipids could be detected with a mass accuracy of less than 2.1 ppm.
Structures of OL
Besides the sphingolipids, OL were isolated from the crude extract of So ce56. They were obtained as a mixed fraction together with PEAGEL (TLC: Rf 0.49 and 0.44 (Fig. 3)), which are discussed separately below. The detailed assignment of 1H- and 13C-NMR shifts are summarized in supplemental Table S5. The integration of the 1H-NMR spectrum showed, that OL were the major compounds (82%) in this fraction (integration of ornithine(OR)-5-H versus two types of glycerol-1-H of PEAGEL).
1H,13C-HMBC-correlations showed unequivocally that the α-amino group of the ornithine formed an amide with a 3-OH fatty acid. Furthermore, it became obvious that the 3-OH fatty acid was esterified with a second fatty acid. For the latter, NMR analysis indicated at least three different species (NMR shifts of the carbonyl carbon atoms: δC 174.6 ppm (two signals) and 174.5 ppm)) and also the presence of unsaturated fatty acids.
The fatty acid composition was determined by GC-MS after methanolysis and peracetylation. Thus, iso-pentadecanoic (iso-15:0; 2%), n-hexadecanoic (n-16:0; 11%), n-hexadecenoic (n-16:1; 20%), iso-heptadecanoic (iso-17:0; 33%), n-octadecanoic (n-18:0; 3%), and n-octadecadienoic (n-18:2; 31%) acid as well as n-3-hydroxy-hexadecanoic (n-16:0(3-OH); 47%) and iso-3-hydroxy-heptadecanoic (iso-17:0(3-OH); 53%) acid were identified. The phenylethylamide derivative showed that both 3-hydroxy fatty acids possessed the d-configuration. To localize the double bonds in unsaturated fatty acids of OL, the pyrrolidide derivatives were analyzed by GC-MS. This revealed n-16:1 (cis-Δ11) and n-18:2 (cis,cis-Δ9,12), respectively. The absolute configuration of ornithine was determined by using the acetylated butyl derivative and identified the l-configuration.
GC-MS analysis and the results of ESI-FT-ICR-MS (Table 1) afforded eleven different OL, differing in the fatty acid composition, whereas OL1b and OL2a (m/z 624.5441), OL1d and OL2b (m/z 638.5597) had the same masses and were indistinguishable from each other. MS/MS analysis did not provide further data to discriminate these lipids, because of overlapping of signals with the isotopic peaks of OL1c and OL2c. OL2d could be distinguished from the hypothetical OL1e by MS/MS analysis. The analysis revealed that only OL2d exist (supplemental Fig. S10). All OL were identified with a mass accuracy of less than 3.8 ppm.
Structures of Ether Lipids
As a third class of lipids, the crude extract contained two types of GEL. PEAGEL were isolated (18%) from the already mentioned mixed fraction together with OL (TLC: Rf 0.49 and 0.44 (Fig. 3)). GGEL were isolated in a fraction devoid of other lipids (Rf 0.74; Fig. 3). For both types of GEL NMR analysis revealed two different variants of substitution pattern besides the respective headgroup (at glycerol-O-1). Here, either glycerol-O-2 and -O-3 were etherified with a long-chain alkylalcohol (hereafter termed as dialkyl) or glycerol-O-2 was esterified with a fatty acid and only -O-3 was etherified (monoalkyl). The characteristic chemical shifts for glycerol-2-H and -C-2 in the dialkyl variant are δH 3.68–3.64 and δC 78.6 (GGEL) as well as δH 3.67–3.60 and δC 78.7 (PEAGEL) in comparison to δH 5.23–5.18 and δC 72.4 (GGEL) as well as 5.18–5.14 and δC 72.5 (PEAGEL) for the monoalkyl variant. Integration of the respective signals for glycerol-2-H revealed a dialkyl:monoalkyl-ratio of 83:17 for PEAGEL and 70:30 for GGEL, respectively. For all compounds the position of O-bound substituents was proven by 1H,13C-HMBC-correlations (for detailed assignments of 1H- and 13C-NMR shifts, see the supporting information (GGEL: supplemental Table S3; PEAGEL: supplemental Table S5)).
GC-MS analysis of fraction GGEL revealed the presence of four dialkyl- and four monoalkyl glycerol ethers. In the first group, 1-O-acetyl-2-O-pentadecyl-3-O-tetradecylglycerol (40%), 1-O-acetyl-2,3-di-(O-pentadecyl)-glycerol (54%), 1-O-acetyl-2-O-pentadecyl-3-O-hexadecenylglycerol (4%), and 1-O-acetyl-2,3-di-(O-hexadecencyl)-glycerol (2%) were identified. The observed acetylated monoalkyl ether were 1,2-di-O-acetyl-3-O-tetradecylglycerol (34%), 1,2-di-O-acetyl-3-O-pentadecylglycerol (46%), 1,2-di-O-acetyl-3-O-hexadecylglycerol (14%), and 1,2-di-O-acetyl-3-O-hexadecenylglycerol (6%). Furthermore, iso-15:0 (41%), n-16:0 (24%), and n-16:1 (35%) were identified as the monoacyl substituents at glycerol-O-2. The glycosyl moiety in GGEL was determined by GC-MS analysis as d-Glc.
All four 1-O-β-d-glucopyranosyl dialkyl glycerol ether structures could be confirmed by ESI-FT-ICR-MS with a mass accuracy less than 3.2 ppm (Table 1). Out of theoretically possible twelve 1-O-β-d-glucopyranosyl-2-O-acyl-3-O-alkylglycerolether eight could be verified by ESI-FT-ICR-MS as listed in Table 1, whereas GGEL1b and GGEL2a, GGEL1d and GGEL3b, as well as GGEL1c and GGEL2b were not distinguishable. By MS/MS analysis these lipid species could not be distinguished from each other because of low intensities and/or overlapping isotopic peaks.
In the fraction PEAGEL+OL the acetylated dialkyl ether 1-O-acetyl-2-O-pentadecyl-3-O-tetradecylglycerol (50%), 1-O-acetyl-2,3-di-(O-pentadecyl)-glycerol (42%), and 1-O-acetyl-2,3-di-(O-hexadecencyl)-glycerol (8%) as well as the acetylated monoalkyl ether 1,2-di-O-acetyl-3-O-pentadecylglycerol (35%), 1,2-di-O-acetyl-3-O-hexadecylglycerol (53%), and 1,2-di-O-acetyl-3-O-hexadecenylglycerol (12%) were identified by GC-MS analysis. As mentioned above, this mixed fraction contained six fatty acids (iso-15:0, n-16:0, n-16:1, iso-17:0, n-18:0, n-18:2). NMR analysis suggested that to a minor amount a methylated double bond was present. The three 1-O-phosphorethanolaminyl dialkyl glycerol ethers PEAGELba, PEAGELbb, and PEAGELdd could be confirmed by ESI-FT-ICR-MS (Table 1). Besides that, six 1-O-phosphorethanolaminyl-2-O-acyl-3-O-alkylglycerolether were ascertained, whereas PEAGEL1d and PEAGEL3b as well as PEAGEL2d and PEAGEL3c were indistinguishable. The discrimination between these lipids was impossible due to the reasons already discussed for GGEL. Because of the small amounts of PEAGEL, it could not be excluded that more combinations were existing. Thus, the compounds listed in Table 1 may represent only the most abundant ones.
DISCUSSION
Although LPS has been identified in several other myxobacterial species (33, 34), the analysis of the genomic data and the absence of specific markers of LPS such as Kdo in phenol/water extracts show that S. cellulosum So ce56 lacks LPS as component of its OM. The absence of LPS in Gram-negative bacteria is still unusual, although proven for several Gram-negative bacteria, i.e. Treponema pallidum, Borrelia burgdorferi, Borrelia hispanica (8, 9), Sphingomonas capsulate, and Sphingomonas paucimobilis (10–12) as well as Thermus thermophilus (13) and Meiothermus taiwanensis (14). Sutherland reported the presence of LPS in the outer leaflet of two strains in the suborder Sorangineae (35). Further on, LPS was detected in Myxococcus xanthus, a myxobacterial species that is less related to Sorangium (5). The absence of LPS in So ce56 could be restricted to a small tree within this group of Gram-negative bacteria, because closely and more distantly related myxobacteria produce LPS. Even though LPS is missing the characteristic Gram-negative cell envelope consisting out of two membranes separated by an periplasmatic space could be confirmed by electron microscopy. As also shown by Lamky (27) the OM is undulated, and the cells are surrounded by numerous OMVs. By analyzing the lipids of the OMVs, three different major lipid classes of the outer membrane of So ce56 with interesting features could be identified, i.e. SL, GEL, and OL.
The first class representing the most abundant lipids of the outer membrane of So ce56 are the sphingolipids. Only two studies about sphingolipids in myxobacteria were reported earlier (36) in which 17-methyl-C18-sphingosine (19:2) was described as the basic structure. The 9-methyl-sphingosine moiety has been unique to fungi and some marine invertebrates until now (37). Here, we report on the structure of the first sphingolipid with a 9-methyl-C20-sphingosine (21:3 and 21:2) moiety discovered in bacteria. Zhang et al. (28) described the 21:2 structure as a lipid of Aspergillus niger with moderate antifungal activity. Interestingly, these sphingolipids were described as predominate mediators of development in fungi (38, 39, 40). Therefore, a similar function of 9-methyl-sphingosine in myxobacterial development could be postulated. Another interesting point is the headgroup 2-aminoethylphosphonate (AEP) of the phosphonosphingolipid, because it is a natural product containing a carbon-phosphorus bond. Such C-P compounds have found widespread use in medicine and agriculture (41), which underlies the great potential of myxobacteria as a wealth for natural bioactive compounds. Phosphonolipids are very rare in nature. They were only described in the sea anemone Metridium senile (42), the bacterium Bacteriovorax stolpii (43), and the protozoan Tetrahymena thermophila (44). Interestingly, phosphonosphingolipids of Bacteriovorax stolpii UKi2 show sensitivity to phospholipase C and resistance to phospholipase D (45). In this context, the close phylogenetic relationship of myxobacteria to Bacteriovorax has to be mentioned (5).
OL are widespread among Gram-negative bacteria and were found in some Gram-positive bacteria as well (46). They possess similarity to bacterial LPS amide-bound 3-OH fatty acids, the hydroxy groups of which are esterified with another fatty acid (acyloxyacyl groups) (47). Furthermore, OL were also described in Sinorhizobium meliloti grown under phosphorous-limiting condition (48). OL have not been identified in myxobacteria before, thus, their role in myxobacterial life is completely unknown up to now.
As a third class of lipids ether lipids could be identified. The major ether lipids are dialkyl ether lipids with a phosphoethanolamin or glucose as headgroup. Ether lipids as monoalkyl moiety have been proposed by Ring et al. (49) as new biomarkers for fruiting bodies formation in Myxococcus xanthus. A basic structure of 1-O-13-methyltetradecyl glycerol has been described. The same moiety was present in So ce56.
The presence of dialkyl ether lipids could be confirmed from vegetative cells of the myxobacterium Stigmatella aurantiaca (50). These findings indicate an occurrence of such ether lipids in myxobacteria and may play an important role in myxobacterial development. Besides phosphoethanolamine, we also identified glucose as a headgroup of the ether lipids. This headgroup together with the dialkyl moiety are described in myxobacteria for the first time. The functions of these ether lipids in myxobacteria are unknown.
Taken together, we isolated and elucidated the complex lipid composition of the OM of S. cellulosum So ce56 with a total number of more than 50 lipids. These data and the fact that LPS is missing give new insights into the complex myxobacterial life cycle with a remarkable eukaryotic character of this bacterium. This is also reflected in marked similarities to eukaryotic regulatory network sytems (5, 51, 52).
Acknowledgments
We thank Regina Engel (Research Center Borstel) for technical assistance and Ulrich Zähringer (Research Center Borstel) for valuable discussion. Furthermore, we would like to thank Sebastian Kusche, Stefan Meyer, Kerstin Frühling, Irena Steinau, Nadine Küpper, and particularly Sandra Markmann.
This work was funded by Grant 32106669 from the German Federal Ministry of Education and Research (BMBF) (GenoMik and GenoMikPlus).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S11 and Tables S1–S6.
- IM
- inner membrane
- LPS
- lipopolysaccharides
- So ce56
- S. cellulosum So ce56
- OM
- outer membrane
- LOS
- lipooligosaccharide
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- OMVs
- outer membrane vesicles
- TLC
- thin-layer chromatography
- SL
- sphingolipids
- GGEL
- glycerol ether lipids
- GSL
- glycosphingolipids
- PEAGEL
- phosphoethanolaminyl glycerol ether lipids
- OL
- ornithine-containing lipids
- AEPnSL
- aminoethyl phosphonyl sphingolipids
- PEASL
- phosphoethanolaminyl sphingolipids
- GC-MS
- gas chromatography-mass spectrometry
- ESI-FT-ICR-MS
- electrospray ionization fourier transform ion cyclotron resonance mass spectrometry
- TEM
- transmission electron microscopy
- COSY
- correlation spectroscopy
- TOCSY
- total correlated spectroscopy
- HSQC
- heteronuclear single quantum coherence
- HMBC
- heteronuclear multiple bond coherence
- HMQC
- heteronuclear multiple quantum coherence.
REFERENCES
- 1. Pradella S., Hans A., Spröer C., Reichenbach H., Gerth K., Beyer S. (2002) Arch. Microbiol. 178, 484–492 [DOI] [PubMed] [Google Scholar]
- 2. Gerth K., Pradella S., Perlova O., Beyer S., Müller R. (2003) J. Biotechnol. 106, 233–253 [DOI] [PubMed] [Google Scholar]
- 3. Gerth K., Bedorf N., Höfle G., Irschik H., Reichenbach H. (1996) J. Antibiot. 49, 560–563 [DOI] [PubMed] [Google Scholar]
- 4. Wenzel S. C., Müller R. (2007) Nat. Prod. Rep. 24, 1211–1224 [DOI] [PubMed] [Google Scholar]
- 5. Schneiker S., Perlova O., Kaiser O., Gerth K., Alici A., Altmeyer M. O., Bartels D., Bekel T., Beyer S., Bode E., Bode H. B., Bolten C. J., Choudhuri J. V., Doss S., Elnakady Y. A., Frank B., Gaigalat L., Goesmann A., Groeger C., Gross F., Jelsbak L., Jelsbak L., Kalinowski J., Kegler C., Knauber T., Konietzny S., Kopp M., Krause L., Krug D., Linke B., Mahmud T., Martinez-Arias R., McHardy A. C., Merai M., Meyer F., Mormann S., Muñoz-Dorado J., Perez J., Pradella S., Rachid S., Raddatz G., Rosenau F., Rückert C., Sasse F., Scharfe M., Schuster S. C., Suen G., Treuner-Lange A., Velicer G. J., Vorhölter F. J., Weissman K. J., Welch R. D., Wenzel S. C., Whitworth D. E., Wilhelm S., Wittmann C., Blöcker H., Pühler A., Müller R. (2007) Nat. Biotechnol. 25, 1281–1289 [DOI] [PubMed] [Google Scholar]
- 6. Reeves P. R., Hobbs M., Valvano M. A., Skurnik M., Whitfield C., Coplin D., Kido N., Klena J., Maskell D., Raetz C. R., Rick P. D. (1996) Trends Microbiol. 4, 495–503 [DOI] [PubMed] [Google Scholar]
- 7. Raetz C. R., Whitfield C. (2002) Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takayama K., Rothenberg R. J., Barbour A. G. (1987) Infect. Immun. 55, 2311–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hardy P. H., Jr., Levin J. (1983) Proc. Soc. Exp. Biol. Med. 174, 47–52 [DOI] [PubMed] [Google Scholar]
- 10. Kawahara K., Seydel U., Matsuura M., Danbara H., Rietschel E. T., Zähringer U. (1991) FEBS Lett. 292, 107–110 [DOI] [PubMed] [Google Scholar]
- 11. Kawasaki S., Moriguchi R., Sekiya K., Nakai T., Ono E., Kume K., Kawahara K. (1994) J. Bacteriol. 176, 284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawahara K., Moll H., Knirel Y. A., Seydel U., Zähringer U. (2000) Eur. J. Biochem. 267, 1837–1846 [DOI] [PubMed] [Google Scholar]
- 13. Leone S., Molinaro A., Lindner B., Romano I., Nicolaus B., Parrilli M., Lanzetta R., Holst O. (2006) Glycobiology 16, 766–775 [DOI] [PubMed] [Google Scholar]
- 14. Yang Y. L., Yang F. L., Huang Z. Y., Tsai Y. H., Zou W., Wu S. H. (2010) Org. Biomol. Chem. 8, 4252–4254 [DOI] [PubMed] [Google Scholar]
- 15. Müller R., Gerth K. (2006) J. Biotechnol. 121, 92–200 [DOI] [PubMed] [Google Scholar]
- 16. Wai S. N., Takade A., Amako K. (1995) Microbiol. Immunol. 39, 451–456 [DOI] [PubMed] [Google Scholar]
- 17. Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 18. Westphal O., Jann K. (1965) Methods Carbohydr. Chem. 5, 83–91 [Google Scholar]
- 19. Engels A., Kahmann U., Ruppel H. G., Pistorius E. K. (1997) Biochim. Biophys. Acta 1340, 33–44 [DOI] [PubMed] [Google Scholar]
- 20. Hübner G., Crone C., Lindner B. (2009) J. Mass Spectrom. 44, 1676–1683 [DOI] [PubMed] [Google Scholar]
- 21. Harvey D. J., Tiffany J. M. (1984) J. Chromatogr. 301, 173–187 [DOI] [PubMed] [Google Scholar]
- 22. Gerwig G. J., Kamerling J. P., Vliegenthart J. F. G. (1979) Carbohydr. Res. 77, 1–7 [DOI] [PubMed] [Google Scholar]
- 23. Rietschel E. T. (1976) Eur. J. Biochem. 64, 423–428 [DOI] [PubMed] [Google Scholar]
- 24. UniProt Consortium (2010) Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McBroom A. J., Kuehn M. J. (2005) in Outer Membrane Vesicles. EcoSal--Escherichia coli and Salmonella: Cellular and Molecular Biology (Curtiss R., III ed) Chapter 2.2.4., ASM Press, Washington, D. C [Google Scholar]
- 27. Lamky J. R. (1976) J. Bacteriol. 126, 1278–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang Y., Wang S., Li X. M., Cui C. M., Feng C., Wang B. G. (2007) Lipids 42, 759–764 [DOI] [PubMed] [Google Scholar]
- 29. Li S., Wilson W. K., Schroepfer G. J., Jr. (1999) J. Lipid Res. 40, 764–772 [PubMed] [Google Scholar]
- 30. Olsen I., Jantzen E. (2001) Anaerobe 7, 103–112 [Google Scholar]
- 31. Gao J. M., Dong Z. L., Liu J. K. (2001) Lipids 36, 175–180 [DOI] [PubMed] [Google Scholar]
- 32. Jin W., Rinehart K. L., Jares-Erijman E. A. (1994) J. Org. Chem. 59, 144–147 [Google Scholar]
- 33. Sutherland I. W., Smith M. L. (1973) J. Gen. Microbiol. 74, 259–266 [Google Scholar]
- 34. Rosenfelder G., Lüderetz O., Westphal O. (1974) Eur. J. Biochem. 44, 411–420 [DOI] [PubMed] [Google Scholar]
- 35. Sutherland I. W. (1979) J. Gen. Microbiol. 111, 211–216 [DOI] [PubMed] [Google Scholar]
- 36. Eckau H., Dill D., Budzikiewicz H. (1984) Z. Naturforsch. C 39, 1–9 [Google Scholar]
- 37. Sperling P., Heinz E. (2003) Biochim. Biophys. Acta 1632, 1–15 [DOI] [PubMed] [Google Scholar]
- 38. Ramamoorthy V., Cahoon E. B., Thokala M., Kaur J., Li J., Shah D. M. (2009) Eukaryot. Cell 8, 217–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kawai G., Ikeda Y. (1982) Biochim. Biophys. Acta 719, 612–618 [Google Scholar]
- 40. Oura T., Kajiwara S. (2010) Microbiology 156, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 41. Metcalf W. W., van der Donk W. A. (2009) Annu. Rev. Biochem. 78, 65–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mason W. T. (1972) Biochim. Biophys. Acta 280, 538–544 [DOI] [PubMed] [Google Scholar]
- 43. Steiner S., Conti S. F., Lester R. L. (1973) J. Bacteriol. 116, 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adosraku R. K., Smith J. D., Nicolaou A., Gibbons W. A. (1996) Biochim. Biophys. Acta 1299, 167–174 [DOI] [PubMed] [Google Scholar]
- 45. Jayasimhulu K., Hunt S. M., Kaneshiro E. S., Watanabe Y., Giner J. L. (2007) J. Am. Soc. Mass Spectrom. 18, 394–403 [DOI] [PubMed] [Google Scholar]
- 46. Geiger O., González-Silva N., López-Lara I. M., Sohlenkamp C. (2010) Prog. Lipid Res. 49, 46–60 [DOI] [PubMed] [Google Scholar]
- 47. López-Lara I. M., Sohlenkamp C., Geiger O. (2003) Mol. Plant Microbe Interact. 16, 567–579 [DOI] [PubMed] [Google Scholar]
- 48. López-Lara I. M., Gao J. L., Soto M. J., Solares-Pérez A., Weissenmayer B., Sohlenkamp C., Verroios G. P., Thomas-Oates J., Geiger O. (2005) Mol. Plant Microbe Interact. 18, 973–982 [DOI] [PubMed] [Google Scholar]
- 49. Ring M. W., Schwär G., Thiel V., Dickschat J. S., Kroppenstedt R. M., Schulz S., Bode H. B. (2006) J. Biol. Chem. 281, 36691–36700 [DOI] [PubMed] [Google Scholar]
- 50. Caillon E., Lubochinsky B., Rigomier D. (1983) J. Bacteriol. 153, 1348–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nariya H., Inouye S. (2006) Mol. Microbiol. 60, 1205–1217 [DOI] [PubMed] [Google Scholar]
- 52. Nariya H., Inouye S. (2005) Mol. Microbiol. 58, 367–379 [DOI] [PubMed] [Google Scholar]