Abstract
Autophagy has recently been implicated in both the prevention and progression of cancer. However, the molecular basis for the relationship between autophagy induction and the initial acquisition of malignancy is currently unknown. Here, we provide the first evidence that autophagy is essential for oncogenic K-Ras (K-RasV12)-induced malignant cell transformation. Retroviral expression of K-RasV12 induced autophagic vacuole formation and malignant transformation in human breast epithelial cells. Interestingly, pharmacological inhibition of autophagy completely blocked K-RasV12-induced, anchorage-independent cell growth on soft agar. Both mRNA and protein levels of ATG5 and ATG7 (autophagy-specific genes 5 and 7, respectively) were increased in cells overexpressing K-RasV12. Targeted suppression of ATG5 or ATG7 expression by short hairpin (sh) RNA inhibited cell growth on soft agar and tumor formation in nude mice. Moreover, inhibition of reactive oxygen species (ROS) with antioxidants clearly attenuated K-RasV12-induced ATG5 and ATG7 induction, autophagy, and malignant cell transformation. MAPK pathway components were activated in cells overexpressing K-RasV12, and inhibition of JNK blunted induction of ATG5 and ATG7 and subsequent autophagy. In addition, pretreatment with antioxidants completely inhibited K-RasV12-induced JNK activation. Our results provide novel evidence that autophagy is critically involved in malignant transformation by oncogenic K-Ras and show that reactive oxygen species-mediated JNK activation plays a causal role in autophagy induction through up-regulation of ATG5 and ATG7.
Keywords: Autophagy, JNK, Oncogene, Reactive Oxygen Species (ROS), Transformation, Tumor, Induction of ATG7, Malignant Transformation, Oncogenic K-Ras, Tumor Formation
Introduction
Ras GTPases act as molecular switches to transduce extracellular signals to the nucleus, where they regulate an overlapping set of cellular responses. Uncontrolled Ras activation is one of the most common genetic alterations associated with human cancers. Point mutations of Ras genes abolish the GTPase-activating protein-dependent GTP hydrolysis activity of Ras, leading to a constitutively activated Ras protein. These mutations occur at high frequency in mammalian cells, resulting in malignant transformation and further progression to cancer. Such point mutations have been found in a broad spectrum of tumors; in fact, oncogenic Ras mutations are found in more than 30% of all human malignancies, suggesting that Ras plays an important role in tumor development (1–6).
Cellular homeostasis depends on a balance between the rates of synthesis and degradation of intracellular components. Eukaryotic cells primarily use two distinct mechanisms, the proteasome pathway and autophagy, for large scale degradation; however, only autophagy has the capacity to degrade entire organelles (7, 8). Autophagy is a dynamic process that prolongs survival for a short time under starvation conditions and is evolutionarily conserved among all eukaryotic cells, from yeast to mammals. It is a highly regulated cellular mechanism in which intracellular double membrane-bound structures, known as autophagosomes, sequester proteins and intracellular organelles, such as mitochondria, and fuse with a lysosome or vacuole. Organelle contents are subsequently degraded and recycled as part of a process that serves to maintain cell viability (9, 10).
A number of recent studies have focused on the importance of autophagy in cancer, but it is still not clear whether autophagy suppresses tumorigenesis or is a mechanism to rescue cancer cells under unfavorable conditions. Autophagy may be involved in regulating both the promotion and prevention of cancer, and its role may be altered during tumor progression. Inhibition of autophagy allows the continuous growth of precancerous cells (11–13), and defects in autophagy are associated with increased tumorigenesis (14, 15), suggesting that autophagy can act as a cancer suppressor. However, autophagy may have another role, as reflected in the correlation between autophagy and promotion of cell survival under stress. As a tumor grows, resident cancer cells may depend on autophagy to survive nutrient-limiting and low oxygen conditions, especially in the internal, poorly vascularized region of the tumor (16). As a corollary, autophagy may favor tumor development, indicating that inhibition of autophagy might be required to block tumorigenesis. Thus, autophagy may be involved not only in the prevention of cancer but also in the initiation and/or promotion of cancer (17–21). However, the molecular basis for the regulatory relationship between autophagy and the initial acquisition of human malignancy has not yet been defined.
In the present study, we investigated whether autophagy is involved in oncogenic K-Ras-induced malignant transformation and, if so, how the Ras signaling pathway is involved in the control of autophagy. We provide novel evidence that autophagy is an essential element in the induction of malignant transformation by oncogenic K-Ras and show that reactive oxygen species (ROS)2-mediated c-Jun N-terminal kinase (JNK) activation plays a causal role in autophagy induction through up-regulation of ATG5 and ATG7 (autophagy-related genes 5 and 7).
EXPERIMENTAL PROCEDURES
Chemical Reagents and Antibodies
3-Methyladenine (3-MA), N-acetyl-l-cysteine (NAC), actinomycin D, and DMSO were purchased from Sigma. Bafilomycin A1, cycloheximide, PD98059, SB203580, and SP600125 were purchased from Calbiochem. LysoTracker Green DND-26 was purchased from Molecular Probes, Inc. (Eugene, OR) (L-7526). Polyclonal antibodies to anti-K-Ras, ATG6 (Beclin 1), ATG7, caspase 8, caspase 9, phospho-ERK, and p38 MAPK were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Polyclonal anti-ATG5, LC3, and LAMP-1 were obtained from Abgent (San Diego, CA). Polyclonal antibodies to cleaved caspase 3, poly(ADP-ribose) polymerase, ERK, JNK, phospho-JNK, and phospho-p38 MAPK were obtained from Cell Signaling Technology (Beverly, MA). Monoclonal anti-β-actin was obtained from Sigma.
Cell Culture and Transfection
The normal human breast epithelial cell line MCF10A was obtained from the American Type Culture Collection (Manassas, VA) and maintained in DMEM/F-12 medium supplemented with 5% heat-inactivated horse serum (Invitrogen), 10 μg/ml insulin, 20 ng/ml EGF, 0.1 μg/ml cholera toxin, 0.5 μg/ml hydrocortisone, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified 5% CO2 atmosphere.
MCF10A cells were transfected with green fluorescent protein (GFP)-LC3, pcDNA-ATG5, pcDNA-ATG7, or pcDNA-catalase using Lipofectamine PLUS reagent (Invitrogen) by following the procedure recommended by the manufacturer.
Production of Retroviral Vector Containing the K-Ras Gene
To generate MFG-K-RasV12, PCR fragments produced against pSPORT-K-RasV12 as templates were cloned into MFG retroviral vector derived from murine Moloney leukemia virus. For retrovirus production, a modified 293T cell line was cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum, 2 mmol/liter GlutaMAX (Invitrogen), 50 units/ml penicillin/streptomycin, 1 μg/ml tetracyclin, 2 μg/ml puromycin, and 0.6 mg/ml G418 sulfate (Calbiochem) and transfected with MFG-K-RasV12 using the Lipofectamine 2000 reagent (Invitrogen). 48 h after the transfection, virus supernatant was harvested every day by replenishing with fresh medium for 5 days and passed through a 0.45-μm filter, and the viral supernatant was frozen at −80 °C. The supernatant was used for infection after adding 4 μg/ml Polybrene (Sigma).
Lentiviral Short Hairpin RNA (shRNA) Experiments
MISSION shRNA plasmids (Sigma) encoding small interfering RNAs (siRNAs) targeting ATG5, ATG7, or control were purchased from Sigma. Plasmids (ATG5, TRCN0000150645; ATG7, TRCN0000000281; control, SHC202V) were effective in knocking down ATG5 or ATG7 expression. To standardize lentiviral transduction assays, viral titers were measured in an MCF10A cell line. For growth assays, titers corresponding to multiplicities of infection of 5 and 1 in MCF10A cells were employed. For ATG5 or ATG7 knockdown, cells were plated on day zero at 1 × 105 cells in a 60-mm dish. Cells were infected, and after 24 h, cells were treated with 1 mg/ml puromycin for 3 days to eliminate uninfected cells. Medium was replaced, and after 2 more days, cells were harvested for Western blot analysis (22–24).
Anchorage-independent Growth Assay
Cells (1 × 105) were mixed with medium containing 0.4% agar and were spread on top of a bottom agar layer (0.8% agar in growth medium). Fresh layer of medium containing 0.4% agar was added weekly on top of the previous layer. Colonies greater than 50 mm in diameter were counted and were photographed after 2 weeks.
Quantification of Cell Death
Flow cytometric analysis using PI (2.5 μg/ml) staining detects cell death by means of the dye entering the cells along with changes in the target cell membrane and DNA damage. For the cell death assessment, the cells were plated in a 60-mm dish with a cell density of 1 × 105 cells/dish. At the indicated time points, cells were harvested by trypsinization, washed in phosphate-buffered saline, and then incubated in PI for 5 min at room temperature. Then cells (10,000 cells/sample) were analyzed on a flow cytometric scan flow cytometer, using Cell Quest software.
Cell Growth Curve
Briefly, cells were seeded individually onto plates at 20,000 cells. Cell counts were determined by counting the viable cells in a hemocytometer using the trypan blue dye exclusion assay. Every 24 h for 6 days, three dishes of each group were counted.
Tumor Xenografts on Nude Mice
All animal procedures and care were approved by the Institutional Animal Care and Usage Committee of Hanyang University. Tumors were formed by subcutaneous inoculation of 5 × 106 cells or into athymic BALB/c female nude mice (5 weeks of age; Charles River Laboratories). Tumor size was measured with a caliper (calculated volume = shortest diameter2 × longest diameter/2) at 3-day intervals. Mice were sacrificed on day 27, and tumors were photographed and harvested immediately.
Transmission Electron Microscopy
Cells incubated as described above were pelleted and fixed with 3% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.4, for 120 min, washed three times, and postfixed in 1% osmium tetroxide for 60 min. The cells were dehydrated in an ascending ethanol battery ranging from 20 to 100% and were later placed in 3:1, 2:1, 1:1, 1:2, and 1:3 ratios of propylene oxide or epon-812 resin for 1 h at room temperature, respectively. Ultrathin sections of 70 nm were made and impregnated with 2% uranyl acetate and Reynolds lead citrate. The sections were visualized in a Zeiss EM-900 transmission electron microscope at 50 kV and photographed. The negatives were scanned at 600 × 600 dpi resolution, and the images obtained were analyzed later with a PC-compatible computer using customized software.
Fluorescence Microscopy
To visualize acidic vacuole in cells overexpressing K-RasV12, cells were stained with 50 nm LysoTracker Green DND-26 fluorescence dye for 10 min and washed with PBS and then changed to culture media. Cells were imaged by fluorescence microscopy. MCF10A cells were transfected with GFP-LC3 to identify autophagosome and then infected with oncogenic K-RasV12. Punctate GFP-LC3 fluorescence were imaged by fluorescence microscopy (Leica Microsystems, Bannockburn, IL).
Western Blot Analysis
Cell lysates were prepared by extracting proteins with lysis buffer (40 mm Tris-HCl (pH 8.0), 120 mm NaCl, 0.1% Nonidet P-40) supplemented with protease inhibitors. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (Amersham Biosciences). The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline and incubated with primary antibodies overnight at 4 °C. Blots were developed with a peroxidase-conjugated secondary antibody, and proteins were visualized by enhanced chemiluminescence (ECL) procedures (Amersham Biosciences), using the manufacturer's protocol.
RNA Isolation and RT-PCR
Total mRNA was isolated using the RNeasy kit (Qiagen, Valencia, CA). RT-PCRs were performed with SuperScript III (Invitrogen) according to the manufacturer's instructions. The expression of autophagy genes was determined by RT-PCR using the following primers: ATG5, 5′-AGCAACTCTGGATGGGATTG-3′ and 5′-CACTGCAGAGGTGTTTCCAA′; ATG6 (Beclin-1), 5′-GGCCAATAAGATGGGTCTGA-3′ and 5′-GCTTTTGTCCACTGCTCCTC-3′; ATG7, 5′-ACCCAGAAGAAGCTGAACGA-3′ and 5′-AGACAGAGGGCAGGATAGCA-3′; GAPDH, 5′-AAGGTCGGAGTCAACGGATT-3′ and 5′-CCATGGGTGGAATCATATTGG-3′.
Measurement of ROS Generation
Briefly, cells were incubated in 10 μm 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA) (Molecular Probes, Inc.) at 37 °C for 15 min and washed with PBS three times. DCFH-DA were analyzed by a flow cytometer with BD CellQuest Pro software (BD Biosciences).
Statistical Analysis
All experimental data are reported as means ± S.E. (error bars). Statistical analysis was performed by non-parametric Student's t test.
RESULTS
Oncogenic K-Ras Induces Cellular Transformation and Autophagy in Human Normal Breast Epithelial Cells
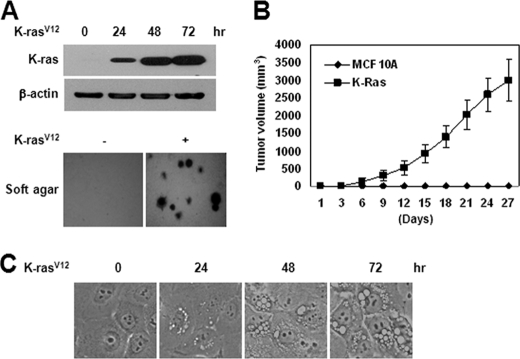
MCF10A, a spontaneously immortalized, normal human breast epithelial cell line, infected with the constitutively active, oncogenic K-Ras mutant G12V (K-RasV12), exhibited anchorage-independent growth in soft agar, forming foci in a monolayer (Fig. 1A), and induced tumor formation in nude mice (Fig. 1B). Transient overexpression of K-RasV12 also induced massive vacuole formation (Fig. 1C), stimulating the time-dependent accumulation of vacuolated cells. At 72 h after infection, at least one vacuole was present in more than 70% of K-RasV12-overexpressed cells.
FIGURE 1.
Oncogenic K-Ras induces cellular transformation in human normal breast epithelial cells. MCF10A cells were infected with retroviral MFG-K-RasV12 (1 × 106 cfu/ml). A (top), after 24, 48, and 72 h, cell lysates were subjected to immunoblot analysis with anti-K-Ras antibody. β-Actin was used as the loading control. The data represent a typical experiment conducted three times with similar results. Bottom, cells (1 × 105) were allowed to grow on soft agar, and colonies were monitored after 2 weeks. Results from three independent experiments are presented as means ± S.E. B, cells (5 × 106 cells) were injected subcutaneously in the right flank of athymic nude mice. Tumor volumes were measured at 3-day intervals as described under “Experimental Procedures.” C, after 24, 48, and 72 h, cells were photographed under a light microscope (magnification, ×400).
Oncogenic K-Ras Induces Autophagic Vacuole Formation in Human Breast Epithelial Cells
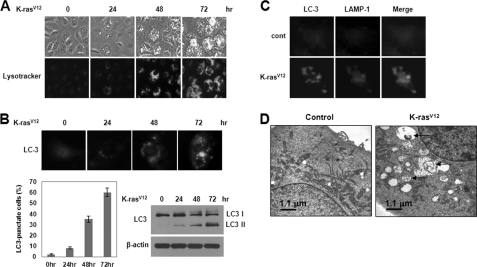
To determine whether vacuolation in cells overexpressing K-RasV12 was associated with autophagy, we first stained cells with the lysosomal marker LysoTracker Green. Staining of cells with LysoTracker followed by analysis by fluorescence microscopy revealed that oncogenic K-Ras induced extensive formation of LysoTracker-positive acidic vacuoles in normal human breast epithelial cell at 48 h after infection (Fig. 2A). Moreover, in cells transfected with a GFP-tagged version of the autophagosome marker, LC3, oncogenic K-Ras promoted an increase in the punctuate distribution of GFP-LC3 (Fig. 2B). Oncogenic K-Ras also induced an increase in LC3-II, a processed form of LC3 (Fig. 2B), indicating that oncogenic K-Ras-induced the accumulation of autophagic vesicles. To further detect lysosomal localization of LC3 protein, we examined co-localization of LC3 and the lysosome marker LAMP-1. As shown in Fig. 2C, infection of cells with oncogenic K-Ras induced co-localization of LC3 and LAMP-1, indicating lysosomal localization of LC3 protein. Electron microscopy of MCF-10A cells overexpressing K-RasV12 also clearly revealed the appearance of large membranous vacuoles in the cytoplasm (Fig. 2D). The formation of these membranous vacuoles increased progressively with increasing duration of exposure to oncogenic K-Ras. Some of the vacuoles resembled autophagosomes and contained remnants of degraded organelles, including mitochondria. In addition, oncogenic K-Ras promoted an increase in the punctuate distribution of GFP-LC3 (supplemental Fig. S1A) and LC3-II, a processed form of LC3 (supplemental Fig. S1B) and induced malignant cell transformation (supplemental Fig. S1C) in normal Rat2 and NIH3T3 fibroblasts.
FIGURE 2.
Oncogenic K-Ras induces autophagic vacuole formation in human normal breast epithelial cells. A, MCF10A cells were infected with MFG-control or MFG-K-RasV12. At the indicated times, cells were stained with LysoTracker Green and imaged by fluorescence microscopy. The percentage of vacuolated cells was calculated under fluorescence microscopy. Results from three independent experiments are presented as means ± S.E. (error bars). B, cells were transfected with GFP-LC3 to identify autophagosome and then infected with MFG-control or MFG-K-RasV12. Top, at the indicated times, punctate GFP-LC3 fluorescence was imaged by fluorescence microscopy. Bottom left, the percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP-positive cells. Results from three independent experiments are presented as means ± S.E. Bottom right, cell lysates were subjected to immunoblot analysis with anti-LC3 antibody. β-Actin was used as a loading control. C, at 72 h after infection, cells were stained with GFP-LC3 (green) and LAMP-1 (red) to identify the autophagosome. D, representative transmission electron micrographs depicting ultrastructures of MFG-control or MFG-K-RasV12 cells at 72 h. The arrows depict autophagosomes in cells containing recognizable cellular materials.
Autophagy Is Required for K-RasV12-induced Cell Transformation in Human Breast Epithelial Cells
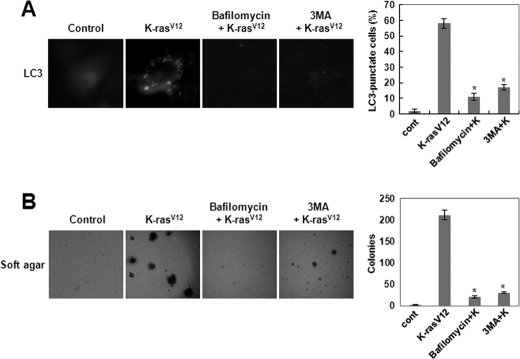
To demonstrate the involvement of the autophagic process in K-RasV12-induced cellular transformation, we pretreated cells that overexpressed oncogenic K-Ras with bafilomycin A1, a selective vacuolar H+-ATPase inhibitor, or with 3-methyladenine, an inhibitor of class III phosphatidylinositol 3-kinases (PI3Ks), both of which are known to be involved in sequestration (i.e. the first step of autophagy). Pretreatment with bafilomycin A1 or 3-MA completely blocked both K-RasV12-induced anchorage-independent cell growth on soft agar (Fig. 3B) and formation of autophagy (Fig. 3A) in the MCF-10A normal human breast epithelial cell line. However, treatment of MCF-10A cells with either inhibitor did not cause significant changes in cell viability (supplemental Fig. S2A) and cell growth (supplemental Fig. S2B). In addition, bafilomycin A1 or 3-MA inhibited K-RasV12-induced autophagy (supplemental Fig. S3A) and cell transformation in Rat2 and NIH3T3 fibroblasts (supplemental Fig. S3B). These results indicate that autophagic vacuole formation may be critical for the induction of cellular transformation by oncogenic K-Ras.
FIGURE 3.
Autophagy is required for K-RasV12-induced cell transformation. A, MCF10A cells were transfected with GFP-LC3 and treated with bafilomycin A1 (1 nm) or 3-MA (10 mm). Then cells were infected with MFG-control or MFG-K-RasV12. After 72 h, punctate GFP-LC3 fluorescence was imaged by fluorescence microscopy. The percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP-positive cells. Results from three independent experiments are presented as means ± S.E. (error bars). *, p < 0.01, significantly different from control. B, cells were treated with DMSO, bafilomycin A1 (1 nm), or 3-MA (10 mm) in the presence of MFG-control or MFG-K-RasV12. Cells (1 × 105) were grown on soft agar, and colonies were monitored after 2 weeks. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control.
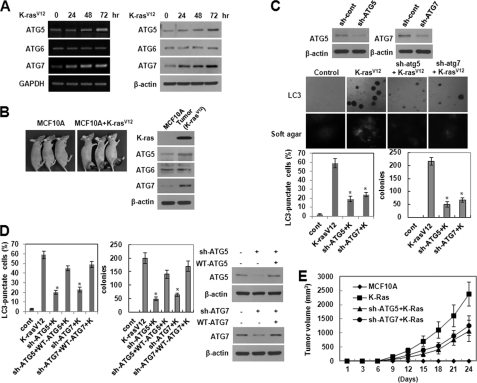
Up-regulation of ATG5 and ATG7 Is Involved in K-RasV12-induced Autophagic Vacuole Formation and Malignant Cell Transformation
We further found that the transcriptional inhibitor actinomycin D and the protein synthesis inhibitor cycloheximide significantly attenuated K-RasV12-induced autophagic vacuole formation (supplemental Fig. S4A), indicating a requirement for transcriptional induction and de novo protein synthesis. Autophagosome formation is mediated by a set of evolutionarily conserved ATG proteins, and studying the expression patterns of ATG genes under specific conditions has provided key information about the autophagic process (25–28). Using RT-PCR and Western blot analyses to examine changes in the level of ATG mRNA and protein expression, respectively, we found that ATG5 and ATG7 were induced at both the transcriptional and translational level in MCF10A cells overexpressing K-RasV12 (Fig. 4A). In contrast, there were no changes in the levels of ATG6. Up-regulation of ATG5 and ATG7 was also found in tumors from nude mice (Fig. 4B). Moreover, shRNA-mediated knockdown of ATG5 or ATG7 effectively attenuated K-RasV12-induced anchorage-independent growth in soft agar as well as autophagic vacuole formation (Fig. 4C). To confirm the direct association between the malignant cell transformation and autophagic vacuole formation induced by oncogenic K-Ras, WT ATG5 or WT ATG7 was reintroduced into ATG5 or ATG7 knockdown cells, respectively, using a pcDNA vector carrying the ATG5 or ATG7 gene. As shown in Fig. 4D, the malignant cell transformation and autophagic vacuole formation were restored to a level similar to that of cells overexpressing K-RasV12. Moreover, shRNA-mediated knockdown of ATG5 or ATG7 effectively attenuated K-RasV12-induced tumor formation in nude mice (Fig. 4E).
FIGURE 4.
Up-regulation of ATG5 and ATG7 is involved in K-RasV12-induced autophagic vacuole formation and malignant cell transformation. A, left, cellular mRNA was isolated 24, 48, and 72 h after infection with MFG or MFG-K-RasV12. RNA expression was examined by RT-PCR toward ATG5, ATG6 (Beclin-1), or ATG7. GAPDH served as an internal standard. The data represent a typical experiment conducted three times with similar results. Right, total cell lysates were isolated 24, 48, and 72 h after infection of MFG-control or MFG-K-RasV12 and were subjected to immunoblot analysis with anti-ATG5, -ATG6, or -ATG7 antibodies. β-Actin was used as a loading control. The data represent a typical experiment conducted three times with similar results. B, cells (5 × 106 cells) were injected subcutaneously in the right flank of athymic nude mice. Tumors in subcutaneous mouse xenografts were subjected to immunoblot analysis with the indicated antibodies. β-Actin was used as a loading control. C, MCF10A cells were transfected with shRNA of ATG5, ATG7, or control and then infected with MFG-K-RasV12. After 72 h, punctate GFP-LC3 fluorescence was imaged by fluorescence microscopy. Cells (1 × 105) were allowed to grow on soft agar, and colonies were monitored after 2 weeks. Bottom left, percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP-positive cells. Results from three independent experiments are presented as means ± S.E. (error bars). *, p < 0.01, significantly different from control. Bottom right, colonies greater than 50 mm in diameter were counted. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control. D, as a rescue experiment, WT ATG5 or WT ATG7 was reintroduced into ATG5 or ATG7 knockdown cells, respectively, using a pcDNA vector carrying the ATG5 or ATG7 gene. Total cell lysates were subjected to immunoblot analysis with anti-ATG5 or -ATG7 antibodies. β-Actin was used as a loading control. Bottom left, After 72 h, the percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP-positive cells. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control. Bottom right, cells (1 × 105) were allowed to grow on soft agar, and colonies were monitored after 2 weeks. Colonies greater than 50 mm in diameter were counted. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control. E, MCF10A cells were transfected with shRNA of ATG5 or ATG7 and then infected with MFG-K-RasV12. Cells (5 × 106 cells) were injected subcutaneous in the right flank of athymic nude mice. Tumor volumes were measured at 3-day intervals as described under “Experimental Procedures.”
In addition, treatment with actinomycin D or cycloheximide significantly inhibited K-RasV12-induced ATG5 and ATG7 expression (supplemental Fig. S4B). These data strongly indicate that up-regulation of ATG5 and ATG7 is involved in both the malignant cell transformation and autophagic vacuole formation induced by oncogenic K-Ras.
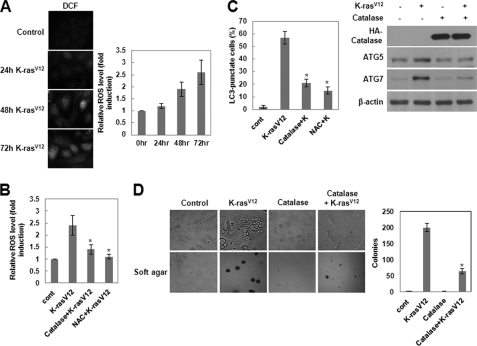
ROS Are Critical Regulators of K-RasV12-induced Autophagy and Malignant Cell Transformation
It has been shown that Ras plays a role in regulating the redox state of the cell and that constitutive production of ROS correlates with Ras-induced cell transformation (29, 30). To investigate whether MCF10A cells overexpressing K-RasV12 induced ROS production, we measured intracellular ROS levels using the fluorescent probe DCFH-DA. As shown in Fig. 5A, cells overexpressing oncogenic K-Ras exhibited a marked increase in fluorescence over time relative to control cells, indicating production of intracellular ROS. To determine whether this increase in ROS is essential for autophagy, we tested the effects of catalase, which specifically decomposes H2O2, and NAC, a chemical antioxidant, on the formation of autophagosomes using LC3 as a marker. Cells treated with these antioxidants did not generate ROS (Fig. 5B) and failed to form autophagosomes (Fig. 5C). Moreover, ectopic expression of catalase clearly attenuated K-RasV12-induced anchorage-independent cell growth on soft agar (Fig. 5D) and expression of ATG5 and ATG7 (Fig. 5C). These results indicate that ROS are critical regulators of K-RasV12-induced development of autophagy and malignant cell transformation.
FIGURE 5.
The ROS is a critical regulator of K-RasV12-induced autophagy and malignant cell transformation. A, MCF10A cells were infected with MFG-control or MFG-K-RasV12. At the indicated times, cells were loaded with DCFH-DA for 15 min. The DCF fluorescence was visualized using fluorescence microscopy, and the amount of retained DCF was measured using flow cytometry as described under “Experimental Procedures.” Results from three independent experiments are presented as means ± S.E. (error bars). B, MCF10A cells were pretreated with catalase or NAC and then infected with MFG-K-RasV12. After 72 h, cells were loaded with DCFH-DA for 15 min. The amount of retained DCF was measured using flow cytometry as described under “Experimental Procedures.” Results from three independent experiments are presented as means ± S.E. *, p < 0.05, significantly different from control. C, MCF10A cells were pretreated with catalase or NAC and then infected with MFG-K-RasV12. Left, after 72 h, punctate GFP-LC3 fluorescence was imaged by fluorescence microscopy. The percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP-positive cells. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control. Right, total cell lysates were subjected to immunoblot analysis with the indicated antibodies. β-Actin was used as a loading control. D, MCF10A cells were transfected with catalase and then infected with MFG-K-RasV12. Cells (1 × 105) were allowed to grow on soft agar, and colonies were monitored after 2 weeks. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control.
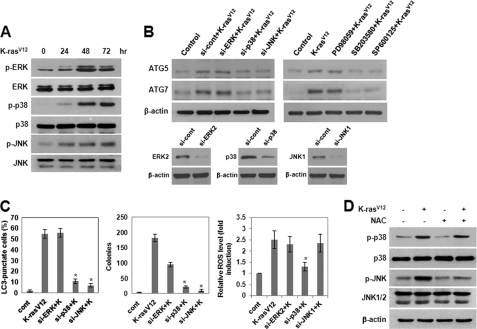
Activation of JNK Plays a Role in K-RasV12-induced Autophagy and Malignant Cell Transformation
Members of the MAPK family of proteins can be phosphorylated and activated in response to various extracellular signals and are the best characterized effectors of Ras. In some cells, certain MAPK family members have been shown to possess ROS-sensitive kinase activity. To examine the potential involvement of MAPKs in K-RasV12-induced autophagy, we analyzed the activation status of ERK, JNK, and p38 MAPK by immunoblot analysis using antibodies specific for the phosphorylated forms of these kinases. As shown in Fig. 6A, the phosphorylated forms of ERK, p38 MAPK, and JNK were up-regulated in cells overexpressing K-RasV12, indicating that these kinases are activated in response to oncogenic K-Ras. To further determine whether activation of MAPK might be required for K-RasV12-induced ROS generation and autophagy, we inhibited MAPK with specific pharmacological inhibitors or siRNAs. As shown in Fig. 6B, pretreatment with a JNK inhibitor or siRNA-mediated JNK knockdown suppressed K-RasV12-induced ATG5 and ATG7 expression. siRNA targeting of JNK also inhibited autophagy and colony formation in soft agar (Fig. 6C) but did not block the associated generation of ROS (Fig. 6C). Moreover, inhibition of ROS by pretreatment with NAC, a general antioxidant, suppressed K-RasV12-induced JNK activation (Fig. 6D) but did not affect ERK or p38 MAPK activation. Interestingly, inhibition of p38 MAPK with a specific chemical inhibitor or p38 MAPK-siRNA markedly suppressed the generation of ROS in cells overexpressing oncogenic K-Ras (Fig. 6C). In addition, inhibition of p38 MAPK clearly suppressed K-RasV12-induced autophagic vacuole formation (Fig. 6C) and anchorage-independent cell growth (Fig. 6C) as well as JNK activation (data not shown). Taken together, these results indicate that p38 MAPK signaling is involved in K-RasV12-induced ROS production and that JNK lies downstream of p38 MAPK-dependent ROS generation and plays a role in ATG5 and ATG7 up-regulation, autophagy, and malignant cell transformation.
FIGURE 6.
Activation of JNK plays a role in K-RasV12-induced autophagy and malignant cell transformation. A, MCF10A cells were infected with MFG-control or MFG-K-RasV12. At the indicated times, total cell lysates were subjected to immunoblot analysis with the indicated antibodies. β-actin was used as a loading control. B, cells were transfected with siRNA of ERK2, p38 MAPK, or JNK1 or were pretreated with PD98059 (25 μmol/liter), SB203580 (10 μmol/liter), or SP600125 (10 μmol/liter) and then infected with MFG-control or MFG-K-RasV12. After 72 h, total cell lysates were subjected to immunoblot analysis with anti-ATG5 or -ATG7 antibodies. β-Actin was used as a loading control. C, MCF10A cells were transfected with siRNA of ERK2, p38 MAPK, or JNK1 and then infected with MFG-K-RasV12. Left, after 72 h, punctate GFP-LC3 fluorescence was imaged by fluorescence microscopy. The percentage of autophagic cells with punctate GFP-LC3 fluorescence was calculated relative to all GFP positive cells. Results from three independent experiments are presented as means ± S.E. *, p < 0.01, significantly different from control. Middle, cells (1 × 105) were allowed to grow on soft agar, and colonies were monitored after 2 weeks. Results from three independent experiments are presented as means ± S.E. (error bars). *, p < 0.01, significantly different from control. Right, after 72 h, cells were loaded with DCFH-DA for 15 min. The amount of retained DCF was measured using flow cytometry as described under “Experimental Procedures.” Results from three independent experiments are presented as means ± S.E. D, MCF10A cells were pretreated with NAC and then infected with MFG-control or MFG-K-RasV12. After 72 h, total cell lysates were subjected to immunoblot analysis with the indicated antibodies. β-Actin was used as a loading control.
DISCUSSION
Recent evidence has indicated that autophagy is associated with both suppression and progression of cancer. However, what has not been clear is whether autophagy is involved in the acquisition of human malignancies in response to activated oncogenes. Here, we show that autophagy is associated with the malignant transformation of mammalian cells induced by oncogenic K-Ras and that cross-talk between ROS and MAPK signaling is required for ATG expression and autophagy induction in cells overexpressing oncogenic K-Ras.
Autophagy has been shown to play important roles in sustaining cellular metabolism and nutrient homeostasis in both normal and tumor cells (7–9). Several recent studies have focused on the importance of autophagy in cancer, but it has remained unclear whether autophagy suppresses tumorigenesis or provides cancer cells with a mechanism for escaping unfavorable conditions. It has been shown that as a tumor grows, autophagy may provide the means for cancer cells to survive nutrient-limiting and low oxygen conditions, especially in internal, poorly vascularized regions of a tumor (16), suggesting that autophagy may play a role in the promotion of cancer. In this study, we further investigated whether autophagy is involved in malignant cell transformation. We provided novel evidence that autophagy is critically involved in K-RasV12-induced malignant cell transformation. Autophagy was induced in cells overexpressing oncogenic K-Ras, and inhibition of autophagy with chemical inhibitors completely blocked K-RasV12-induced anchorage-independent cell growth on soft agar and development of autophagic vacuoles. The chemical inhibitors did not cause significant changes in cell viability and cell growth. These results indicate roles for autophagy in malignant cell transformation as well as cancer progression.
The formation of autophagosomes is a complex process regulated by multiple molecules and mediated by a set of evolutionarily conserved proteins of the ATG family (25–28). An example of this is a conjugation system involving ATG3 and ATG7 that is responsible for the addition of a phospholipid moiety to LC3-I that triggers LC3-I membrane localization. In this study, we found that ATG5 and ATG7 mRNA and protein levels were increased in cells overexpressing oncogenic K-Ras. Importantly, inhibition of autophagy with siRNAs targeting ATG5 or ATG7 attenuated K-RasV12-mediated cell growth on soft agar and tumor formation in nude mice. siRNA against ATG5 or ATG7 did not significantly alter the cell growth and survival (data not shown). Overexpression of ATG alone did not induce anchorage-independent cell growth (i.e. cell transformation), indicating the collaborative involvement of other effector pathways; however, down-regulation of ATG expression clearly reduced tumorigenic growth. These results indicate that autophagy alone is not sufficient to induce malignant transformation but is absolutely necessary for the tumorigenic response to oncogenic K-Ras. However, we do not know how autophagy is involved in the regulation of cellular signaling associated with malignant transformation induced by oncogenic K-Ras. The precise molecular mechanisms governing the cross-talk between autophagy and cell transformation remain to be elucidated.
Autophagy is a unique intracellular trafficking pathway activated in response to extracellular signals (31–34). Although many of the proteins involved in this process have been identified, the signaling pathway leading to activation of autophagy is not fully resolved. In this study, we demonstrated that ROS are involved as signaling molecules in K-RasV12-induced autophagy. Not only did overexpression of K-RasV12 in normal human breast epithelial cells induce a marked increase in intracellular ROS levels, but inhibition of ROS with antioxidants also clearly attenuated induction of autophagy and formation of anchorage-independent colonies on soft agar, suggesting that ROS are critical regulators of K-RasV12-induced autophagy and malignant cell transformation. These findings are in agreement with several recent reports implicating ROS in autophagosome formation and autophagic cell death in response to various stimuli (35–45). However, in the current study, we did not observe any changes in cell viability in K-RasV12-overexpressing MCF-10A human breast epithelial cells (supplemental Fig. S5).
Numerous studies using various experimental systems have shown that MAPKs, particularly JNK and p38 MAPK, are strongly activated by ROS and play important roles in various ROS-related cellular events (46–49). In the present study, we found that the increase in intracellular ROS induced by oncogenic K-Ras was involved in the activation of JNK and that inhibition of JNK attenuated ATG5 and ATG7 expression, autophagy, and formation of colonies in soft agar. Interestingly, we further found that K-RasV12-induced increases in intracellular ROS were attenuated by p38 MAPK inhibition, which also suppressed autophagy and subsequent cellular transformation. Collectively, these results suggest that p38 MAPK signaling is involved in ROS production in response to oncogenic K-Ras and suggest that JNK acts downstream of ROS to play a functional role in K-RasV12-induced autophagy.
In normal fibroblasts, Ras has shown to be involved in the negative control of autophagy through class I PI3K activation (50). In this study, we also found dramatic activation of PI3K in cells overexpressing oncogenic K-Ras (data not shown). However, inhibition of PI3K did not significantly affect ATG expression and autophagy induction. A possible explanation for this observation is that oncogenic K-Ras activates both positive (ROS-JNK) and negative (class I PI3K) signaling pathways that control autophagy, and that oncogenic Ras influences regulation of autophagy through either of these pathways dependent on the cell types. A recent study also provides evidence that oncogenic Ras-induced down-regulation of autophagy mediator Beclin-1 and is required for malignant transformation of intestinal epithelial cells (51). In this study, however, in breast epithelial cells, the expression level of Beclin-1 was not changed by overexpression of oncogenic K-Ras. Moreover, in contrast to the previous finding, we found that siRNA targeting of Beclin-1 effectively attenuated oncogenic K-Ras-induced cell transformation as well as autophagy (data not shown). The exact reason for the different observation between two studies is currently unknown. Cell type-specific activation of signaling pathways could explain the variability of the effect of oncogenic Ras on autophagy.
In summary, our results provide novel evidence that autophagy is critically involved in malignant cell transformation by oncogenic K-Ras and show that ROS-mediated JNK activation plays a causal role in autophagy induction through up-regulation of ATG5 and ATG7. The molecular mechanisms that link oncogenic K-Ras to induction of autophagy and subsequent malignant transformation determined in this study may provide new insights into the relationship between autophagy and oncogenesis.
This work was supported by the Korea Science and Engineering Foundation (KOSEF) and Ministry of Education, Science and Technology (MEST), Korean government, through its National Nuclear Technology Program (2009-0081812) and the Program of Basic Atomic Energy Research Institute (BAERI).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ROS
- reactive oxygen species
- 3-MA
- 3-methyladenine
- NAC
- N-acetyl-l-cysteine
- DCFH-DA
- 2,7-dichlorodihydrofluorescein diacetate
- ATG
- autophagy-specific gene.
REFERENCES
- 1. Karnoub A. E., Weinberg R. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Downward J. (2003) Nat. Rev. Cancer 3, 11–22 [DOI] [PubMed] [Google Scholar]
- 3. Castagnola P., Giaretti W. (2005) Biochim. Biophys. Acta 1756, 115–125 [DOI] [PubMed] [Google Scholar]
- 4. Malumbres M., Barbacid M. (2003) Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 5. Konstantinopoulos P. A., Karamouzis M. V., Papavassiliou A. G. (2007) Nat. Rev. Drug Discov. 6, 541–555 [DOI] [PubMed] [Google Scholar]
- 6. Schubbert S., Shannon K., Bollag G. (2007) Nat. Rev. Cancer 7, 295–308 [DOI] [PubMed] [Google Scholar]
- 7. Levine B., Klionsky D. J. (2004) Dev. Cell 6, 463–477 [DOI] [PubMed] [Google Scholar]
- 8. Yorimitsu T., Klionsky D. J. (2005) Cell Death Differ. 12, 1542–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dunn W. A., Jr. (1994) Trends Cell Biol. 4, 139–143 [DOI] [PubMed] [Google Scholar]
- 10. Lum J. J., DeBerardinis R. J., Thompson C. B. (2005) Nat. Rev. Mol. Cell Biol. 6, 439–448 [DOI] [PubMed] [Google Scholar]
- 11. Mathew R., White E. (2007) Autophagy 3, 502–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang X. H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. (1999) Nature 402, 672–676 [DOI] [PubMed] [Google Scholar]
- 13. Gozuacik D., Kimchi A. (2004) Oncogene 23, 2891–2906 [DOI] [PubMed] [Google Scholar]
- 14. Degenhardt K., Mathew R., Beaudoin B., Bray K., Anderson D., Chen G., Mukherjee C., Shi Y., Gélinas C., Fan Y., Nelson D. A., Jin S., White E. (2006) Cancer Cell 10, 51–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen N., Karantza-Wadsworth V. (2009) Biochim. Biophys. Acta 1793, 1516–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Todde V., Veenhuis M., van der Klei I. J. (2009) Biochim. Biophys. Acta. 1792, 3–13 [DOI] [PubMed] [Google Scholar]
- 17. Kondo Y., Kanzawa T., Sawaya R., Kondo S. (2005) Nat. Rev. Cancer 5, 726–734 [DOI] [PubMed] [Google Scholar]
- 18. Shintani T., Klionsky D. J. (2004) Science 306, 990–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ng G., Huang J. (2005) Mol. Carcinog. 43, 183–187 [DOI] [PubMed] [Google Scholar]
- 20. Cuervo A. M. (2004) Trends Cell Biol. 14, 70–77 [DOI] [PubMed] [Google Scholar]
- 21. Lum J. J., Bauer D. E., Kong M., Harris M. H., Li C., Lindsten T., Thompson C. B. (2005) Cell 120, 237–248 [DOI] [PubMed] [Google Scholar]
- 22. Singh A., Greninger P., Rhodes D., Koopman L., Violette S., Bardeesy N., Settleman J. (2009) Cancer Cell 15, 489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 24. Naldini L., Blömer U., Gallay P., Ory D., Mulligan R., Gage F. H., Verma I. M., Trono D. (1996) Science 272, 263–267 [DOI] [PubMed] [Google Scholar]
- 25. Klionsky D. J., Cregg J. M., Dunn W. A., Jr., Emr S. D., Sakai Y., Sandoval I. V., Sibirny A., Subramani S., Thumm M., Veenhuis M., Ohsumi Y. (2003) Dev. Cell 5, 539–545 [DOI] [PubMed] [Google Scholar]
- 26. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. (2004) Mol. Biol. Cell 15, 1101–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reggiori F., Klionsky D. J. (2005) Curr. Opin. Cell Biol. 17, 415–422 [DOI] [PubMed] [Google Scholar]
- 29. Kopnin P. B., Agapova L. S., Kopnin B. P., Chumakov P. M. (2007) Cancer Res. 67, 4671–4678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alexandrova A. Y., Kopnin P. B., Vasiliev J. M., Kopnin B. P. (2006) Exp. Cell Res. 312, 2066–2073 [DOI] [PubMed] [Google Scholar]
- 31. Botti J., Djavaheri-Mergny M., Pilatte Y., Codogno P. (2006) Autophagy 2, 67–73 [DOI] [PubMed] [Google Scholar]
- 32. Stromhaug P. E., Klionsky D. J. (2001) Traffic 2, 524–531 [DOI] [PubMed] [Google Scholar]
- 33. Yang Y. P., Liang Z. Q., Gu Z. L., Qin Z. H. (2005) Acta Pharmacol. Sin. 26, 1421–1434 [DOI] [PubMed] [Google Scholar]
- 34. Wang C. W., Klionsky D. J. (2003) Mol. Med. 9, 65–76 [PMC free article] [PubMed] [Google Scholar]
- 35. Delgado M. A., Deretic V. (2009) Cell Death Differ. 16, 976–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Codogno P., Meijer A. J. (2005) Cell Death Differ. 12, 1509–1518 [DOI] [PubMed] [Google Scholar]
- 37. Levine B., Yuan J. (2005) J. Clin. Invest. 115, 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsujimoto Y., Shimizu S. (2005) Cell Death Differ. 12, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 39. Baehrecke E. H. (2005) Nat. Rev. Mol. Cell Biol. 6, 505–510 [DOI] [PubMed] [Google Scholar]
- 40. Maiuri M. C., Zalckvar E., Kimchi A., Kroemer G. (2007) Nat. Rev. Mol. Cell Biol. 8, 741–752 [DOI] [PubMed] [Google Scholar]
- 41. Yu L., Alva A., Su H., Dutt P., Freundt E., Welsh S., Baehrecke E. H., Lenardo M. J. (2004) Science 304, 1500–1502 [DOI] [PubMed] [Google Scholar]
- 42. Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E. H., Lenardo M. (2006) Proc. Natl. Acad. Sci. 103, 4952–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scherz-Shouval R., Elazar Z. (2007) Trends Cell Biol. 17, 422–427 [DOI] [PubMed] [Google Scholar]
- 45. Scott R. C., Juhász G., Neufeld T. P. (2007) Curr. Biol. 17, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sahu R. P., Zhang R., Batra S., Shi Y., Srivastava S. K. (2009) Carcinogenesis 30, 1744–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang C. S., Shin D. M., Lee H. M., Son J. W., Lee S. J., Akira S., Gougerot-Pocidalo M. A., El-Benna J., Ichijo H., Jo E. K. (2008) Cell Microbiol. 10, 741–754 [DOI] [PubMed] [Google Scholar]
- 48. Noguchi T., Ishii K., Fukutomi H., Naguro I., Matsuzawa A., Takeda K., Ichijo H. (2008) J. Biol. Chem. 283, 7657–7665 [DOI] [PubMed] [Google Scholar]
- 49. Rockwell P., Martinez J., Papa L., Gomes E. (2004) Cell. Signal. 16, 343–353 [DOI] [PubMed] [Google Scholar]
- 50. Furuta S., Hidaka E., Ogata A., Yokota S., Kamata T. (2004) Oncogene 23, 3898–3904 [DOI] [PubMed] [Google Scholar]
- 51. Yoo B. H., Wu X., Li Y., Haniff M., Sasazuki T., Shirasawa S., Eskelinen E. L., Rosen K. V. (2010) J. Biol. Chem. 285, 5438–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]