Abstract
Although stress-activated protein kinases/c-Jun N-terminal kinases (SAPK/JNK) are rapidly activated by genotoxins, the role of DNA damage in this response is not well defined. Here we show that the SEK1/MKK4-mediated dual phosphorylation of SAPK/JNK (Thr-183/Tyr-185) correlates with the level of cisplatin-DNA adducts at late times (16–24 h) after drug treatment in both human and mouse cells. Transfection of platinated plasmid DNA also caused SAPK/JNK activation. A defect in transcription-coupled nucleotide excision repair resting on a mutation in Cockayne syndrome group B protein promoted the late SAPK/JNK activation following cisplatin exposure. Signaling to SAPK/JNK was accompanied by activation of Ataxia telangiectasia mutated- and Rad3-related kinase, replication protein A, and checkpoint kinases as well as by the formation of DNA double strand breaks (DSBs). Ionizing radiation-induced DSBs did not provoke SAPK/JNK activation, and inhibition of transcription also failed to provoke this response. Late activation of SAPK/JNK stimulated by cisplatin-induced DNA lesions was reduced in the absence of specific DNA repair proteins, such as xeroderma pigmentosum protein C, pointing to an essential function of individual repair factors in DNA damage signaling to SAPK/JNK. Collectively, the data indicate that late SAPK/JNK activation is triggered by non-repaired cisplatin adducts in transcribed genes and involves replication-associated events, DSBs, tyrosine kinases, Rho GTPases, and specific repair factors.
Keywords: Anticancer Drug, DNA Damage, DNA Repair, DNA Nucleotide Excision Repair, MAP Kinases (MAPKs), Cisplatin, DNA Damage Response, Stress-activated Protein Kinases
Introduction
Stress-activated protein kinases/c-Jun N-terminal kinases (SAPK/JNK) belong to the mitogen-activated protein kinase family, which is stimulated by different types of stress factors, including genotoxins (1, 2). SAPK/JNK regulates the activity of various transcription factors, including AP-1-like transcription factors (i.e. Jun/Fos and Jun/Activating transcription factor heterodimers) (3) that affect genomic stability and survival after genotoxin exposure (4, 5). The majority of the currently available data indicate that SAPK/JNK-triggered mechanisms promote apoptosis (6, 7), although opposing reports also exist (8). Proapoptotic functions ascribed to SAPK/JNK signaling rest on the expression of FAS ligand, which is regulated in an AP-1-dependent manner (9), and modulation of the activity of members of the Bcl-2 protein family (10). Protective mechanisms of SAPK/JNK-regulated signaling are thought to be due to the stimulation of DNA repair functions (8, 11, 12).
It is still a matter of controversy whether SAPK/JNK are stimulated by receptor-related mechanisms only or whether DNA damage-related mechanisms are involved as well. It is well established that genotoxins, such as UV light or alkylating agents, rapidly activate mitogen-activated protein kinase pathways by stimulation of receptors for growth factors and cytokines (13–15) very likely via mechanisms involving reactive oxygen species (ROS)2 formation and subsequent inhibition of tyrosine phosphatases (16). Membrane-bound small GTPases of the Ras and Rho family transduce this signal to SAPK/JNK (17, 18). Apart from causing damage to membranes, genotoxins severely attack the genomic DNA, thereby triggering carcinogenesis and cell death. Hence, it is tempting to speculate that DNA damage also contributes to the activation of SAPK/JNK. Evidence that DNA damage-dependent functions might impact genotoxin-induced signaling to SAPK/JNK rests on the analysis of cell lines compromised in particular DNA repair functions, such as nucleotide excision repair (NER) or mismatch repair (19–21). Mechanisms related to DNA damage are suggested to be of particular relevance for a delayed and sustained activation of stress kinases (22–24). However, data supporting the view that SAPK/JNK are regulated in a DNA damage-dependent manner are still limited. Hence, in contrast to p38 kinases, SAPK/JNK are not yet fully established as part of the eukaryotic DNA damage response (25).
UV-C light, the monofunctional alkylating agent methyl methanesulfonate (MMS), and the anticancer drug cisplatin are powerful and therefore prototypical activators of SAPK/JNK (26–28). These agents induce different types of DNA adducts. Although the SN2 alkylating agent MMS exclusively forms monoadducts, in particular N7-methylguanine, N3-methylguanine, and N3-methyladenine, UV-C light specifically forms DNA intrastrand cross-links, in particular cyclobutane pyrimidine dimers and (6-4) photoproducts (29). The anticancer drug cisplatin initially generates monoadducts that pass into DNA intra- or interstrand cross-links with a half-time of 6 and 14 h, respectively (30). The majority (>95%) of cisplatin-induced DNA adducts are intrastrand cross-links (31, 32). A major drawback of most of the currently available studies claiming that DNA damage contributes to the activation of SAPK/JNK following genotoxin exposure is the lack of qualitative and quantitative detection of DNA damage. Although the genomic DNA is a most important cellular target for the mutagenic, recombinogenic, and carcinogenic effects of genotoxic compounds, genotoxins can also provoke stress signaling and death by targeting multiple cellular structures other than DNA, such as membrane receptors (15, 33), membrane-associated structures (34), endoplasmic reticulum (35–37), and others (2, 34, 38). Therefore, not each cellular response to genotoxins can be classified as a DNA damage-induced response. Bearing this in mind, the aim of the present study was to ascertain whether DNA damage is involved in the late activation of SAPK/JNK following cisplatin treatment and to characterize the molecular mechanisms involved.
EXPERIMENTAL PROCEDURES
Materials
Cisplatin was provided by the pharmaceutical department of the Medical Center of the University of Mainz (Mainz, Germany). Transplatin and transcription inhibitors α-amanitin and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole as well as wortmannin, MEK1 inhibitor PD98059, and c-Src inhibitor PP2 were purchased from Sigma-Aldrich. Fotemustine originated from Servier Laboratories (Neuilly sur Seine, France). Phosphospecific antibodies detecting activated forms of protein kinases, i.e. SAPK/JNK (Thr-183/Tyr-185), SEK1/MKK4 (Thr-261), checkpoint kinase-1 (Chk-1) (Ser-345), and checkpoint kinase-2 (Chk-2) (Thr-68), were obtained from Cell Signaling Technology (Beverly, MA), and phosphospecific H2AX antibody (Ser-139) was from Upstate (Lake Placid, NY). ERK2, Cockayne syndrome B protein (CSB), replication protein A (RPA), β-actin, C/EBP homologous protein, and mitogen-activated kinase phosphatase 1 (MKP-1) antibodies were from Santa Cruz Biotechnology (Heidelberg, Germany). Antibody detecting the major DNA adduct formed by cisplatin (1,2-GG intrastrand cross-link) was generated and provided by J. Thomale (Essen, Germany) (39). Rac1 and Rho antibody, Bcr-Abl inhibitor type II, JNK inhibitor II (SP600125), ATM inhibitor Ku55933, and Akt inhibitor (catalog number 124005) were from Calbiochem. EGFR inhibitor Iressa was purchased from AstraZeneca (London, UK), dichlorodihydrofluorescein diacetate was from Invitrogen, and validated siRNA for down-regulation of CSB protein was from Qiagen (Hilden, Germany).
Cell Culture and Drug Treatment
Cell lines used in this study were routinely grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum at 37 °C in a humidified atmosphere. Mouse embryonic fibroblasts (MEFs) lacking ATM (ATM−/−) were provided by P. Leder (Boston, MA), CSB−/− MEFs were provided by G. van der Horst (Rotterdam, The Netherlands), and p53−/− MEFs are described elsewhere (40). Human cells lacking functional CSB protein originated from T. Stevnsner (Aarhus, Denmark) (41). Mouse DNA-PKcs-deficient fibroblasts derived from severe combined immunodeficiency mice were kindly provided by K. Bidermann (42). Primary human fibroblasts (HIFB-D) were purchased from Provitro (Berlin, Germany). Primary human fibroblasts defective in xeroderma pigmentosum protein C (XPC) were provided by L. H. Mullenders (Leiden, The Netherlands). The origin of the human XP12RO cell line deficient in xeroderma pigmentosum protein A (XPA) and complemented cell line (XP129) is described elsewhere (43). ATR-defective (ATR-Seckel) fibroblasts were generously provided by M. O'Driscoll (East Sussex, UK). FANCD2-defective human cells and corresponding primary fibroblasts were obtained from D. Schindler (Würzburg, Germany). Hamster ERCC1-defective 43-3B cells and corresponding CHO wild type cells were reported before (44). If not stated otherwise, cisplatin exposure was performed as a chronic treatment, meaning that the medium was not replaced after drug addition.
In Vitro Preparation of Platinated Plasmid DNA and Cell Transfection
For preparation of platinated plasmid DNA, we used a modified method from Hansson and Wood (45). pEGFP-C1 plasmid (Clontech) (100 μg/ml in Tris-EDTA buffer) was incubated overnight at 37 °C in the absence or presence of 100 μm cisplatin. The reaction was stopped by adding 0.5 m NaCl. Plasmid DNA was precipitated with 2 volumes of ethanol at −70 °C in the presence of 2.5 m CH3COONH4, washed in 70% ethanol, dried, and redissolved in Tris-EDTA buffer. The transfection of platinated and non-platinated control plasmid was performed using Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. 24 h after the beginning of transfection, the transfected cells were harvested for analysis of their SAPK/JNK phosphorylation status.
Analysis of Cisplatin-induced DNA Adduct Formation
DNA adduct formation by cisplatin was analyzed by two different methods, namely Southwestern analysis (slot blot) and measurement of DNA platination. For Southwestern analysis, genomic DNA was isolated from subconfluent cells by the use of the DNeasy Blood and Tissue kit (Qiagen). DNA (2 μg) was directly transferred to a positively charged nylon membrane (Hybond Plus) (Amersham Biosciences) by vacuum slot blotting, denatured with 0.3 m NaOH, neutralized with 5× SSC, and fixed by heat treatment (incubation for 2 h at 80 °C). Monoclonal antibody specifically detecting platinum adducts (1,2-GG intrastrand cross-links) (39) was used at a dilution of 1:200. Blocking, hybridization, and detection procedures were as described below. To quantify the level of DNA platination, DNA was isolated as described before and DNA concentration was measured photometrically (E260/E280 ratio). The amount of platinum atoms bound to DNA was quantified by inductively coupled plasma mass spectometry using an Element 2 high resolution ICP-MS instrument (Thermo Finnigan, Bremen, Germany). The platinum content per nucleotide was calculated using the relative molar masses of platinum and nucleotides as described (24).
Analysis of Kinase Activation by Western Blot Analysis
Dual phosphorylation of SAPK/JNK by upstream regulatory kinase SEK1/MKK4, which results in SAPK/JNK activation, was measured by Western blot analysis using phosphospecific SAPK/JNK (Thr-183/Tyr-185) antibody. Activation status of checkpoint kinases was analyzed in the same way using phospho-Chk-1 (Ser-345)- and phospho-Chk-2 (Thr-68)-specific antibodies. Cell extracts were prepared by lysing an identical number of cells in SDS sample buffer as recommended by the manufacturer (Cell Signaling Technology). Proteins were separated by SDS gel electrophoresis (10% gels) and transferred onto nitrocellulose membrane. After blocking the membrane (5% dry milk in TBS, 0.2% Tween 20 for 2 h at room temperature (RT)), the phosphorylation status of kinases was analyzed using the corresponding phosphospecific antibody (1:1000 in 5% BSA in TBS, 0.2% Tween; overnight incubation at 4 °C). After washing (TBS, 0.2% Tween 20), blots were incubated with secondary horseradish peroxidase-coupled antibody (1:5000; 2 h at RT). Phosphorylated proteins were visualized by chemiluminescence using ECLTM detection reagent (Amersham Biosciences/GE Healthcare). As an internal protein loading control, ERK2 or β-actin protein expression was determined by reprobing the membranes with the corresponding antibody. For comparative analysis of SAPK/JNK activation in repair-defective versus wild type cells, protein extracts were usually separated on the same gel or on different gels run in parallel. Also, incubation with primary and secondary antibodies as well as autoradiography was performed in parallel, enabling a direct comparison of the signal intensities of phosphorylated SAPK/JNK between repair-proficient and -deficient cells.
Down-regulation of CSB by siRNA
siRNA was used to specifically knock down the expression of the CSB protein in primary human fibroblasts. Validated CSB-specific siRNA (Qiagen) (5 nm) was transfected into primary human fibroblasts cells using Hyperfect transfection reagent (Qiagen) according to the manufacturer's protocol. Successful down-regulation of CSB protein was confirmed by Western blot analysis.
Analysis of DNA Damage
For detection of DNA double strand breaks (DSBs), the neutral comet assay was used as described (46). 50 nuclei were evaluated per treatment, and means ± S.D. were determined. Alternatively, phosphorylation of histone H2AX (γH2AX) by protein kinases ATM, ATR, or DNA-PKcs, which play a key role in the eukaryotic DNA damage response (47, 48), is also considered a specific marker of DSB formation (49, 50) and other DNA distortions (47, 48). H2AX phosphorylation was analyzed by Western blot analysis and additionally by the detection of γH2AX foci by immunohistochemistry (see below).
Immunohistochemical Analysis of Formation of Foci
Cells were fixed in 4% paraformaldehyde for 15 min, washed with PBS, and incubated in methanol at −20 °C for ≥2 h. Permeabilization and blocking of the cells were achieved by incubation with 5% BSA in PBS, 0.3% Triton X-100 for 2 h at RT. Afterward, cells were incubated with a mouse monoclonal anti-γH2AX antibody or anti-RPA antibody (1:1000) overnight at 4 °C. Cells were washed with PBS and incubated for 2 h at room temperature with Alexa Fluor anti-mouse IgG antibody (1:500) (Invitrogen). Counterstaining of nuclei was performed with DAPI for another 15 min. Formation of γH2AX and RPA foci was visualized by fluorescence microscopy.
Analysis of ROS Formation and Endoplasmic Reticulum Stress
ROS formation and an increase in the expression of C/EBP homologous protein are prototypical markers of endoplasmic reticulum stress (35). The level of ROS was analyzed by FACS using the fluorescent dye dichlorodihydrofluorescein diacetate (Invitrogen). Expression of C/EBP homologous protein was analyzed by Western blot.
Analysis of Rho Activity Status
As the active, i.e. GTP-bound, form of the Ras-homologous GTPase Rac1 is located on the plasma membrane, we investigated its expression level on membrane fractions by Western blot. To this end, cells were disrupted by sonication on ice in buffer (10 mm Tris (pH 7.4), 10 mm NaCl, 3 mm MgCl2, 1 mm PMSF, protease inhibitor mixture). Cell debris were removed by centrifugation (10 min at 400 × g at 4 °C). After protein determination by Bradford assay, an identical amount of total cell protein was subjected to a further centrifugation step (15 min at 10,000 × g at 4 °C). The membrane fraction (i.e. the pellet) obtained was solubilized in SDS buffer, and proteins were processed for Western blot analysis as described above. Additionally, the amount of GTP-bound Rac1 protein was determined by GST-PAK pulldown assay as described (51).
RESULTS
DNA Platination but Not ROS Formation Coincides with Cisplatin-induced SAPK/JNK Phosphorylation
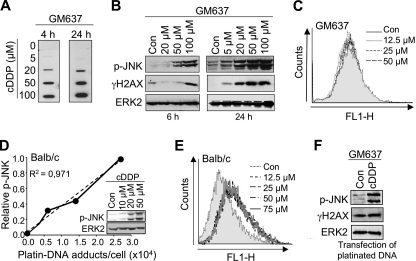
Cisplatin (cDDP) is well known to cause a sustained activation of SAPK/JNK (23, 24, 28, 52). As a first step to substantiate the role of DNA damage in triggering this response, we investigated whether the level of SAPK/JNK phosphorylation coincides with cisplatin-induced DNA adduct formation. To this end, human fibroblasts (GM637 cells) were exposed to increasing doses of cisplatin (continuous treatment), and the amount of 1,2-GG intrastrand cross-links and the dual phosphorylation status of SAPK/JNK were determined at early and late times after drug addition. As shown in Fig. 1A, steady-state cDDP adduct levels were clearly detectable both 4 and 24 h after treatment with 20 μm cDDP and increased with dose. The level of cDDP adducts observed 24 h after drug addition was slightly higher than after 4 h (Fig. 1A). At early times after treatment with 20 μm cDDP, the observed increase in cDDP adduct levels (Fig. 1A, left panel) was not paralleled by an increased SAPK/JNK phosphorylation, and the γH2AX level also was not enhanced (Fig. 1B, left panel). Apparently, cDDP-induced DNA adducts do not cause rapid activation of SAPK/JNK. At later times after exposure, the cDDP-induced increase in SAPK/JNK phosphorylation (Fig. 1B, right panel) was accompanied by higher cDDP adduct and γH2AX levels (Fig. 1, A and B, right panel). This finding supports the view that cDDP-induced DNA damage triggers SAPK/JNK activation at late times. The level of ROS, as measured by FACS analysis, was not enhanced in GM637 cells at late times following cisplatin exposure (Fig. 1C). To substantiate the data in a second experimental model system, MEFs derived from BALB/c mice were treated with increasing doses of cisplatin, and dual phosphorylation of SAPK/JNK and DNA platination were measured 24 h later. Here overall DNA platination was assayed by mass spectometry (i.e. ICP-MS). In line with the data obtained with GM637 cells, we found that the DNA platination level quantitatively correlated with SAPK/JNK phosphorylation (Fig. 1D). In MEFs, cDDP treatment caused ROS production (Fig. 1E). However, unlike the cDDP-stimulated SAPK/JNK phosphorylation, the ROS formation observed after cDDP exposure was not dose-dependent (Fig. 1E). Overall, the data indicate that cDDP-induced DNA adducts, rather than cDDP-induced ROS, can trigger late signaling to SAPK/JNK. This view gains support by the finding that transfection of platinated plasmid DNA also gives rise to SAPK/JNK phosphorylation (Fig. 1F). As expected, H2AX phosphorylation (γH2AX) was not simulated under these experimental conditions (Fig. 1F).
FIGURE 1.
Level of cisplatin-DNA adducts quantitatively correlates with dual phosphorylation status of SAPK/JNK. A, human fibroblast cells (GM637) were exposed to increasing concentrations of cDDP. 4 and 24 h after continuous cDDP treatment, DNA was isolated, and the level of 1,2-GG intrastrand cross-links was determined by Southwestern analysis as described under “Experimental Procedures.” B, GM637 cells were exposed to increasing concentrations of cisplatin. After incubation periods of 6 (left panel) and 16 h (right panel), cells were harvested, and the dual phosphorylation status of SAPK/JNK (p-JNK) was analyzed by Western blot analysis. In addition, Ser-139 phosphorylation of histone H2AX (γH2AX) was determined. As a loading control, protein expression of ERK2 was monitored. Con, untreated cells. C, GM637 cells were left untreated (Con) or were exposed to different concentrations of cisplatin for 16 h. Afterward, fresh medium was added, and ROS levels were determined by FACS as described under “Experimental Procedures.” D, Balb/c mouse fibroblasts were treated with different concentrations of cDDP for 24 h. DNA platination was measured by ICP-MS as described under “Experimental Procedures.” In parallel, the dual phosphorylation status of SAPK/JNK (p-JNK) was analyzed. After densitometric analysis of the autoradiography, the relative JNK phosphorylation level observed with the highest dose of cisplatin was set to 1.0. E, Balb/c mouse fibroblasts were exposed to different concentrations of cisplatin for 16 h. Afterward, fresh medium was added, and the level of ROS was measured by FACS. F, human fibroblast cells (GM637) were transfected with platinated pEGFP plasmid DNA (cDDP) or non-platinated plasmid (Con). 16 h after transfection, cells were harvested, and the phosphorylation status of SAPK/JNK (p-JNK) and H2AX (γH2AX) was analyzed by Western blot analysis.
Lack of CSB Facilitates Late SAPK/JNK Phosphorylation Induced by Cisplatin
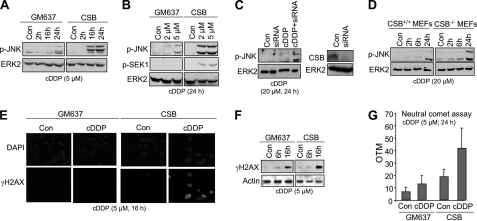
Mutants defective in NER are unable to repair cisplatin-induced adducts in DNA (29, 53). Assuming that cisplatin-induced DNA damage is responsible for late SAPK/JNK activation, NER mutants are expected to differ from wild type cells with respect to the phosphorylation status of SAPK/JNK. As transcription blockage is of importance for cellular stress responses (54–56) and transcription-coupled nucleotide excision repair (TC-NER) is a major determinant of cisplatin sensitivity (57), we investigated cells that are defective in TC-NER due to lack of CSB. Performing time course analyses, we found that human CSB-defective cells displayed a pronounced dual phosphorylation of SAPK/JNK 16 and 24 h after cisplatin administration compared with GM637 wild type cells (Fig. 2A). At an earlier time point (i.e. 6 h) after high dose (i.e. 100 μm) cisplatin treatment, CSB-deficient cells did not differ from wild type cells with respect to SAPK/JNK activation (data not shown). The increase in late SAPK/JNK activation was paralleled by elevated SEK1/MKK4 activity (Fig. 2B). Cisplatin-induced SAPK/JNK phosphorylation was also increased after siRNA-mediated down-regulation of CSB in primary human cells (Fig. 2C). Furthermore, CSB-deficient (CSB−/−) MEFs also exhibited an enhanced late SAPK/JNK phosphorylation after cisplatin treatment (Fig. 2D). Collectively, the data show that in the absence of functional CSB protein cisplatin treatment increases the level of phosphorylated SAPK/JNK at late times after drug addition. Obviously, CSB protects against SAPK/JNK activation following cDDP treatment.
FIGURE 2.
TC-NER-defective CSB cells show increased SAPK/JNK phosphorylation at late times after cisplatin treatment. A, SV40 immortalized human cells defective in TC-NER due to lack of CSB protein (CBS) and GM637 wild type cells were treated with 5 μm cDDP for 2–24 h before dual phosphorylation of SAPK/JNK (p-JNK) was analyzed. ERK2 protein expression was determined as an internal loading control. Con, untreated cells. B, 24 h after exposure of CSB-deficient (CSB) or -proficient (GM637) cells to low dose cisplatin, the phosphorylation status of SAPK/JNK (p-JNK) and SEK1/MKK4 (p-SEK1) were analyzed. C, primary human fibroblasts (HFIB-D) were transfected with CSB-specific siRNA (5 nm). 24 h later, residual CSB protein expression was analyzed by Western blot analysis (right panel). In parallel experiments, siRNA-transfected and mock-transfected (Con) HFIB-D cells were treated with cDDP (20 μm) for 24 h before SAPK/JNK phosphorylation status (p-JNK) was analyzed. D, MEFs lacking the csb gene (CSB−/− MEFs) and corresponding wild type MEFs (CSB+/+ MEFs) were treated with 20 μm cisplatin. Up to 24 h later, the phosphorylation status of stress kinases (p-JNK) was determined. E, formation of γH2AX foci was assayed 16 h after addition of cDDP (5 μm) by immunohistochemistry using phosphospecific H2AX (Ser-139) antibody. DAPI staining was performed to visualize the nucleus. F, 6 and 16 h after chronic cisplatin (5 μm) treatment of wild type (GM637) and CSB cells, phosphorylation of H2AX (γH2AX) was investigated by Western blot analysis. Expression of β-actin (Actin) was taken as an internal loading control. G, 24 h after chronic treatment of GM637 cells or CSB-defective cells (CSB) with 5 μm cDDP, formation of DNA double strand breaks was assayed by the neutral comet assay as described under “Experimental Procedures.” OTM, olive tail moment. Data shown are means ± S.D.
As shown by immunohistochemical analysis, the phosphorylation of histone H2AX (γH2AX), which is indicative of DSB formation (58), is higher in CSB-defective cells than in wild type cells (Fig. 2E). Analysis of γH2AX phosphorylation by Western blot confirmed these results, although the differences observed were weaker (Fig. 2F). In line with the H2AX assay, analysis of DSB levels using the neutral comet assay also revealed a higher number of DSBs in cisplatin-treated CSB-defective cells compared with wild type cells (Fig. 2G). Apparently, enhanced SAPK/JNK phosphorylation in CSB-deficient cells at late times after cDDP treatment coincides with an elevated level of DSBs. Both ICP-MS and Southwestern analysis showed similar steady-state levels of DNA platination and 1,2-GG intrastrand cross-links, respectively, in wild type and CSB-defective cells 24 h after cisplatin administration (see supplemental Fig. S1). These data support the view that secondary DNA lesions (such as DSBs), rather than primary cDDP adducts, are of importance for signaling to SAPK/JNK at late times after cDDP exposure.
Replication-associated Mechanisms Contribute to Late SAPK/JNK Activation in CSB-defective Cells
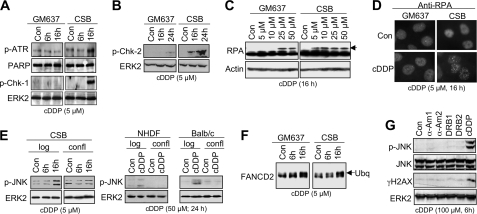
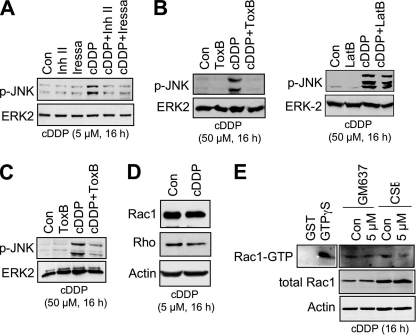
To determine whether replication-associated mechanisms are involved in late activation of SAPK/JNK by cDDP, the activation status of checkpoint control mechanisms was investigated. As shown in Fig. 3A, CSB cells revealed a stronger activation of ATR and Chk-1 than did wild type cells (Fig. 3A). Also, activation of Chk-2 was enhanced in cDDP-treated CSB-defective cells (Fig. 3B). In line with these data, the phosphorylation of RPA, which is triggered by ATR, was observed after low dose cisplatin treatment in CSB-defective cells but not in the wild type (Fig. 3C). Preferential activation of RPA in the absence of CSB protein was also demonstrated by analyzing the formation of RPA foci (Fig. 3D). The involvement of replication-associated mechanisms gains additional support by the finding that cisplatin-induced activation of SAPK/JNK was mitigated in non-growing human and murine cells compared with logarithmically growing cells (Fig. 3E). Interestingly, ubiquitination of FANCD2 protein occurred in both GM637 and CSB cells (Fig. 3F). This indicates that the enhanced SAPK/JNK activation in the absence of CSB is not due to a preferential activation of the FANC pathway, which plays a key role in intra-S phase checkpoint activation especially by interstrand cross-linking agents (59–61). Importantly, pharmacological inhibition of transcription by the RNA polymerase II inhibitors α-amanitin and 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole, which are known to activate p53 (62), did not stimulate SAPK/JNK phosphorylation (Fig. 3G). This indicates that transcription inhibition per se is not sufficient for evoking signaling to SAPK/JNK.
FIGURE 3.
Cisplatin-induced checkpoint response is increased in TC-NER-defective CSB cells. A, 6 and 16 h after cisplatin (5 μm) treatment of GM637 wild type and CSB cells, phosphorylation of ATR (p-ATR) and Chk-1 (p-Chk-1) was investigated by Western blot analysis. The expression of poly(ADP-ribose) polymerase (PARP) and ERK2 was determined as internal loading controls. B, the phosphorylation status of Chk-2 (p-Chk-2) was analyzed 16 and 24 h after cisplatin treatment (5 μm). C, CSB-proficient (GM637) and-defective cells (CSB) were treated with 5–50 μm cisplatin for 16 h. Afterward, cells were harvested, and the phosphorylation status of RPA was analyzed by Western blot. D, formation of RPA foci was analyzed 16 h after addition of cisplatin (5 μm) by immunohistochemistry. E, logarithmically growing (log) or non-growing confluent (confl) CSB-defective cells (CSB) were treated with 5 μm cisplatin for 6 or 16 h before the phosphorylation status of SAPK/JNK (p-JNK) was determined by Western blot. Growing (log) or non-growing (confl) primary human fibroblasts (NHDF) and mouse Balb/c fibroblasts were treated with cisplatin. After an incubation period of 24 h, the phosphorylation status of SAPK/JNK was analyzed. F, 6 and 16 h after addition of cisplatin (5 μm), ubiquitination of FANCD2 protein was determined in GM637 and CSB cells by Western blot analysis (90). The arrow points to the ubiquitinated form of FANCD2. G, GM637 cells were treated with RNA polymerase II inhibitors α-amanitin (0.5 (α-Am1) or 2.0 μg/ml (α-Am2)) and 5,6-dichloro-1-b-d-ribofuranosylbenzimidazole (40 (DRB1) or 80 μm (DRB2)), both of which are known to activate p53 (62). As a control, cDDP exposure was performed (100 μm). After an incubation period of 6 h, cells were harvested for the analysis of SAPK/JNK (p-JNK) and H2AX (γH2AX) phosphorylation. Con, untreated cells.
DSBs Are Not Sufficient for Dual Phosphorylation of SAPK/JNK
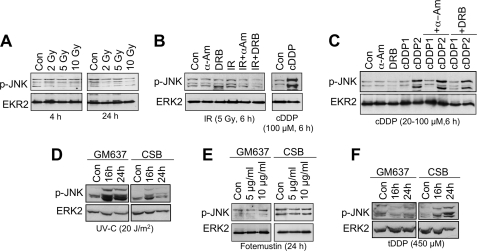
To elucidate the relevance of DSBs for cDDP-induced signaling to SAPK/JNK, we investigated the effect of ionizing radiation (IR), which is a prototypical inducer of DSBs, on dual phosphorylation of SAPK/JNK. Irradiation with doses of up to 10 grays did not provoke SEK1/MKK4-catalyzed dual SAPK/JNK phosphorylation at either 4 or 24 h after exposure (Fig. 4A). Also in combination with RNA polymerase II inhibitors, IR failed to stimulate SAPK/JNK phosphorylation (Fig. 4B). Furthermore, RNA polymerase inhibitors also failed to potentiate the cDDP-induced signaling to SAPK/JNK (Fig. 4C). Overall, these findings indicate that DSBs on their own or in combination with transcription blockage are not sufficient to activate SAPK/JNK. Of note, late activation of SAPK/JNK in CSB-defective cells is strikingly agent-specific. Neither UV-C light (Fig. 4D) nor fotemustine (Fig. 4E), which is an anticancer drug mainly generating interstrand cross-links, provoked the same response in CSB cells as cisplatin. Only transplatin caused preferential phosphorylation of SAPK/JNK in CSB-defective cells (Fig. 4F). In this context, we would like to note that, at the same level of platin-DNA adducts, cisplatin was observed to be 10–15 times more effective than transplatin in stimulating SAPK/JNK activity (supplemental Fig. S2). This indicates that cisplatin-induced intrastrand cross-links, rather than interstrand cross-links, are most effective in activating SAPK/JNK. Overall, the data show that late signaling to SAPK/JNK in TC-NER-defective CSB cells is specific for platinum compounds and that DSBs, although likely not sufficient to trigger this response on their own, might be involved in this process.
FIGURE 4.
Dual phosphorylation of SAPK/JNK is agent-specific and is not stimulated by IR-induced DNA double strand breaks. A, GM637 cells were left untreated (Con) or were irradiated with 2–10 grays (Gy). After incubation periods of 4 and 24 h, cells were harvested for the analysis of the phosphorylation status of SAPK/JNK (p-JNK). Con, untreated cells. B, GM637 cells were left untreated or were pretreated with the RNA polymerase inhibitors α-amanitin (2.0 μg/ml) (α-Am) or 5,6-dichloro-1-b-d-ribofuranosylbenzimidazole (80 μm) (DRB) for 2 h. Afterward, cells were irradiated with 5 grays, and the phosphorylation status of SAPK/JNK (p-JNK) was analyzed 6 h later. As a positive control, cells were treated with cisplatin (100 μm) for 6 h. C, GM637 cells were left untreated (Con) or were pretreated with the RNA polymerase inhibitors α-amanitin (2.0 μg/ml) (α-Am) or 5,6-dichloro-1-b-d-ribofuranosylbenzimidazole (80 μm) (DRB) for 2 h. Afterward, cells were exposed to 20 (cDDP1) or 100 μm (cDDP2) cisplatin. The phosphorylation status of SAPK/JNK (p-JNK) was analyzed 6 h later. D, human cells proficient (GM637) or deficient in CSB (CSB) protein were irradiated with UV-C light (20 J/m2). 16–24 h after irradiation, the phosphorylation status of SAPK/JNK (p-JNK) was determined. E, human cells either proficient (GM637) or deficient in CSB (CSB) protein were exposed to the interstrand cross-linking agent fotemustine (5 and 10 μg/ml). After an incubation period of 24 h, the phosphorylation status of SAPK/JNK (p-JNK) was determined by Western blot analysis. F, CSB-proficient (GM637) or -deficient (CSB) human cells were treated with transplatin (tDDP) (450 μm). After an incubation period of 16–24 h, the phosphorylation status of SAPK/JNK (p-JNK) was analyzed.
Various DNA Repair Deficiencies Specifically Affect Late SAPK/JNK Activation after Cisplatin Treatment
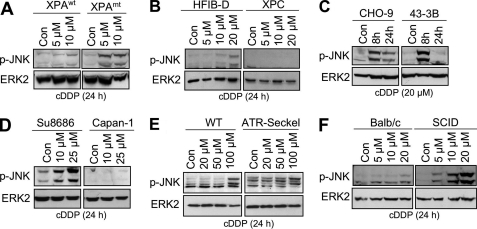
To elucidate whether late activation of SAPK/JNK by cisplatin is specific for CSB cells or is a more general phenomenon associated with DNA repair deficiency, various repair-defective mutants were investigated. Similar to CSB cells, XPA-mutated cells also displayed a higher SAPK/JNK phosphorylation status after cDDP exposure (Fig. 5A). The data indicate that insufficient repair of cisplatin-formed DNA damage, likely in transcribed genes, promotes stress signaling to SAPK/JNK. In contrast, human cells defective in XPC, which are specifically impaired in global genome NER, revealed a reduced late SAPK/JNK phosphorylation compared with wild type cells after cisplatin exposure (Fig. 5B). Also ERCC1-defective hamster 43-3B cells showed a reduced SAPK/JNK phosphorylation level 24 h after cDDP treatment (Fig. 5C). Collectively, NER deficiency can both promote (CSB and XPA) and inhibit (XPC and ERCC1) SAPK/JNK activity at late times after cDDP exposure.
FIGURE 5.
DNA repair defects differently affect cisplatin-induced late SAPK/JNK activation. A, SV40 immortalized human cells defective in TC-NER due to lack of XPA protein (XPAmt) and XPRO12 cells (XPAWT), which are reconstituted for XPA expression by stable transfection of xpa cDNA (43), were treated with 5 or 10 μm cisplatin. After an incubation period of 24 h, the dual phosphorylation status of SAPK/JNK (p-JNK) was determined. B, primary human wild type fibroblasts (HFIB-D) and XPC-defective human fibroblasts (XPC) were exposed to cisplatin (5–20 μm). After an incubation period of 24 h, SAPK/JNK phosphorylation (p-JNK) was analyzed by Western blot. C, ERCC1-proficient (CHO-9) and -defective (43−3B) hamster cells were treated with 20 μm cisplatin. Up to 24 h later, the phosphorylation status of SAPK/JNK (p-JNK) was analyzed. D, pancreatic carcinoma cells proficient (Su8686) or deficient (Capan-1) for BRCA2 protein were exposed to cisplatin (10 and 25 μm). After an incubation period of 24 h, the phosphorylation status of SAPK/JNK (p-JNK) was analyzed. E, human fibroblasts proficient (WT) or defective (ATR-Seckel) in ATR were treated with different concentrations of cisplatin. After an incubation period of 24 h, cells were harvested, and the phosphorylation status of SAPK/JNK (p-JNK) was determined. F, DNA-PKcs-deficient mouse fibroblasts cells (SCID) and Balb/c wild type fibroblasts were treated with different doses of cisplatin for 24 h before the phosphorylation status of SAPK/JNK (p-JNK) was analyzed. Con, untreated cells.
Interestingly, pancreatic Capan-1 carcinoma cells, which are defective in DSB repair due to lack of functional BRCA2 protein (63), do not respond to cDDP treatment with an increase in phosphorylated SAPK/JNK (Fig. 5D). Human ATR-defective ATR-Seckel cells were slightly impaired in their late response to cisplatin damage (Fig. 5E). Mouse SCID cells, which are compromised in non-homologous end joining due to a mutated DNA-PKcs, revealed an elevated phosphorylation level of SAPK/JNK at late times (i.e. 24 h) following cisplatin treatment (Fig. 5F). At an earlier time (i.e. 6 h) after high dose (i.e. 100 μm) cDDP addition, SAPK/JNK phosphorylation was similar in DNA-PKcs-mutated and wild type cells (data not shown). Similar to CSB-defective cells, DNA-PKcs-mutated SCID cells also showed increased H2AX and Chk-1 phosphorylation compared with the wild type. Moreover, the steady-state level of DNA-platin adducts was not affected in the absence of DNA-PKcs (supplemental Fig. S3). On the other hand, human cells defective in FANCD2 protein or ATM-deficient MEFs (ATM−/−) responded to cisplatin similarly to wild type cells (Table 1). Also, MEFs deficient (p53−/−) or proficient for p53 (p53+/+) (Table 1) showed identical responses, indicating that the p53 status does not determine the cisplatin response regarding SAPK/JNK. Collectively, the data show that a lack of specific DNA repair factors can either promote or mitigate SAPK/JNK phosphorylation at late times after cisplatin exposure (Table 1). Apparently, individual repair factors have an essential function for DNA damage-triggered signaling to SAPK/JNK.
TABLE 1.
Influence of cellular repair capacity on cDDP-induced late activation of SAPK/JNK
Dual phosphorylation of SAPK/JNK was analyzed by Western blot analysis 24 h after treatment of repair-defective and wild type cells of human or rodent origin with cisplatin (5–100 μm). Autoradiographies were densitometrically analyzed. Differences in the level of phosphorylated SAPK/JNK (p-SAPK/JNK) of ≥2-fold (or ≤0.5-fold) between repair-deficient and -proficient cells were considered as relevant. +, o, and −, increased (≥2-fold), unchanged, and reduced (≤0.5-fold) compared with repair-proficient cells, respectively.
| Repair deficiency | Late cDDP-induced p-SAPK/JNK level |
|---|---|
| CSBa | + |
| XPAa | + |
| XPCa | − |
| ERCC1b | − |
| BRCA2a | − |
| ATRa | − |
| FANCD2a | o |
| p53b | o |
| DNA-PKcsb | + |
| ATMb | o |
a Human cell lines.
b Rodent cell lines.
Typrosine Kinases and Rho GTPases Are Involved in Late Activation of SAPK/JNK by Cisplatin
Cisplatin is known to activate EGF receptor signaling (33), which influences DNA repair by affecting non-homologous end joining (64). Besides, the non-receptor tyrosine kinase c-Abl is required for SAPK/JNK activation in response to genotoxin treatment (65). On the basis of these reports, we investigated whether pharmacological inhibition of receptor and non-receptor tyrosine kinases impacts the late cisplatin-triggered activation of SAPK/JNK. To this end, tyrosine kinase inhibitors were added 8 h after the onset of cisplatin treatment. After a further incubation period of 8 h, the phosphorylation status of SAPK/JNK was analyzed. As shown in Fig. 6A, inhibition of both c-Abl tyrosine kinase and EGFR kinase mitigated SAPK/JNK phosphorylation. Apparently, both receptor and non-receptor tyrosine kinases are required for cisplatin-induced late signaling to SAPK/JNK. Investigating the effect of other protein kinases on cisplatin-induced signaling, we found that inhibition of phosphatidylinositol 3-kinase-related kinases by wortmannin and inhibition of ATM by Ku55933 also mitigated the late cisplatin response. In contrast, inhibition of MEK1, Akt, and c-Src kinases was ineffective in SAPK/JNK activation following cisplatin treatment (Table 2).
FIGURE 6.
Cisplatin-induced activation of SAPK/JNK requires tyrosine kinases and Rho GTPases. A, human CSB-defective cells were left untreated (Con) or were exposed to cisplatin (5 μm). 8 h after addition of cisplatin, tyrosine kinase inhibitors targeting Bcr-Abl (Inh II) (10 μm) (91) or EGFR (Iressa) (10 μm) were added. After a further incubation period of 8 h in the presence of inhibitor and cisplatin, cells were harvested for analysis of SAPK/JNK phosphorylation (p-JNK). B, human GM637 fibroblasts were pretreated for 2 h with either Rho-inactivating ToxB (2.5 ng/ml) (left panel) or latrunculin B (LatB) (1 μm) (right panel), which disrupts F-actin filaments. Afterward, cDDP was added (50 μm), and cells were further incubated for 16 h before the phosphorylation status of SAPK/JNK (p-JNK) was analyzed by Western blot analysis. C, primary human fibroblasts were pretreated with Rho-inactivating clostridial ToxB (1 ng/ml) for 2 h. Afterward, cDDP (50 μm) was added, and cells were harvested 16 h later for determination of SAPK/JNK activation (p-JNK) by Western blot. D, 16 h after continuous cisplatin treatment (5 μm) of human CSB-defective cells (CSB), the expression of the Rho GTPase Rac1 and RhoA-like GTPases (Rho) was analyzed in the membrane fractions. E, 16 h after permanent cisplatin (5 μm) treatment of CSB-proficient (GM637) and CSB-defective human cells (CSB), the GTP binding status of Rac1 (Rac1-GTP) was analyzed by GST-PAK pulldown experiments as described under “Experimental Procedures.” As internal controls, protein expression of β-actin (Actin) and total Rac1 were determined in total cell extracts. GST, pulldown using GST (negative control); GTPγS, GST-PAK pulldown using cell extract preincubated with GTPγS (positive control).
TABLE 2.
Influence of various protein kinase inhibitors on cDDP-induced late activation of SAPK/JNK
The dual phosphorylation status of SAPK/JNK (p-SAPK/JNK) was analyzed by Western blot analysis 24 h after the onset of cisplatin treatment of TC-NER-defective CSB cells or wild type GM637 cells. The cisplatin concentration used was 5 and 20 μm for CSB and GM637 cells, respectively. Inhibitors were added 4–8 h after cDDP addition and were left on the cells until harvest. The following widely used protein kinase inhibitors were applied: Bcr-Abl inhibitor type II (10 μm), c-Src inhibitor PP2 (10 μm), EGFR inhibitor Iressa (10 μm), Akt kinase inhibitor (5 μm), MEK1 kinase inhibitor PD98059 (10 μm), PI3K-related kinase (PI3K-RK) inhibitor wortmannin (1 μm), and ATM kinase inhibitor Ku55933 (5 μm). An inhibitor was considered as effective if it reduced the cDDP-stimulated SAPK/JNK phosphorylation by ≥50% compared with the non-inhibitor-treated control. +, inhibition observed (≥50%); o, no inhibition observed; ND, not determined.
| Protein kinase inhibitor | Late cDDP-induced p-SAPK/JNK level |
|
|---|---|---|
| CSB | GM637 | |
| Bcr-Abl | + | + |
| c-Src | o | o |
| EGFR | + | + |
| Akt | o | o |
| MEK1 | o | o |
| PI3K-RK | + | + |
| ATM | + | ND |
Membrane-bound Rho GTPases are essential for SEK1/MKK4-mediated early activation of SAPK/JNK (17, 18, 66). Correspondingly, specific inhibition of Rho functions by Clostridium difficile toxin B (ToxB) abrogates both UV light- and MMS-induced early (i.e. DNA damage-independent) activation of SAPK/JNK (22, 66). As shown in Fig. 6, ToxB also blocked cisplatin-induced late activation of SAPK/JNK in GM637 cells (Fig. 6B, left panel) as well as in primary human fibroblasts (Fig. 6C). The inhibitory effect of ToxB is not due to its interference with the actin cytoskeleton (67) because disruption of actin filament structure by latrunculin B did not block SAPK/JNK phosphorylation stimulated by cDDP (Fig. 6B, right panel). Among the family of Rho GTPases, Rac1 is of particular importance for UV light-induced signaling to SAPK/JNK (66, 68). Cisplatin did not increase the amount of membrane-bound Rac1 (Fig. 6D) and did not alter the GTP binding status of Rac1 protein (Fig. 6E), indicating that cDDP does not cause de novo stimulation of Rac1 activity. Altogether, the data point to a permissive function of Rho GTPases in the late DNA damage-triggered SAPK/JNK activation.
Late SAPK/JNK Activation after Cisplatin Treatment Promotes Cell Survival
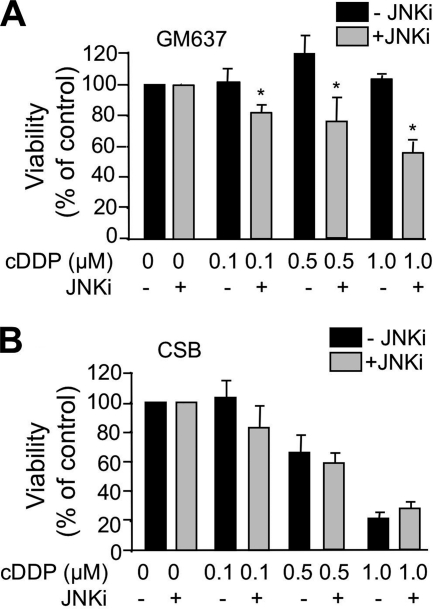
SAPK/JNK are suggested to have a protective function after cDDP exposure (11, 24, 69). To determine the relevance of SAPK/JNK-regulated mechanisms at late times after cDDP exposure, the JNK inhibitor SP600125 was added 16 h after cisplatin treatment for a total time period of 8 h. After further incubation over a period of 40 h, cell viability was monitored. Under this experimental condition, JNK inhibition increased the sensitivity of GM637 wild type cells to cisplatin (Fig. 7A), supporting the view of a protective function of SAPK/JNK in cisplatin-treated wild type cells. SP600125 treatment did not affect the cisplatin sensitivity of CSB cells (Fig. 7B).
FIGURE 7.
Activation of SAPK/JNK by cisplatin promotes survival of GM637 cells. A and B, logarithmically growing wild type (GM637) (A) and CSB-deficient (CSB) (B) cells were treated with different doses of cisplatin (0.1–1 μm) for 16 h. Afterward, the medium was replaced by fresh medium containing JNK inhibitor (JNKi) SP600125 (10 μm). After a further incubation period of 8 h, the medium was replaced again, and cell viability was assayed 40 h later using the water soluble tetrazolium salt 1 (WST-1) assay. Data shown are the mean ± S.D. from triplicate determinations. The cell viability of non-cisplatin-treated controls was set to 100%. *, p < 0.05.
DISCUSSION
Early activation of SAPK/JNK is a key response of eukaryotic cells following genotoxin exposure and is believed to originate mainly from the activation of membrane receptors (1, 15). However, as DNA repair-defective mutants differ from wild type cells with respect to genotoxin-triggered SAPK/JNK activation (19–21), it is reasonable to hypothesize that DNA damage also impacts genotoxin-stimulated signaling to SAPK/JNK. In the present study, we provide evidence that DNA damage following cisplatin treatment contributes to the activation of SAPK/JNK. We also elucidated the molecular mechanisms involved. We found that the dose-dependent stimulation of SAPK/JNK activity at late times after drug addition correlates with the steady-state level of 1,2-GG intrastrand cross-links and overall DNA platination. There was no correlation with ROS formation. We also showed that transfection of platinated plasmid DNA gives rise to activation of SAPK/JNK. We therefore suggest that cisplatin-DNA adducts can provoke dual phosphorylation of SAPK/JNK. This is in line with a recent finding demonstrating that a single DNA interstrand cross-link is able to trigger checkpoint signaling in cell-free extracts (70).
To further substantiate the contribution of cisplatin-induced DNA adducts for signaling to SAPK/JNK, we analyzed the cisplatin response of cells that are defective in TC-NER, a major repair pathway dictating stress responses, DNA repair, and death (55, 56, 71, 72). TC-NER-defective CSB cells showed enhanced phosphorylation of SAPK/JNK specifically at late times (i.e. 16–24 h) after cisplatin treatment compared with the wild type. siRNA-mediated knockdown of CSB in human cells and CSB-defective MEFs provided identical results. Similar to CSB cells, human XPA cells also showed elevated SAPK/JNK phosphorylation at late times after drug exposure. By contrast, cells that harbor a specific defect in global genome NER (i.e. XPC cells) displayed a reduced SAPK/JNK phosphorylation after cisplatin exposure. Based on the data, we suggest that defective TC-NER triggers a strong late SAPK/JNK activation following the formation of cisplatin-DNA adducts. Notably, apart from cells affected in XPC, ERCC1- and BRCA2-defective cells also revealed a reduced late SAPK/JNK phosphorylation. Apparently, individual repair factors are required to different extents for late signaling to SAPK/JNK following cisplatin treatment. Of note, XPC protein has also been reported to be essential for SAPK/JNK activation after exposure to UV light (20, 73). Collectively, it appears that individual DNA repair proteins have repair-independent crucial functions in the DNA damage response.
DSBs and replication-blocking lesions are potent triggers of the eukaryotic DNA damage response (25, 74). Regarding SAPK/JNK, pharmacological inhibition of replicative DNA polymerase was found to be sufficient for stimulating its dual phosphorylation (73). Bearing this in mind, we speculated that replication-associated events, together with DSBs generated in the absence of TC-NER, might provide the ultimate signal leading to SAPK/JNK activation after long lasting cisplatin treatment. This hypothesis gained support by the observation that the level of DSBs, as monitored by the neutral comet assay and by ATM/ATR-catalyzed phosphorylation of H2AX (γH2AX) (50), largely increased in TC-NER-defective cells. The phosphorylation status of ATR, ATR-catalyzed phosphorylation of RPA, formation of RPA foci, and ATM/ATR-regulated activation of checkpoint kinases, all of which are indicative of replicative stress, were also enhanced in cisplatin-treated CSB cells. Moreover, logarithmically growing human and murine cells revealed a higher SAPK/JNK activation after cisplatin treatment than did non-growing confluent cells. A reasonable hypothesis inferred from these data is that persistent cisplatin-DNA adducts in transcriptionally active regions of the genome elicit replicative stress, thereby leading to SAPK/JNK activation. This response is enhanced if TC-NER is absent and all essential signaling factors (including XPC) are present. Bearing in mind that ATR plays a key role in replication fork progression (75, 76) and is involved in formation of γH2AX foci after cisplatin treatment (77), it is reasonable to assume that this kinase has a major role in signaling to SAPK/JNK. Notably, SAPK/JNK activation also occurred after transfection of non-replicating platinated plasmid DNA. This finding indicates that replication facilitates the recognition of cDDP adducts inside the chromatin, thereby allowing signaling. Transcription inhibition by cisplatin is not sufficient for stimulating signaling to SAPK/JNK. This is concluded from the finding that pharmacological inhibition of RNA polymerase II, which stimulates p53 (62), does not provoke activation of SAPK/JNK. Besides, inhibitors of RNA polymerase II did not further augment the cisplatin-stimulated activation of SAPK/JNK.
The significance of DSBs for cisplatin-stimulated signaling to SAPK/JNK is ambiguous. On the one hand, DSBs are not enough to stimulate SEK1/MKK4-catalyzed dual SAPK/JNK phosphorylation on their own. This assumption rests on the observation that IR, alone or in combination with RNA polymerase II inhibitors, was not able to provoke this response. On the other hand, cells harboring a defect in the non-homologous end joining pathway of DSB repair due to a defect in DNA-PKcs revealed elevated levels of phosphorylated SAPK/JNK as well as an enhanced H2AX phosphorylation and checkpoint activation at late times after cisplatin exposure. Furthermore, SAPK/JNK has been reported to co-precipitate with H2AX after IR exposure (78), to phosphorylate H2AX upon UV-A irradiation (79), and to interact with DNA-PKcs after MMS exposure (22). These findings are suggestive of a connection between DSBs and SAPK/JNK. We suppose that DSBs are essential for triggering SAPK/JNK activity after cDDP exposure, but they are not sufficient to provoke it on their own. The presumption of a solely permissive, yet not dominant, function of DSBs in signaling to SAPK/JNK gains support from the finding that CSB-defective cells exhibit a high number of DSBs 24 h after UV-C light exposure but do not show an enhanced dual SAPK/JNK phosphorylation (data not shown). The startling agent specificity of the cellular stress response is highlighted by the fact that, as opposed to cisplatin, the activation of SAPK/JNK by the alkylating agent MMS is even reduced in repair-defective CSB and SCID cells (22). It is evident that the absence of individual repair factors can result in both abrogation and improvement of signaling with respect to SAPK/JNK. Thus, the actual outcome of SAPK/JNK activation seems to depend on both the genotoxic agent and the repair components present at the time of DNA damage formation.
Late cisplatin-induced activation of SAPK/JNK in CSB cells is accompanied by an increased SEK1/MKK4 activity. By contrast, SEK1/MKK4 is not relevant for the UV light response of these cells (20). Reduced expression of MKP-1 was demonstrated to enhance UV light-induced signaling to SAPK/JNK in CSB and c-Fos deficient MEFs (20, 80). However, under our experimental conditions, cisplatin did not influence MKP-1 protein expression either in human wild type or in CSB cells (data not shown). Hence, in contrast to UV light, cisplatin-triggered activation of SAPK/JNK coincides with SEK1/MKK4 activation and is independent of MKP-1. It should be noted that the alkylating genotoxin MMS is able to induce MKP-1 protein expression (22). UV light stimulates the secretion of molecules, which in turn evoke receptor-mediated signaling processes by autocrine/paracrine mechanisms (81, 82). However, supernatants of cisplatin-treated repair-defective cells (CSB and SCID) did not elicit SAPK/JNK phosphorylation in untreated cells (data not shown). Therefore, autocrine/paracrine mechanisms are likely not important for cisplatin-induced late activation of SAPK/JNK in TC-NER- or non-homologous end joining-defective cells, again pointing to the high agent specificity of genotoxin-induced stress responses. Endoplasmatic reticulum (ER) stress is also not involved (data not shown).
Membrane-bound small GTPases of the Rho family, in particular Rac1, are important regulators of SAPK/JNK (17, 18). They have been shown to be essential for the activation of SAPK/JNK by the alkylating agent MMS (22) and UV light (66, 68). As shown here, Rho GTPases are also required for early (data not shown) and late activation of SAPK/JNK by cisplatin. Because cisplatin does not stimulate a translocation of Rac1 to the membrane, which would be indicative of its de novo activation, we propose that this GTPase, which has recently been found in the nucleus (83), has a permissive function in the cisplatin-induced signaling cascade. The same appears to be true for receptor (i.e. EGFR) and non-receptor tyrosine kinases (i.e. c-Abl). An intriguing question that still remains to be solved is which player links the JNK pathway with the DNA damage response (25). A candidate might be c-Abl, which is known to be activated by genotoxins, including cisplatin (84, 85), and is regulated in an ATM-dependent manner (86, 87). Furthermore, it activates the MEK kinase 1 (65), which is the upstream kinase of the SEK1-SAPK/JNK pathway. A converging role of c-Abl has recently been reported for the ATM and DNA-PKcs pathways in response to ionizing radiation (88).
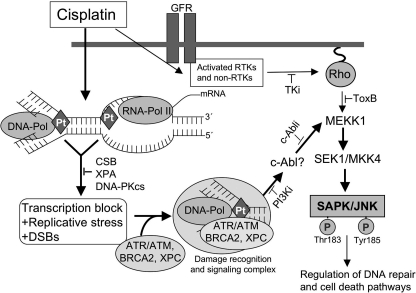
A model linking cisplatin-induced DNA damage, its repair, and SAPK/JNK activation is shown in Fig. 8. We propose that DNA damage triggers late activation of SAPK/JNK following cisplatin treatment of TC-NER- and DNA-PKcs-defective cells. We posit that late SAPK/JNK activation is the consequence of an inaccurate processing of cisplatin adducts in transcribed genes and involves the formation of DSBs, which might result from a sustained blockade of the transcription machinery that collides with DNA replication in TC-NER-defective cells. The evolving replicative stress, together with platin-DNA adducts, DSBs, and/or distortions of the DNA helix, in turn attracts multiple repair and signaling factors forming a multifunctional protein complex. This complex finally triggers SAPK/JNK activation in an SEK1/MKK4-dependent manner. We also hypothesize that the protein-tyrosine kinase c-Abl provides a link between DNA damage processing and the SEK1-SAPK/JNK pathway. In consequence of SAPK/JNK activation, DNA repair and/or cell death pathways (89) are regulated.
FIGURE 8.
Model of DNA damage-triggered activation of SAPK/JNK at late times following cisplatin treatment. Apart from DNA damage-independent mechanisms that can cause an early activation of SAPK/JNK after cisplatin treatment, cisplatin-induced DNA damage is also able to trigger signaling to SAPK/JNK at late times (i.e. 16–24 h) after drug exposure. Late SAPK/JNK activation is pronounced in TC-NER- (i.e. CSB- and XPA-) and DNA-PKcs-defective cells. It is mitigated under the condition of XPC, ERCC1, and BRCA2 deficiency, indicating that these repair factors have additional functions as part of a signaling complex. Transcription blockage and DSBs are not sufficient for stimulating SAPK/JNK activity. The response appears to be specific for platinum compounds. Replication-associated ATR-regulated mechanisms are likely involved in signaling to SAPK/JNK triggered by cisplatin-induced DNA lesions. Furthermore, Rho GTPases (Rho) and tyrosine kinases (i.e. EGFR and c-Abl) seem to have a permissive function. Bearing in mind that c-Abl tyrosine kinase is activated by genotoxins, including cisplatin (84), is regulated in an ATM-dependent manner (86, 87), and furthermore regulates MEK kinase 1 (65), this kinase is a rational candidate to converge the DNA damage response with the SAPK/JNK pathway. Late activation of SAPK/JNK by cisplatin is suggested to promote survival. c-Abli, inhibitor of the c-Abl tyrosine kinase; GFR, growth factor receptor; PI3Ki, phosphatidylinositol 3-kinase-related kinase inhibitors (i.e. wortmannin and Ku55933); Rho, Ras-homologous GTPases; RTK, receptor tyrosine kinases; TKi, tyrosine kinase inhibitors (i.e. Iressa and c-Abl inhibitor); JNKi, JNK inhibitor type II; Pol, polymerase; Pt, platinum.
Acknowledgments
We are particularly grateful to J. Thomale (Essen, Germany) for generously providing the antibody specifically detecting cisplatin-DNA intrastrand cross-links and S. Hülsenbeck (Hannover, Germany) for performing the GST-PAK pulldown assay.
This work was supported by Deutsche Forschungsgemeinschaft Grant Fr 1541/5-3.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- ROS
- reactive oxygen species
- ATM
- Ataxia telangiectasia mutated
- FANC
- Fanconi anemia
- ATR
- ATM- and Rad3-related
- RPA
- replication protein A
- CSB
- Cockayne syndrome B protein
- DSB
- double strand break
- XPC
- xeroderma pigmentosum protein C
- SCID
- severe combined immunodeficiency
- NER
- nucleotide excision repair
- MMS
- methyl methanesulfonate
- EGFR
- EGF receptor
- MEF
- mouse embryonic fibroblast
- DNA-PKcs
- DNA-dependent protein kinase, catalytic subunit
- XPA
- xeroderma pigmentosum protein A
- Chk
- checkpoint kinase
- cDDP
- cisplatin
- ICP
- inductively coupled plasma
- TC-NER
- transcription-coupled nucleotide excision repair
- ToxB
- C. difficile toxin B
- MKP-1
- mitogen-activated kinase phosphatase 1
- GTPγS
- guanosine 5′-3-O-(thio)triphosphate
- PAK
- p21-associated protein kinase.
REFERENCES
- 1. Canman C. E., Kastan M. B. (1996) Nature 384, 213–214 [DOI] [PubMed] [Google Scholar]
- 2. Benhar M., Engelberg D., Levitzki A. (2002) EMBO Rep. 3, 420–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Karin M. (1996) Philos. Trans. R. Soc. Lond. B Biol. Sci. 351, 127–134 [DOI] [PubMed] [Google Scholar]
- 4. Kaina B., Haas S., Kappes H. (1997) Cancer Res. 57, 2721–2731 [PubMed] [Google Scholar]
- 5. Shaulian E., Karin M. (2002) Nat. Cell Biol. 4, E131–E136 [DOI] [PubMed] [Google Scholar]
- 6. Xia Z., Dickens M., Raingeaud J., Davis R. J., Greenberg M. E. (1995) Science 270, 1326–1331 [DOI] [PubMed] [Google Scholar]
- 7. Verheij M., Bose R., Lin X. H., Yao B., Jarvis W. D., Grant S., Birrer M. J., Szabo E., Zon L. I., Kyriakis J. M., Haimovitz-Friedman A., Fuks Z., Kolesnick R. N. (1996) Nature 380, 75–79 [DOI] [PubMed] [Google Scholar]
- 8. Hayakawa J., Depatie C., Ohmichi M., Mercola D. (2003) J. Biol. Chem. 278, 20582–20592 [DOI] [PubMed] [Google Scholar]
- 9. Kolbus A., Herr I., Schreiber M., Debatin K. M., Wagner E. F., Angel P. (2000) Mol. Cell. Biol. 20, 575–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng X., Xiao L., Lang W., Gao F., Ruvolo P., May W. S., Jr. (2001) J. Biol. Chem. 276, 23681–23688 [DOI] [PubMed] [Google Scholar]
- 11. Potapova O., Haghighi A., Bost F., Liu C., Birrer M. J., Gjerset R., Mercola D. (1997) J. Biol. Chem. 272, 14041–14044 [DOI] [PubMed] [Google Scholar]
- 12. Christmann M., Tomicic M. T., Origer J., Aasland D., Kaina B. (2006) Nucleic Acids Res. 34, 6530–6539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coffer P. J., Burgering B. M., Peppelenbosch M. P., Bos J. L., Kruijer W. (1995) Oncogene 11, 561–569 [PubMed] [Google Scholar]
- 14. Kitagawa D., Tanemura S., Ohata S., Shimizu N., Seo J., Nishitai G., Watanabe T., Nakagawa K., Kishimoto H., Wada T., Tezuka T., Yamamoto T., Nishina H., Katada T. (2002) J. Biol. Chem. 277, 366–371 [DOI] [PubMed] [Google Scholar]
- 15. Rosette C., Karin M. (1996) Science 274, 1194–1197 [DOI] [PubMed] [Google Scholar]
- 16. Gross S., Knebel A., Tenev T., Neininger A., Gaestel M., Herrlich P., Böhmer F. D. (1999) J. Biol. Chem. 274, 26378–26386 [DOI] [PubMed] [Google Scholar]
- 17. Minden A., Lin A., Claret F. X., Abo A., Karin M. (1995) Cell 81, 1147–1157 [DOI] [PubMed] [Google Scholar]
- 18. Coso O. A., Chiariello M., Yu J. C., Teramoto H., Crespo P., Xu N., Miki T., Gutkind J. S. (1995) Cell 81, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 19. Nehmé A., Baskaran R., Aebi S., Fink D., Nebel S., Cenni B., Wang J. Y., Howell S. B., Christen R. D. (1997) Cancer Res. 57, 3253–3257 [PubMed] [Google Scholar]
- 20. Hamdi M., Kool J., Cornelissen-Steijger P., Carlotti F., Popeijus H. E., van der Burgt C., Janssen J. M., Yasui A., Hoeben R. C., Terleth C., Mullenders L. H., van Dam H. (2005) Oncogene 24, 7135–7144 [DOI] [PubMed] [Google Scholar]
- 21. Dhar V., Adler V., Lehmann A., Ronai Z. (1996) Cell Growth Differ. 7, 841–846 [PubMed] [Google Scholar]
- 22. Fritz G., Kaina B. (2006) Mol. Biol. Cell 17, 851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mansouri A., Ridgway L. D., Korapati A. L., Zhang Q., Tian L., Wang Y., Siddik Z. H., Mills G. B., Claret F. X. (2003) J. Biol. Chem. 278, 19245–19256 [DOI] [PubMed] [Google Scholar]
- 24. Brozovic A., Fritz G., Christmann M., Zisowsky J., Jaehde U., Osmak M., Kaina B. (2004) Int. J. Cancer 112, 974–985 [DOI] [PubMed] [Google Scholar]
- 25. Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 26. Wilhelm D., Bender K., Knebel A., Angel P. (1997) Mol. Cell. Biol. 17, 4792–4800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Dam H., Wilhelm D., Herr I., Steffen A., Herrlich P., Angel P. (1995) EMBO J. 14, 1798–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sánchez-Perez I., Murguía J. R., Perona R. (1998) Oncogene 16, 533–540 [DOI] [PubMed] [Google Scholar]
- 29. Friedberg E. C., Wallner G. C., Siede W. (1995) DNA Repair and Mutagenesis, pp. 25–29, ASM Press, Washington, D. C [Google Scholar]
- 30. Zamble D. B., Lippard S. J. (1995) Trends Biochem. Sci. 20, 435–439 [DOI] [PubMed] [Google Scholar]
- 31. Jamieson E. R., Lippard S. J. (1999) Chem. Rev. 99, 2467–2498 [DOI] [PubMed] [Google Scholar]
- 32. Malinge J. M., Giraud-Panis M. J., Leng M. (1999) J. Inorg. Biochem. 77, 23–29 [DOI] [PubMed] [Google Scholar]
- 33. Benhar M., Engelberg D., Levitzki A. (2002) Oncogene 21, 8723–8731 [DOI] [PubMed] [Google Scholar]
- 34. Rebillard A., Lagadic-Gossmann D., Dimanche-Boitrel M. T. (2008) Curr. Med. Chem. 15, 2656–2663 [DOI] [PubMed] [Google Scholar]
- 35. Rao R. V., Ellerby H. M., Bredesen D. E. (2004) Cell Death Differ. 11, 372–380 [DOI] [PubMed] [Google Scholar]
- 36. Mandic A., Hansson J., Linder S., Shoshan M. C. (2003) J. Biol. Chem. 278, 9100–9106 [DOI] [PubMed] [Google Scholar]
- 37. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., Ron D. (2000) Science 287, 664–666 [DOI] [PubMed] [Google Scholar]
- 38. Benhar M., Dalyot I., Engelberg D., Levitzki A. (2001) Mol. Cell. Biol. 21, 6913–6926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liedert B., Pluim D., Schellens J., Thomale J. (2006) Nucleic Acids Res. 34, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lackinger D., Kaina B. (2000) Mutat. Res. 457, 113–123 [DOI] [PubMed] [Google Scholar]
- 41. Stevnsner T., Nyaga S., de Souza-Pinto N. C., van der Horst G. T., Gorgels T. G., Hogue B. A., Thorslund T., Bohr V. A. (2002) Oncogene 21, 8675–8682 [DOI] [PubMed] [Google Scholar]
- 42. Biedermann K. A., Sun J. R., Giaccia A. J., Tosto L. M., Brown J. M. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 1394–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Köberle B., Roginskaya V., Wood R. D. (2006) DNA Repair 5, 641–648 [DOI] [PubMed] [Google Scholar]
- 44. Dunkern T. R., Wedemeyer I., Baumgärtner M., Fritz G., Kaina B. (2003) DNA Repair 2, 49–60 [DOI] [PubMed] [Google Scholar]
- 45. Hansson J., Wood R. D. (1989) Nucleic Acids Res. 17, 8073–8091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ochs K., Kaina B. (2000) Cancer Res. 60, 5815–5824 [PubMed] [Google Scholar]
- 47. Cortez D., Guntuku S., Qin J., Elledge S. J. (2001) Science 294, 1713–1716 [DOI] [PubMed] [Google Scholar]
- 48. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 49. Rothkamm K., Löbrich M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5057–5062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 51. Sander E. E., van Delft S., ten Klooster J. P., Reid T., van der Kammen R. A., Michiels F., Collard J. G. (1998) J. Cell Biol. 143, 1385–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nehmé A., Baskaran R., Nebel S., Fink D., Howell S. B., Wang J. Y., Christen R. D. (1999) Br. J. Cancer 79, 1104–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. de Boer J., Hoeijmakers J. H. (2000) Carcinogenesis 21, 453–460 [DOI] [PubMed] [Google Scholar]
- 54. Lainé J. P., Egly J. M. (2006) Trends Genet. 22, 430–436 [DOI] [PubMed] [Google Scholar]
- 55. Fousteri M., Mullenders L. H. (2008) Cell Res. 18, 73–84 [DOI] [PubMed] [Google Scholar]
- 56. Ljungman M. (2007) Cell Cycle 6, 2252–2257 [DOI] [PubMed] [Google Scholar]
- 57. Furuta T., Ueda T., Aune G., Sarasin A., Kraemer K. H., Pommier Y. (2002) Cancer Res. 62, 4899–4902 [PubMed] [Google Scholar]
- 58. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 59. Patel K. J., Joenje H. (2007) DNA Repair 6, 885–890 [DOI] [PubMed] [Google Scholar]
- 60. Pichierri P., Rosselli F. (2004) EMBO J. 23, 1178–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. D'Andrea A. (2003) Curr. Biol. 13, R546. [DOI] [PubMed] [Google Scholar]
- 62. Ljungman M., Zhang F., Chen F., Rainbow A. J., McKay B. C. (1999) Oncogene 18, 583–592 [DOI] [PubMed] [Google Scholar]
- 63. Wang H., Zeng Z. C., Bui T. A., DiBiase S. J., Qin W., Xia F., Powell S. N., Iliakis G. (2001) Cancer Res. 61, 270–277 [PubMed] [Google Scholar]
- 64. Rodemann H. P., Dittmann K., Toulany M. (2007) Int. J. Radiat. Biol. 83, 781–791 [DOI] [PubMed] [Google Scholar]
- 65. Kharbanda S., Pandey P., Yamauchi T., Kumar S., Kaneki M., Kumar V., Bharti A., Yuan Z. M., Ghanem L., Rana A., Weichselbaum R., Johnson G., Kufe D. (2000) Mol. Cell. Biol. 20, 4979–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Gnad R., Aktories K., Kaina B., Fritz G. (2000) Mol. Pharmacol. 58, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 67. Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Nature 375, 500–503 [DOI] [PubMed] [Google Scholar]
- 68. von Bardeleben R., Kaina B., Fritz G. (2003) Biochem. Biophys. Res. Commun. 307, 401–407 [DOI] [PubMed] [Google Scholar]
- 69. Hayakawa J., Mittal S., Wang Y., Korkmaz K. S., Adamson E., English C., Ohmichi M., McClelland M., Mercola D. (2004) Mol. Cell 16, 521–535 [DOI] [PubMed] [Google Scholar]
- 70. Ben-Yehoyada M., Wang L. C., Kozekov I. D., Rizzo C. J., Gottesman M. E., Gautier J. (2009) Mol. Cell 35, 704–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ljungman M., Lane D. P. (2004) Nat. Rev. Cancer 4, 727–737 [DOI] [PubMed] [Google Scholar]
- 72. Ljungman M. (2005) Mutat. Res. 577, 203–216 [DOI] [PubMed] [Google Scholar]
- 73. Damrot J., Helbig L., Roos W. P., Barrantes S. Q., Kaina B., Fritz G. (2009) J. Mol. Biol. 385, 1409–1421 [DOI] [PubMed] [Google Scholar]
- 74. Durocher D., Jackson S. P. (2001) Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 75. Stiff T., Walker S. A., Cerosaletti K., Goodarzi A. A., Petermann E., Concannon P., O'Driscoll M., Jeggo P. A. (2006) EMBO J. 25, 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Paulsen R. D., Cimprich K. A. (2007) DNA Repair 6, 953–966 [DOI] [PubMed] [Google Scholar]
- 77. Pabla N., Huang S., Mi Q. S., Daniel R., Dong Z. (2008) J. Biol. Chem. 283, 6572–6583 [DOI] [PubMed] [Google Scholar]
- 78. Cook P. J., Ju B. G., Telese F., Wang X., Glass C. K., Rosenfeld M. G. (2009) Nature 458, 591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lu C., Zhu F., Cho Y. Y., Tang F., Zykova T., Ma W. Y., Bode A. M., Dong Z. (2006) Mol. Cell 23, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Christmann M., Tomicic M. T., Aasland D., Kaina B. (2007) Carcinogenesis 28, 183–190 [DOI] [PubMed] [Google Scholar]
- 81. Yoshizumi M., Nakamura T., Kato M., Ishioka T., Kozawa K., Wakamatsu K., Kimura H. (2008) Cell Biol. Int. 32, 1405–1411 [DOI] [PubMed] [Google Scholar]
- 82. Krämer M., Sachsenmaier C., Herrlich P., Rahmsdorf H. J. (1993) J. Biol. Chem. 268, 6734–6741 [PubMed] [Google Scholar]
- 83. Sandrock K., Bielek H., Schradi K., Schmidt G., Klugbauer N. (2010) Traffic 11, 198–209 [DOI] [PubMed] [Google Scholar]
- 84. Kharbanda S., Ren R., Pandey P., Shafman T. D., Feller S. M., Weichselbaum R. R., Kufe D. W. (1995) Nature 376, 785–788 [DOI] [PubMed] [Google Scholar]
- 85. Liu Z. G., Baskaran R., Lea-Chou E. T., Wood L. D., Chen Y., Karin M., Wang J. Y. (1996) Nature 384, 273–276 [DOI] [PubMed] [Google Scholar]
- 86. Baskaran R., Wood L. D., Whitaker L. L., Canman C. E., Morgan S. E., Xu Y., Barlow C., Baltimore D., Wynshaw-Boris A., Kastan M. B., Wang J. Y. (1997) Nature 387, 516–519 [DOI] [PubMed] [Google Scholar]
- 87. Shafman T., Khanna K. K., Kedar P., Spring K., Kozlov S., Yen T., Hobson K., Gatei M., Zhang N., Watters D., Egerton M., Shiloh Y., Kharbanda S., Kufe D., Lavin M. F. (1997) Nature 387, 520–523 [DOI] [PubMed] [Google Scholar]
- 88. Shangary S., Brown K. D., Adamson A. W., Edmonson S., Ng B., Pandita T. K., Yalowich J., Taccioli G. E., Baskaran R. (2000) J. Biol. Chem. 275, 30163–30168 [DOI] [PubMed] [Google Scholar]
- 89. Roos W. P., Kaina B. (2006) Trends Mol. Med. 12, 440–450 [DOI] [PubMed] [Google Scholar]
- 90. Bogliolo M., Lyakhovich A., Callén E., Castellà M., Cappelli E., Ramírez M. J., Creus A., Marcos R., Kalb R., Neveling K., Schindler D., Surrallés J. (2007) EMBO J. 26, 1340–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Radi M., Crespan E., Botta G., Falchi F., Maga G., Manetti F., Corradi V., Mancini M., Santucci M. A., Schenone S., Botta M. (2008) Bioorg. Med. Chem. Lett. 18, 1207–1211 [DOI] [PubMed] [Google Scholar]