Abstract
Acyl-CoA:diacylglycerol acyltransferase (EC 2.3.1.20) is a membrane protein present mainly in the endoplasmic reticulum. It catalyzes the final and committed step in the biosynthesis of triacylglycerol, which is the principal repository of fatty acids for energy utilization and membrane formation. Two distinct family members of acyl-CoA:diacylglycerol acyltransferase, known as DGAT1 and DGAT2, have been characterized in different organisms, including mammals, fungi, and plants. In this study, we characterized the functional role and topological orientation of signature motifs in yeast (Saccharomyces cerevisiae) DGAT2 using mutagenesis in conjunction with chemical modification. Our data provide evidence that both the N and C termini are oriented toward the cytosol and have different catalytic roles. A highly conserved motif, 129YFP131, and a hydrophilic segment exclusive to yeast DGAT2 reside in a long endoplasmic reticulum luminal loop following the first transmembrane domain and play an essential role in enzyme catalysis. In addition, the strongly conserved His195 within the motif HPHG, which may play a role in the active site of DGAT2, is likely embedded in the membrane. These results indicate some similarities to the topology model of murine DGAT2 but also reveal striking differences suggesting that the topological organization of DGAT2 is not ubiquitously conserved.
Keywords: Enzyme Mutation, Enzymes, Lipid, Membrane Enzymes, Membrane Proteins, Chemical Modification, Cysteine, Membrane Topology, Saccharomyces cerevisiae, Site-directed Mutagenesis
Introduction
In eukaryotes, triacylglycerol (TG)2 serves as both a source of energy and a reservoir of fatty acids for membrane formation and maintenance. Acyl-CoA:diacylglycerol acyltransferase (DGAT, EC 2.3.1.20) is found mainly in the endoplasmic reticulum (ER) and catalyzes the final and committed step in acyl-CoA-dependent TG biosynthesis (1, 2).
There are at least two known microsomal DGAT family members, DGAT1 and DGAT2, that do not share substantial sequence homology and are proposed to have distinct physiological roles in TG metabolism. DGAT1 is a member of a superfamily of membrane-bound O-acyltransferases (3), whereas DGAT2 belongs to a family that includes acyl-CoA:monoacylglycerol acyltransferase and acyl-CoA:wax alcohol acyltransferase (4). Orthologues of both DGAT1 and DGAT2 have been widely identified in animals and plants. Reverse genetic studies in DGAT knock-out mice have demonstrated that DGAT2 plays a dominant role in TG biosynthesis in mammals (5). Down-regulation of DGAT2 expression in mice resulted in increased resistance to obesity as well as significant improvement in hepatic steatosis and insulin sensitivity (6–8). DGAT2 expression levels are also considered to be related to the skin disease psoriasis (9). Therefore, DGAT2 represents an important therapeutic target for management or treatment of these disorders. In certain plant species, DGAT2 plays a role in the selective accumulation of unusual fatty acids into TG (10, 11). Overexpression of these DGAT2s in crop plants represents a potential means of producing novel value-added oils to meet the growing demand from industry (12). In fungi such as the yeast Saccharomyces cerevisiae, DGAT2 (also known as Dga1p) is the only characterized DGAT and contributes to most of the TG synthesis (13). Yet a detailed molecular understanding of DGAT2 function is lacking due to the intrinsic technical difficulties associated with purification and crystallization of this membrane protein (14). To date, no three-dimensional structural data have been reported for DGAT-related proteins.
Recently, there have been few reports providing insights into the molecular aspects of DGAT2-related enzymes. Experimental data from murine DGAT2 have led to a topology model in which DGAT2 is proposed to possess one or two adjacent transmembrane domains (TMDs) at the N terminus, with both termini residing in the cytosol. In this model, the bulk of the protein at the C terminus faces toward the cytosolic side and likely contains the active site of the enzyme (15). Studies using tung tree DGAT2 expressed in tobacco cells also demonstrated that both the N and C termini were exposed to the cytosol (10). Although the location of the putative TMDs was consistent with the predicted result, it remains unclear whether the N terminus of the protein consists of two TMDs or one long domain embedded in the membrane bilayer. Site-directed mutagenesis showed that two conserved regions are catalytically essential for murine DGAT2 (15). One region in the first TMD is proposed to be a putative neutral lipid-binding domain, and the other region is the tetrapeptide HPHG, facing the cytosol.
As an initial step toward functional and structural analysis of this enzyme, we previously used yeast DGAT2 (ScDGAT2) as a model to probe the role of cysteines in enzyme catalysis and structural support; we found that cysteines were not essential to either of these processes, but a widely conserved motif at the C terminus of yeast DGAT2 was identified that may include an active site or be involved in protein folding (16).
In a continuing investigation to better understand the molecular mechanism of this enzyme, we have probed the functional and structural importance of signature motifs in ScDGAT2. Through in silico analyses, we uncovered unique sequence features and conserved motifs in ScDGAT2. Mutagenesis revealed that some of the studied motifs are functionally important for ScDGAT2 activity, including a previously undisclosed hydrophilic stretch found only in this enzyme. We then evaluated their topological orientation using chemical modification of single cysteines. Our work provides new experimental evidence that ScDGAT2 has a different topology compared with murine DGAT2.
EXPERIMENTAL PROCEDURES
Materials
Zymolase 100T was obtained from Seikagaku Corp. (East Falmouth, MA). Complete protease inhibitor mixture tablets was purchased from Roche Applied Science. [14C]Oleoyl-CoA was from GE Healthcare. Diolein was from Avanti Polar Lipids (Alabaster, AL). mPEG5000-maleimide (PEG-mal, 5 kDa) was obtained from SunBio (Orinda, CA). Anti-V5-HRP antibodies were purchased from Invitrogen. Rabbit anti-Kar2p antibodies were from Santa Cruz Biotechnology (Redwood City, CA). The goat anti-rabbit-HRP secondary antibodies were from Zymed Laboratories Inc.
Construction of DGAT2 Mutants
The coding region for the yeast S. cerevisiae DGAT2 was PCR-amplified and ligated into the yeast expression vector pYES2.1-TOPO (Invitrogen) and fused with a C-terminal V5 tag (referred to as ScDGAT-V5). N- and C-terminal truncation mutants N1(Δ1–62), N2 (Δ1–33), C1 (Δ374–418), C2 (Δ391–418), C3 (Δ413–418), C4 (Δ413–418, 413::A6), as well as N3, in which V5 tag was fused to N terminus instead, were similarly constructed. Other deletion, insertion, and point mutants, including TM (Δ70–91), DS1 (Δ150–187), DS2 (Δ156–174), DS3 (162::163–172), DS4 (162::GA5), H193A, H195A, EPHS (H193E/G196S), YFP (Y129A/F130A/P131A), F71A, and L73A, were constructed by PCR-driven overlapping extension according to the method of Heckman and Pease (17) using ScDGAT2-V5 as a template. Plasmids coding for various single Cys-containing mutants were generated by the same method using a previously constructed Cys-less mutant (C0) of ScDGAT2-V5 as the template, in which all seven endogenous cysteines were mutated to alanines (16). A list of primers used for the construction of various mutants is given in supplemental Table S1. A schematic representation of each mutant is also shown in supplemental Fig. 1. All constructs were sequenced to verify their integrity and that the genes were cloned in-frame with the epitope tag.
Strains and Culture Conditions
A yeast quadruple knock-out strain H1246 (MATα are1-Δ::HIS3, are2-Δ::LEU2, dga1-Δ::KanMX4, lro1-Δ::TRP1 ADE2), which is devoid of DGAT activity and TG biosynthesis, was used as the host strain for protein expression experiments (provided by Dr. S. Stymne and U. Ståhl) (13). Constructs were transformed into the yeast cells using the lithium acetate/single-stranded carrier DNA/PEG method (18). Yeast cells were also transformed with an empty vector or pYESLacZ as a control. The transformants were selected on plates lacking uracil and cultivated in yeast nitrogen base with 2% (w/v) dextrose (YNBD) medium. Cells were then harvested, washed with water, and inoculated in induction medium where dextrose was replaced by 2% (w/v) galactose and 1% (w/v) raffinose to induce the gene expression.
Yeast Microsome Preparation
Right side-out microsomes were prepared as described elsewhere with the following modifications (19). Cells were collected, washed with water, and resuspended in spheroplasting buffer (1.2 m sorbitol, 50 mm potassium acetate, 20 mm Hepes, pH 7.5, 2 mm dithiothreitol). Cells were then converted to spheroplasts by incubating at 30 °C for 15 min in the presence of 0.75 mg/ml zymolase 100T. The spheroplasts were collected by centrifugation at 1000 × g for 5 min at 4 °C and resuspended in lysis buffer (100 mm sorbitol, 50 mm potassium acetate, 20 mm Hepes, pH 7.4) supplemented with complete protease inhibitor mixture. Spheroplasts were lysed by 20 strokes in a Dounce homogenizer. Unlysed cells and cell debris were pelleted by another centrifugation at 1000 × g. The supernatant was subjected to centrifugation at 100,000 × g for 30 min at 4 °C to recover the microsomes. The resulting pellets were resuspended in 10 mm Hepes-NaOH, pH 7.4.
DGAT Assay
Equivalent amounts of microsomal protein as determined by BCA assay (Pierce) were used for in vitro DGAT activity assays that were performed using a previously described method (16). Briefly, DGAT activity was measured as the production of [14C]TG from [14C]oleoyl-CoA and unlabeled dioleoyl-DAG in a 10-min assay. TG was separated from the reaction mixture by TLC, and the radioactivity was quantified using a Beckman-Coulter LS6500 liquid scintillation counter.
Nile Red Fluorescence Detection of TG in Yeast
The Nile red fluorescence assay was performed as described previously (20, 21). Briefly, yeast cells expressing recombinant ScDGAT2 and ensuing mutants were cultivated in induction medium for 24 h. Ninety five microliters of cell suspension were placed in a 96-well dark flat-bottom plate (UNIPLATE, Whatman). The background fluorescence was measured with 485 and 538 nm emission and excitation filters, respectively, using a plate fluorimeter (Fluoroscan Accent, Thermo, Milford, MA). Five microliters of Nile Red solution (0.8 mg/ml) were added to the culture, and a second fluorescence measurement was conducted under the same conditions. The first measurement was subtracted from the second measurement, and the results were divided by A600 nm with appropriate dilutions. The assays were conducted with three individual cultures for each ScDGAT2 mutant and three technical replicates.
Fractionation and Extraction of Membrane Proteins
Preparation of microsomes was performed as described above. An equal volume of 2% SDS (w/v) or 0.2 m Na2CO3, pH 11, was added to the suspended microsomal proteins. After incubation at 4 °C for 1 h, samples were centrifuged at 100,000 × g for 30 min at 4 °C to separate the supernatant and pellet fractions. The pellets were brought up to the same volume as the supernatant with resuspension buffer (10 mm Hepes-NaOH, pH 7.4). Samples from supernatant and pellet were mixed with SDS loading buffer and subjected to SDS-PAGE and immunoblotting.
Thiol-specific Chemical Blocking and Modification
Thiol-specific chemical blocking with N-ethylmaleimide (NEM) followed by modification with PEG-mal under nondenaturing and denaturing conditions was performed using modifications of the procedures described by Wang et al. (22). For modification under nondenaturing conditions, microsomal protein was diluted to 1 mg/ml and aliquoted equally for one of three treatments as follows: 1) 1 h of incubation on ice with 0.5 mm PEG-mal (to modify cysteines facing the cytoplasm), followed by addition of dithiothreitol (DTT) to 10 mm and further incubation for 10 min to quench the reaction; 2) 30 min of incubation with 5 mm NEM to block accessible sulfhydryl groups, followed by reacting with PEG-mal and quenching with DTT as above; 3) untreated control.
For modification under denaturing conditions, diluted microsomal vesicles were divided into 2 aliquots. One was directly treated with 0.5 mm PEG-mal in the presence of 2% SDS followed by addition of DTT and incubation for 10 min as described above. The other half was first incubated with 5 mm NEM on ice for 30 min in the absence of SDS followed by immediate ultracentrifugation (100,000 × g, 30 min). The pellet was washed once with 1 ml of ice-cold resuspension buffer to remove excess NEM. After another ultracentrifugation, the pellet was resuspended in 50 μl of resuspension buffer. The sample was then incubated with 0.5 mm PEG-mal in the presence of 2% SDS at room temperature for 20 min. The reaction was finally quenched by incubation with 10 mm DTT for 10 min. All aliquots were mixed with sample buffer and subjected to SDS-PAGE.
SDS-PAGE and Immunoblotting Analysis
Protein samples were resolved by 10% SDS-PAGE and transferred to a PVDF membrane. V5-tagged ScDGAT2 mutants were detected using anti-V5-HRP antibodies according to the protocol described by the manufacturer. Control protein chaperone Kar2p was detected using anti-Kar2p as the primary antibody followed by incubation with goat anti-rabbit-HRP secondary antibody. The bound antibodies were detected using the ECL detection system (GE Healthcare). Densitometry analysis of immunoblots was performed using ImageQuant TL software (GE Healthcare).
Computational Methods
The following algorithms were used for predicting the topological organization of DGAT2: HMMTOP (23), TMHMM (24), TopPred (25), SVMtm (26), SOSUI (27), Conpred II (28), TMpred (29). Sequence alignments were conducted with Geneious (Biomatters Ltd.) using the ClustalW algorithm. Sequence alignments were performed using homologous DGAT2 polypeptides obtained by BLAST search. Sequences from the different organisms with corresponding accession numbers are as follows: Ac, Ajellomyces capsulatus, XP_001540241; Acl, Aspergillus clavatus, XP_001273210; Ao, Aspergillus oryzae, XP_001822244; At, Arabidopsis thaliana, NP_566952; Bf, Branchiostoma floridae, XP_002208225; Bt, Bos taurus, CAD58968; Ce, Caenorhabditis elegans, CeDGAT2a, CAB04533, CeDGAT2b, AAB04969; Ci, Coccidioides immitis, XP_001240299; Cn, Cryptococcus neoformans, EAL20089; Dd, Dictyostelium discoideum, XP_635762; Gz, Gibberella zeae, XP_381525; Hs, Homo sapiens, AAK84176; Lb, Laccaria bicolor, EDR14458; Mg, Magnaporthe grisea, XP_368741; Mm, Mus musculus, AAK84175; Mt, Medicago truncatula, ACJ84867; Nc, Neurospora crassa, CAE76475; Nf, Neosartorya fischeri, XP_001261291; Os, Oryza sativa, NP_001057530; Pm, Penicillium marneffei, XP_002146410; Pn, Phaeosphaeria nodorum, EAT89076; Pp, Physcomitrella patens, XP_001777726; Pt, Populus trichocarpa, XP_002317635; Rc, Ricinus communis, AAY16324; Sc, S. cerevisiae, NP_014888; Sp, Schizosaccharomyces pombe, XP_001713160; Ts, Talaromyces stipitatus, EED21737; Um, Ustilago maydis, XP_760084; Ur, Umbelopsis ramanniana, UrDGAT2a, AAK84179, UrDGAT2b, AAK84180; Vf, Vernicia fordii, ABC94474; Vv, Vitis vinifera, CAO68497; and Zm, Zea mays, ACG38122.
RESULTS
Sequence Motifs in ScDGAT2
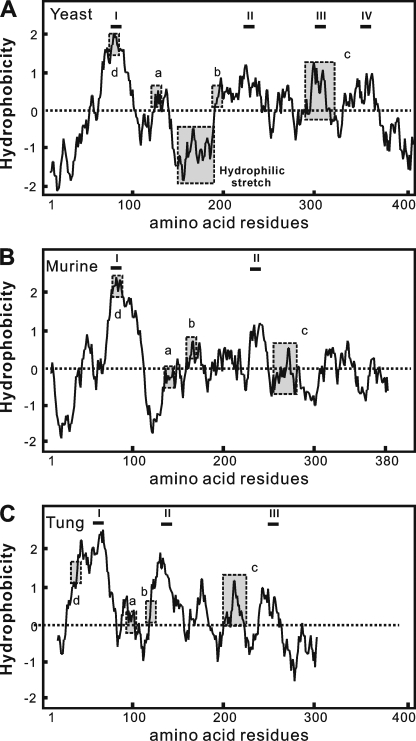
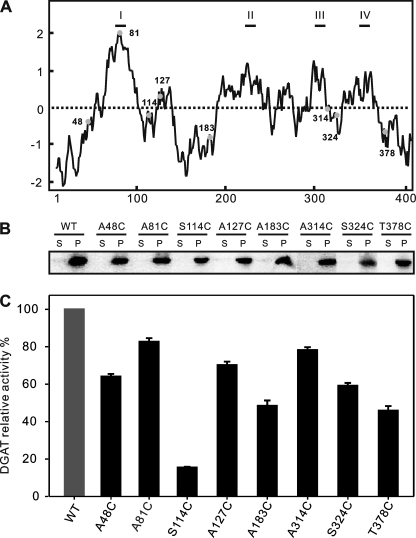
Multiple sequence alignment is an essential tool for predicting important structural features of proteins. To identify DGAT2 conserved motifs in silico, 34 representative polypeptides from a wide range of organisms were aligned using Clustal W algorithm. Three regions with remarkable sequence conservation were located (Fig. 1). First, the motif YFP (129YFP131 in ScDGAT2) is completely conserved in DGAT2 from different organisms with the exception of castor bean, where it corresponds to HFP. Second, the motif HPHG (193HPHG196 in ScDGAT2) is conserved in sequences from animals and fungi and is essential for DGAT2 activity in mice (15). In plant DGAT2s, the corresponding motif is EPHS. The third motif consisting of RXGFX(K/R)XAXXXGXXX(L/V)VPXXXFG(E/Q) spans the longest conserved region in DGAT2-homologous proteins. This motif is found between residues 288 and 311 in ScDGAT2 and is in close proximity to Cys314, which has been previously demonstrated to be the locus of NEM-mediated enzyme inhibition (16). It is noteworthy that a putative neutral lipid-binding domain, FLXLXXXn (where n is a nonpolar amino acid), earlier reported in murine DGAT2 (15), could not be identified in ScDGAT2. Instead, the corresponding motif 71FVLF74 is found between positions 71 and 74. This domain was not identified in plant orthologues as well. The sequence alignment also showed a segment of 38 amino acids situated between YFP and HPHG motifs (positions 150–187 in ScDGAT2), which is unique to ScDGAT2 and absent in plant, animal, and other fungal DGAT2s.
FIGURE 1.
Sequence motifs in DGAT2 family. Thirty four DGAT2 polypeptides from animals, plants, and fungi were aligned by Geneious (Biomatters Ltd.). The conserved motif YFP and HPHG are indicated. Abbreviations for each DGAT2 polypeptide indicating the organism of origin and corresponding accession numbers are described under “Experimental Procedures.”
Hydropathy Profile Analysis and TMD Prediction of DGAT2s
To further explore the potential structural aspects using in silico approaches, we evaluated the hydropathy profile and topology distribution of ScDGAT2 in comparison with murine and tung tree DGAT2s, whose topological orientations were experimentally tested (15). The distribution of sequence motifs in yeast, murine, and tung DGAT2 was compared in Kyte-Doolittle (30) hydropathy plots (Fig. 2, dashed boxes). Generally, these three proteins show similar plots. The N-terminal region of each protein contains a hydrophilic stretch followed by a hydrophobic region except that the hydrophilic tail in tung DGAT2 is shorter. ScDGAT2 and tung DGAT2, however, contain other segments that are more hydrophobic than murine DGAT2. In particular, ScDGAT2 has a contiguous sequence of mostly hydrophobic amino acids between residues 192 and 250 that contain the motif HPHG (Fig. 2A). A highly hydrophobic stretch, including the motif EPHS, was also observed in tung DGAT2 between positions 117 and 157 (Fig. 2C). In addition, the 38-residue segment found exclusively in ScDGAT2 (Fig. 2A) resides in a hydrophilic region and is positioned between the conserved motifs 129YFP131 and 193HPHG196.
FIGURE 2.
Hydropathy analysis of yeast murine and tung DGAT2s. Aligned hydropathy plots of yeast (A), murine (B), and tung tree (C) DGAT2 proteins were generated by the method of Kyte and Doolittle (30) using a window size of 19. The most hydrophobic segments, which may contain putative TMDs, are indicated by roman numerals. Motifs studied here are boxed and labeled as follows: a, YFP; b, HPHG; c, RXGFX(K/R)XAXXXGXXX(L/V)VPXXXFG(E/Q); and d, putative lipid-binding site.
We then subjected three DGAT2 sequences to seven common topological prediction programs based on different algorithms (Table 1). Murine DGAT2 was predicted to have one or two abutting TMDs located at the N-terminal region and an N terminus facing the cytosol, which is in agreement with the experimentally determined topology. The N terminus of tung DGAT2 was also predicted to face the cytosolic side that is consistent with previous experimental evidence. However, six out of seven programs predicted that tung DGAT2 has at least three TMDs, which include two adjoining TMDs at N-terminal region and another TMD (between positions 112 and 139) in the hydrophobic region described above. In the case of ScDGAT2, the orientation of the N terminus was generally predicted to be toward the cytosol with three to four TMDs in the polypeptide. The first TMD (residues 66–97) was predicted by all algorithms and is comparable with the model of murine DGAT2. There was more variability, however, in the predictions for the location of putative TMDs in the region between residues 188 and 236. Two additional TMDs were predicted between amino acids 290–313 and 341–359. These predictions corroborate the hydropathy plot analysis suggesting that DGAT2s from other kingdoms may have a different topology compared with murine DGAT2, which could affect the orientation of signature motifs discussed above.
TABLE 1.
Predicted TMDs in yeast, murine, and tung tree DGAT2s
The polypeptides corresponding to yeast (S. cerevisiae), murine (M. musculus), and tung tree (V. fordii) DGAT2s were submitted to seven transmembrane prediction algorithms.
| HMMTOP | TMHMM | TopPred | SVMtm | SOSUI | ConPred II | TMpred | |
|---|---|---|---|---|---|---|---|
| ScDGAT2 | 68–92 | 70–92 | 68–88 | 66–97 | 75–97 | 73–93 | 72–92 |
| 193–207 | 188–210 | 189–209 | 196–214 | ||||
| 200–224 | 215–235 | 217–231 | 213–234 | 215–235 | 216–236 | ||
| 294–310 | 290–310 | 293–313 | |||||
| 341–359 | |||||||
| No. of TMD | 4 | 1 | 3 | 3 | 3 | 4 | 3 |
| Orientationa | IN | IN | IN | NA | NA | IN | IN |
| MmDGAT2 | 68–92 | 73–95 | 76–96 | 73–93 | 66–88 | 70–90 | 76–96 |
| 95–109 | 93–115 | ||||||
| 230–250 | 224–248 | ||||||
| No. of TMD | 1 | 1 | 2 | 2 | 2 | 1 | 2 |
| Orientationa | IN | IN | IN | NA | NA | IN | IN |
| VfDGAT2 | 14–36 | ||||||
| 29–47 | 24–46 | 28–48 | 27–52 | 28–48 | 25–46 | ||
| 52–70 | 50–72 | 53–73 | 54–69 | 47–69 | 51–71 | 50–72 | |
| 121–139 | 112–134 | 119–139 | 122–136 | 119–139 | 121–139 | ||
| 170–187 | 168–188 | 169–189 | 170–193 | ||||
| 208–226 | 211–231 | 211–231 | |||||
| 247–265 | 211–231 | 246–266 | 250–273 | ||||
| No. of TMD | 6 | 3 | 6 | 3 | 2 | 5 | 6 |
| Orientationa | IN | IN | IN | NA | NA | IN | IN |
a Orientation was of the N terminus. Cytosol, IN.
Site-directed Mutagenesis of Signature Motifs
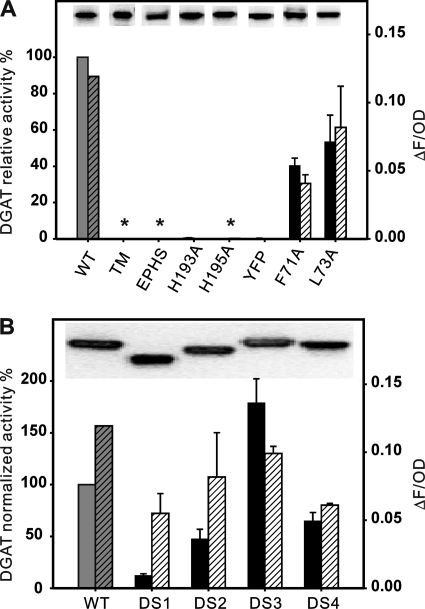
To determine the functional significance of these motifs in ScDGAT2, we conducted site-directed mutagenesis using recombinant in S. cerevisiae strain H1246. The impact of these mutations was evaluated by measuring the in vitro enzyme activity and protein expression level using isolated microsomes as well as through in vivo detection of TG produced by Nile Red assay of the yeast cultures. Because the strain H1246 is devoid of DGAT activity, results are a direct measurement of the performance of each mutant. The expression of wild-type (WT) ScDGAT2 and the ensuing mutants was verified using immunoblotting by taking advantage of the C-terminal V5 tag. As shown in Fig. 3A, substitution of 129YFP131 to 129AAA131 (mutant YFP) and His193 to Ala193 (mutant H193A) led to almost complete loss of enzyme activity. Similarly, substitution of His195 to Ala195 and replacement of 193HPHG196 to corresponding motif EPHS found in plant DGAT2s (mutants H195A and EPHS, respectively) both abolished the enzyme activities. Deletion of the first putative TMD between residues 70 and 91 also resulted in the total loss of activity (mutant TM). The Phe71 to Ala71 and Leu73 to Ala73 substitution in the sequence corresponding to the putative lipid-binding motif in murine DGAT2 (mutant F71A and L73A, respectively) retained significant activity (over 40%) compared with the WT ScDGAT2. These effects correlated with the quantity of TG produced by yeast cells determined by Nile Red fluorescence. Immunoblotting analysis of microsomes used for the enzyme assays revealed similar accumulation of each mutant protein in comparison to WT (Fig. 3A, insets), indicating that the observed changes in activity were not a result of differential expression levels.
FIGURE 3.
Site-directed mutagenesis of signature motifs. A, performance of mutants TM (Δ70–91), H193A, H195A, EPHS (H193E/G196S), YFP (Y129A/F130A/P131A), F71A, and L73A. In vitro enzyme activities are expressed as picomoles of TG formed per min per mg of microsomal protein and quoted relative to the activity of WT protein (257 pmol/mg protein/min, defined as 100%). Solid bars represent in vitro activity conducted with microsomes from yeast cells expressing V5-tagged ScDGAT2 (WT) and mutants, incubated in the presence of diolein and [14C]oleoyl-CoA, and assayed as described under “Experimental Procedures.” Hatched bars represent the levels of TG accumulation quantified by Nile Red fluorescence (ΔF), normalized by the optical density (OD) at 600 nm, and subtracted from the value obtained for a negative control (LacZ, data not shown). Values represent average ± S.D. (n = 3), and asterisks indicate mutants that did not present activity. Immunoblots of the microsomal proteins prepared from cells expressing these mutants are shown in the insets. B, performance of mutants DS1 (Δ150–187), DS2 (Δ156–174), DS3 (162::163–172), and DS4 (162:: GA5). The in vitro enzyme relative activities (as described in Fig. 3A) are normalized by expression level of the protein that was measured through comparative densitometry analyses of immunoblots. Specific activity of WT protein is 270 pmol of TG/mg protein/min.
The fact that the hydrophilic stretch between motif 129YFP131 and 193HPHG196 (Fig. 2A) is unique for ScDGAT2 raised the question of whether this segment plays a role in enzyme function. To address this, we performed deletion and insertion mutagenesis on this segment. Deletion of the whole segment (mutant DS1, Δ150–187) resulted in an enzyme with less than 20% of WT activity (Fig. 3B), whereas removal of a smaller segment (mutant DS2, Δ156–174) preserved around 50% of the original activity. In view of these results, we hypothesized that insertions in this region might have a positive effect on enzyme activity. Thus, we duplicated a stretch of 10 amino acids from residues 163 to 172 at position 162 creating mutant DS3 (162::163–172). This mutant showed an 80% increase in enzyme activity compared with the WT. To investigate whether this effect was specific to the sequence duplicated, we inserted a decapeptide composed of (GA)5 in the same position. In contrast to DS3, the resulting mutant, (DS4, 162::GAGAGAGAGA) presented a decrease of activity of about 40% compared with WT, suggesting that the positive effect on the activity in DS3 was sequence-related. The level of protein expression of each mutant is comparable with WT (Fig. 3B, inset); therefore, the trend in activity was not due to the different protein accumulation. As expected, no DGAT activity was detected in H1246 yeast cells transformed with empty vector or the control pYESLacZ (data not shown). Detection of TG accumulation in these yeast cultures showed a similar trend between mutants DS1 to DS4, but interestingly, the relationship between mutants and the WT was different when determining in vivo TG levels. Nonetheless, these results show that the unique hydrophilic stretch in ScDGAT2 may play a key role in modulating enzyme activity.
ScDGAT2 Is an Integral Membrane Protein
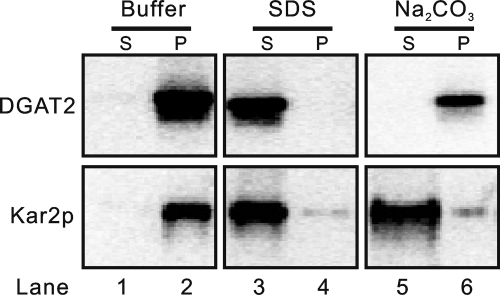
Our previous studies have indicated that ScDGAT2 can be copiously detected in the microsomal membrane fraction (16), which was also observed in the immunoblots in Fig. 3. Hydropathy analysis and topology prediction suggest that ScDGAT2 is likely an integral membrane protein and probably has a different topology compared with murine DGAT2. Before exploring topological details, we sought to experimentally determine the nature of membrane association of ScDGAT2. Microsomes containing V5-tagged ScDGAT2 were extracted with either detergent (SDS) or alkaline salt (Na2CO3) and then separated by ultracentrifugation into supernatant and pellet, which were analyzed by immunoblotting. As shown in Fig. 4 (lanes 3 and 4), ScDGAT2 could be extracted from the membrane and was present in the supernatant only when microsomes were treated with 1% SDS detergent, a condition that disrupts the integrity of membrane bilayers. In contrast, when the membrane was treated with 0.1 m Na2CO3, an alkaline salt commonly used to strip peripheral membrane proteins, also releasing luminal proteins, DGAT2 remained in the pellet (Fig. 4, lanes 5 and 6). The same experiment was conducted using Kar2p, a well characterized chaperone present in the ER lumen (31). As expected, this control protein was detected in the soluble fractions after treatment with either SDS or Na2CO3 (Fig. 4, lanes 3–6). These results clearly showed that ScDGAT2 is strongly associated with microsomal membranes.
FIGURE 4.
Yeast DGAT2 is an integral membrane protein. Microsomes were prepared from yeast cells expressing ScDGAT2-V5 (see “Experimental Procedures”). The membrane was incubated with buffer (10 mm Hepes-NaOH, pH 7.4) alone (lanes 1 and 2) or buffer containing 1% SDS (lanes 3 and 4) or 0.1 m Na2CO3 at pH 11 (lanes 5 and 6). After incubation, samples were separated by ultracentrifugation into the supernatant (S) or pellet (P) that was subjected to immunoblotting analysis with anti-V5 or anti-Kar2p antibodies.
Integrity and Sidedness of Microsomal Vesicles
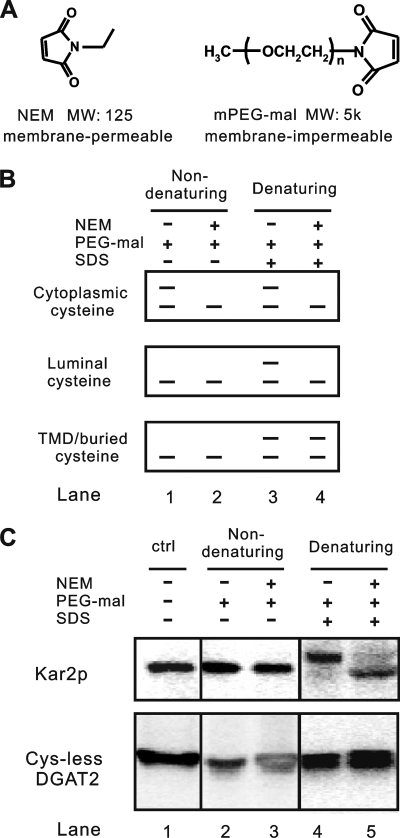
Topology is a fundamental aspect of the structure of membrane proteins. Determination of the topological orientation of signature motifs in ScDGAT2 is an important step toward understanding the catalytic mechanism of this protein. Previously, we constructed a Cys-less ScDGAT2 in which all seven native cysteine residues were substituted with alanines. This mutant retained significant activity and was expressed at comparable levels to the WT (16), indicating that it likely has a similar structure to the native ScDGAT2. These findings provide the basis for mapping membrane topology by probing the accessibility of reintroduced single cysteine residues in the protein to thiol-modifying reagents. Modification of each single cysteine can be detected by distinct band shift patterns on an immunoblot by using thiol-specific reagents with different membrane permeabilities and molecular weights (i.e. NEM and PEG-mal in Fig. 5A). The position of reintroduced single cysteine residues relative to a TMD can yield three different immunoblot patterns under four modification conditions, as schematically represented in Fig. 5B. If the target cysteine is exposed to the cytosol, PEG-mal, which has a large and membrane-impermeable carbohydrate moiety, can be attached, leading to a detectable band shift (∼5 kDa) in both SDS denaturing and nondenaturing conditions (Fig. 5B, lanes 1 and 3). This band shift will not appear if NEM, which has smaller size (125 Da), is used to block the cysteine prior to PEG-mal modification (Fig. 5B, lanes 2 and 4). Detection of a cysteine exposed to the luminal side is characterized by a band shift with PEG-mal treatment that occurs only after membranes are disrupted under SDS-denaturing conditions (Fig. 5B, lanes 1 and 3). This shift, however, is prevented by prior exposure of protein to NEM that can permeate membranes and thus block cysteine residues on the luminal side (Fig. 5B, lanes 2 and 4). Because the reaction between the NEM and the ionized form of thiol requires a water molecule as a proton acceptor, NEM is virtually unreactive to cysteine residues embedded in the membrane bilayer or at a protein-protein interface (32, 33). This property can be used to detect buried cysteine residues that can be characterized by a band shift when NEM is removed following PEG-mal treatment in the presence of SDS (Fig. 5B, lane 4).
FIGURE 5.
Thiol-specific modification to map membrane topology. A, chemical structures and properties of NEM and PEG-mal. B, schematic depiction of immunoblot patterns from the cysteine accessibility assay. C, membrane integrity and orientation as determined by thiol-specific modification. Microsomal membranes were prepared as detailed under “Experimental Procedures” and treated or not with NEM under nondenaturing conditions, followed by treatment with PEG-mal in the presence or absence of SDS. In the condition that NEM-treated sample was used for PEG-mal modification in the presence of SDS, the NEM was removed. The reaction mixtures were analyzed by immunoblotting using anti-Kar2p and anti-V5 antibodies to detect Kar2p and Cys-less DGAT2, respectively. The results are representative of two independent experiments. ctrl, control.
This methodology, originally developed from the gel-shift assay described by Lu and Deutsch (34), has been successfully applied for membrane proteins in yeast (22). One major assumption with the use of this approach is that topological orientation of microsome vesicles is identical to that of the ER in vivo. To test this hypothesis, we examined the accessibility of the ER luminal protein Kar2p, which contains a single native cysteine at the N terminus, under four different conditions depicted in Fig. 5B. Immunoblotting analysis of cell fractions showed that Kar2p was detected only in the microsomal fraction (supplemental Fig. 2), indicating that the vesicles remained sealed during preparation and can be used for accessibility assay. When the yeast microsomes were incubated with PEG-mal in the presence or absence of NEM under nondenaturing conditions, a band shift was not observed (Fig. 5C, lanes 2 and 3), indicating that Kar2p was not exposed to the cytosol. Under denaturing conditions with SDS, a band shift resulting from the PEG-mal attachment could be detected (Fig. 5C, lane 4). The band representing unmodified protein was not evident, suggesting that efficient modification by PEG-mal occurred. A prior treatment with NEM substantially blocked PEG-mal modification under denaturing conditions (Fig. 5C, lane 5), agreeing with the expected immunoblot pattern for the luminal cysteine residue in Fig. 5B. The same treatment was repeated with Cys-less ScDGAT2 as an additional control (Fig. 5C), and as expected, no band shifts were detected. Thus, we validated the thiol modification methodology, showing also that the microsomal vesicles retained correct orientation.
Activities of Single Cys-containing Mutants
Having demonstrated the utility of the accessibility assay for mapping membrane topology, we constructed a series of single cysteine mutants of ScDGAT2. To minimize structural perturbation, we individually re-introduced three cysteine residues back to their original positions in the native ScDGAT2 using the Cys-less version as the template, creating mutants A48C, A127C, and A183C. In addition, the mutants A81C, S114C, S324C, and T378C were created to evaluate topological organization of motifs that are distant from a native cysteine. In this case, substitutions were performed in positions having low conservation yet residing in close proximity to the desired motif, mitigating possible structural disturbance. The N and C termini were located by using mutants A48C and T378C, respectively. Mutants S114C, A127C, and A183C were used to map the topological organization of 129YFP131 and 193HPHG196 motifs as well as the unique hydrophilic stretch, respectively. Mutant A81C served as a control because it resides in the most hydrophobic segment (position 66–97), universally predicted to be a TMD (Table 1). Together with the previously constructed mutant A314C (16), S324C was created to determine the orientation of the highly conserved domain preceding Cys314. The position of each introduced cysteine residue in the hydropathy plot is shown in Fig. 6A and supplemental Fig. 1.
FIGURE 6.
Expression of various single Cys-containing mutants. A, hydropathy plot of ScDGAT2 showing the position and hydrophobicity environments of each introduced cysteine residue (represented by filled circles with the positions numbered). B, immunoblot showing expression of each mutant. The microsomal pellet (P) was separated from the cytosolic protein in the supernatant (S) by 100,000 × g centrifugation. Equal amounts of protein from each fraction were separated by 10% SDS-PAGE and probed with anti-V5 antibodies. C, relative DGAT activities of the single Cys-containing mutants and WT ScDGAT2. Activities were measured as described under “Experimental Procedures.” Data represent means ± S.D. (n = 3). All activities are quoted relative to that of the WT enzyme (set to 100%). The activity of expressed WT enzyme was 600 pmol/mg protein/min.
The constructs encoding the single cysteine residue forms of ScDGAT2 were expressed in yeast strain H1246 as described previously. To determine whether replacement of a single cysteine residue produced deleterious effects on the protein folding or structure, the ScDGAT expression level and DGAT activity for each mutant were measured. The immunoblotting analysis suggested all single Cys proteins were expressed at levels comparable with WT, localized exclusively in the microsomal fraction with the expected molecular mass of ∼48 kDa (Fig. 6B). In addition, results from in vitro enzyme assays showed that all mutants were functional, although S114C retained only around 20% activity compared with the WT (Fig. 6C). Therefore, we concluded that the effect of cysteine residue replacement in each mutant is tolerated and does not cause gross structural changes in the protein.
N and C Termini Are Located in the Cytosol and Have Different Roles in Enzyme Catalysis
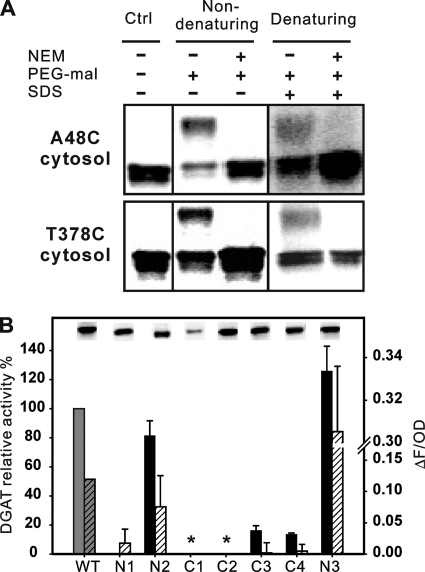
Microsomes were prepared from H1246 cells expressing A48C and T378C and subjected to thiol-specific modification and accessibility analysis as described in Fig. 5B. The results showed that PEG-mal can modify both mutants without NEM blocking under nondenaturing or denaturing conditions (Fig. 7A), indicating a cytoplasmic labeling pattern for both termini of ScDGAT2. To examine the role of the N- and C-terminal tails in DGAT2 function, deletion mutagenesis was carried out. Hydrophilic tails of N and C termini are composed of 62 and 44 amino acid residues, respectively (Table 1 and Fig. 2A). Sequential deletion was performed to remove the entire N terminus (N1) and the first 33 amino acid residues (N2). Similar deletions along the C terminus were also accomplished to remove the entire hydrophilic tail (C1), the last 28 amino acids (C2), and the last six amino acid residues (C3). The impact of these mutations was evaluated by measuring the enzyme activity in vitro and protein expression level using isolated microsomes. Recombinant proteins were expressed with a C-terminal V5 tag to facilitate the detection in immunoblotting. As demonstrated in Fig. 7B, the mutant lacking the entire hydrophilic N terminus (N1) presented minimal activity detected only by Nile Red fluorescence while maintaining a substantial expression level. Removal of the first 33 amino acid residues in the N terminus (N2) resulted in minor decrease in enzyme activity (Fig. 7B). However, deletion of the last six amino acid residues from the C terminus (C3) caused a decrease in the enzyme activity of more than 80%. Deletion of the whole C terminus (C1) completely abolished the enzyme activity and had a substantial impact on the protein accumulation (Fig. 7B, inset); some expression was recovered in the C2 mutant in which about half of the hydrophilic C terminus was deleted, but this mutant also exhibited a complete loss of enzyme activity. To further investigate the importance of the last six amino acid residues, they were replaced with six alanine residues to generate mutant C4. This mutant retained similar activity and expression levels to C3, suggesting that the effect of these variations is related to the last six residues in the C terminus. Considering the recombinant protein used in this study was tagged with a C-terminal V5 epitope, the influence of capping the C terminus was investigated by instead fusing the epitope to the N terminus (mutant N3). The enzyme activity of this mutant was ∼20% higher compared with the WT in which the epitope was attached at the C terminus. These observations were also reflected in in vivo quantification of TG levels using Nile Red. These results indicate that the hydrophilic C-terminal tail of ScDGAT2 is more sensitive to modifications than the N terminus.
FIGURE 7.
Sidedness and mutagenesis analysis of N and C termini. A, accessibility of mutant A48C and T378C to thiol-specific modification. Modification was performed under the same conditions described in Fig. 5. After modification, samples were subjected to immunoblotting analysis using anti-V5 antibodies. The results shown are from one of two independent experiments. Ctrl, control. B, performance of mutants N1 (Δ1–62), N2 (Δ1–33), C1 (Δ374–418), C2 (Δ391–418), C3 (Δ413–418), C4 (Δ413–418, 413::A6), and N3 (N terminus V5 tag and free C terminus). Solid bars represent in vitro enzyme activities quoted relative to the activity of WT enzyme (135 pmol/mg protein/min, defined as 100%) as described in Fig. 3. Hatched bars represent TG accumulation detected by Nile Red fluorescence as described in Fig. 3. Values represent means ± S.D. of experiments conducted in triplicate, and asterisks indicate mutants that did not present activity. Immunoblotting analysis of the microsomes prepared from cells expressing these mutants is shown in the insets.
Topological Analysis of Signature Motifs
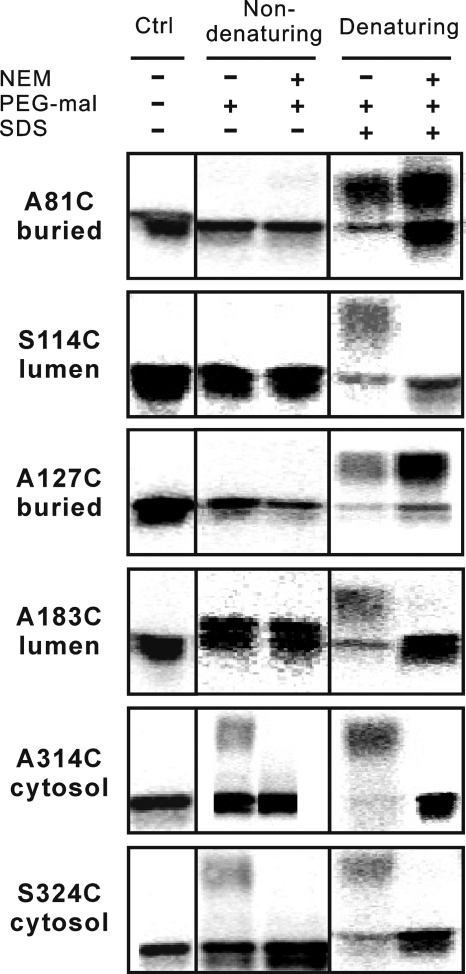
The previous results indicated that both termini are located in the cytosol, which is in agreement with earlier studies on mammalian and plant DGAT2s (10, 15). In silico analyses, however, suggested the existence of additional TMDs in ScDGAT2. To further investigate the topological orientation of signature motifs in ScDGAT2, the accessibility of single cysteine residues in mutants A81C, S114C, A127C, and A183C was analyzed. Thiol-modification experiments showed a buried modification pattern for mutant A81C, which was characterized by band shifts with both treatments under denaturing conditions (Fig. 8). This result indicates a TMD that is likely spanning residues 66–97, which includes the sequence 71FVLF74 corresponding to the putative lipid-binding site in murine DGAT2 (15). For mutant S114C, a luminal modification pattern was observed, which was evidenced by a band shift only under denaturing conditions in the absence of NEM. Although mutant A127C showed a buried modification pattern, it is unlikely that a TMD resides between positions 114 and 183, as it was not predicted with any of algorithms used in Table 1. In fact, only short hydrophobic stretches with a low hydrophobic index are present in this region (Fig. 2A). This is also supported by the fact that a luminal modification pattern was observed for mutant A183C. Therefore, these data indicate that motif 129YFP131 is in the lumen, probably in close contact with the membrane bilayer or in a highly structured region of the protein. Taken together, these results indicate that the hydrophilic segment after the first TMD is an ER luminal loop that contains motif 129YFP131.
FIGURE 8.
Sidedness of signature motifs. Accessibility of single Cys-containing mutants A81C, S114C, A127C, A183C, A314C and S324C to thiol-modifying reagents. Accessibility assay was conducted and detected as described previously (Fig. 5). The results shown are from one of two independent experiments. Ctrl, control.
Topology predictions indicate two putative TMDs from positions 290–313 and 341–359 (Table 1). To map this region, PEG-mal accessibility assays were conducted with mutants A314C and S324C. As shown in Fig. 8, both cysteine residues in these mutants face the cytosolic side, excluding the possibility of a TMD from residues 341 to 359, which was predicted only by one algorithm (HMMTOP). The shifted bands representing the mutant protein modified by PEG-mal were weak, indicating that residues 314 and 324 could be in close contact with the membranes or protected in a protein-protein interface with limited accessibility to PEG-mal. Taking into account in silico analyzes (Table 1 and Fig. 2A), these results suggest that the conserved motif 288RXGFX(K/R)XAXXXGXXX(L/V)VPXXXFG(E/Q)311 preceding Cys314 is the most likely region to be a TMD, returning the C terminus of the polypeptide to the cytosol.
DISCUSSION
In this study, we explored the functional importance and topological orientation of sequence motifs in ScDGAT2 using mutagenesis in conjunction with chemical modification.
Proposed Model and Topological Distribution of Important Signature Motifs in ScDGAT2
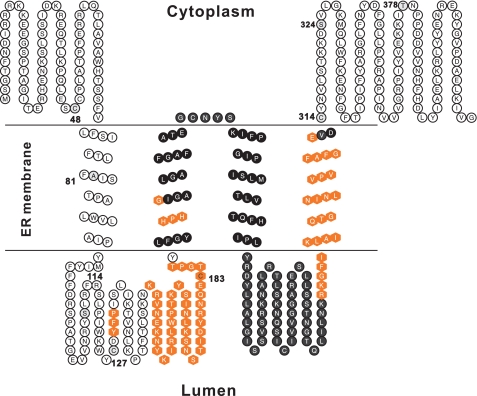
Based on experimental data and in silico analyzes, our current view of the topological distribution of signature motifs in ScDGAT2 is shown in Fig. 9. In this model, ScDGAT2 is proposed to have four TMDs with both termini oriented toward the cytosol, satisfying experimental and computational constraints. In Fig. 9, the residues highlighted in black (from the second to fourth TMD) indicate the orientation of regions established by computational analyses, whereas the other regions were supported by experimental evidence. The exact borders of TMDs were predicted using Conpred II, which applies a consensus technique and is reported to have high prediction accuracy (35).
FIGURE 9.
Proposed topology model of ScDGAT2. This model proposes that ScDGAT2 contains four TMDs with both termini oriented toward the cytosol. Important functional motifs are represented by filled hexagons. Black-filled residues represent regions where topology was established based on in silico analysis only. Topology of other regions was experimentally determined. Positions where single cysteines were introduced are numbered.
Both termini have the same sidedness, but the C terminus may play a more important role in maintaining enzyme activity (Fig. 7B). An ER retrieval motif LKLEI has been previously identified in the extreme C terminus of tung tree DGAT2 (10). The similar motif 413ELKIVG418 is also present in the corresponding region of ScDGAT2 as shown in the multiple sequence alignment (supplemental Fig. 3). Subcellular targeting experiments would help to determine whether 413ELKIVG418 constitutes an ER retrieval motif for ScDGAT2. It is worth noting that the N terminus is not essential for enzyme catalysis in either murine DGAT2 (36) or ScDGAT2, and similar results were recently reported for murine DGAT1 (37). A mitochondrial targeting signal in the N terminus of murine DGAT2 was characterized between amino acids 61 and 66. This sequence is conserved in mammalian DGAT2s but could not be identified in ScDGAT2.
The first TMD likely spans from residue 66 to 97, followed by an ER luminal loop that contains 129YFP131 and the distinctive hydrophilic stretch (Fig. 2A). Following this luminal region, two TMDs were predicted between residues 188 and 236. These adjacent TMDs could also include a single segment embedded in the membrane bilayer. The second TMD contains the conserved domain 193HPHG196. We demonstrate that His195 is essential for the enzyme activity, indicating that it could be directly involved with the active site of the enzyme. This finding is consistent with previous results from murine DGAT1 and DGAT2 in which mutation of a conserved histidine residue also led to a substantial loss in enzyme activity (15, 37). In addition, an invariant histidine residue in a DGAT1-related enzyme, type 1 acyl-CoA:cholesterol acyltransferase (EC 2.3.1.26), has also been proposed to function as a base to facilitate enzyme catalysis in the hydrophobic membrane bilayer (38). Although evidence for His195 being involved in the active site is compelling, it is still possible that the loss of enzyme activity could result from gross structural perturbation caused by substitution of histidine. The fourth TMD, bringing the polypeptide to the cytosol, contains the motif RXGFX(K/R)XAXXXGXXX(L/V)VPXXXFG(E/Q), preceding Cys314.
Differences in Topological and Functional Features between the Yeast and Murine DGAT2
The topological distribution that we suggest for ScDGAT2 resembles the model proposed for murine DGAT2, with both termini facing the cytosol (15). But our work reveals important differences in the topology of yeast and murine DGAT2, supported by several lines of computational and experimental evidence.
A relatively low level of sequence homology and differences in hydrophobic profiles of certain regions were observed between murine and yeast DGAT2s. ScDGAT2 has more hydrophobic segments and was predicted to have more TMDs compared with murine DGAT2 (Fig. 2 and Table 1). In addition, deletion of the first TMD did not affect the association of ScDGAT with microsomal membranes, supporting the idea that other segments in yeast DGAT2 could possibly mediate interaction with membrane bilayers (Fig. 3A, insets). Furthermore, the region following the first TMD, from position 114 to 183, was experimentally proven to reside in the lumen, which was not the case in the model of murine DGAT2. Such divergence in topology between homologous proteins from different categories of organisms has been reported for DGAT1 and serine palmitoyltransferase (37, 39).
These structural differences were also reflected in the unique functional features of ScDGAT2. For instance, the unique hydrophilic stretch in the ER luminal loop, absent in murine DGAT2, was shown to influence enzyme activity in a sequence-dependent fashion (Fig. 3B). It would be worthwhile to mimic this insertion in other DGAT2s to further clarify the role of this unique sequence. Moreover, ScDGAT2 lacks the consensus FLXLXXXn proposed to be the putative lipid-binding site in murine DGAT2, showing sequence 71FVLFSIF77 instead. Thus, it is not surprising that point mutations on Phe71 and Leu73 did not cause significant loss of the enzyme activity. Removal of the first TMD, including these residues, abolished activity (Fig. 3A), indicating that a refinement of this binding domain is needed for yeast DGAT2.
Some of the functional and structural features of DGAT2 discussed here might have resulted from evolutionary adjustments specific to mammals, fungi, and plants. Indeed, site-directed mutagenesis of HPHG to the plant-specific EPHS motif resulted in complete loss of ScDGAT2 activity, corroborating this hypothesis (Fig. 3). Moreover, attempts to recover the activity of nonfunctional Arabidopsis DGAT2 (10) by replacing the EPHS motif with the mammalian/fungal HPHG motif were unsuccessful.3 Furthermore, hydropathy analysis of functional DGAT2s showed divergences between animal, plant, and fungal proteins (supplemental Fig. 4).
Advantages and Limitations of Chemical Modification
In this study, the topological orientation of ScDGAT2 was accessed by chemical modification combined with Cys-scanning mutagenesis that has been used for topology studies of other membrane proteins (22, 38). This methodology has several attractive advantages. Introduction of a single cysteine residue in many positions is tolerated because the cysteine is nonbulky. The chemical modifications were performed on active enzymes that likely retain similar structures to the native protein. Additionally, the library of functional singe Cys mutants could be used for more detailed structural analysis using biophysical approaches such as NMR spectroscopy to yield both static and dynamic information (40). An inherent limitation of this methodology is that when the cysteine residue is in a highly protected region of the protein or located close to the protein-lipid interfaces, it might have limited access to a thiol-modifying reagent (33, 41). Under this condition, careful analysis of the hydrophobicity of the segment is needed for the correct interpretation of results and might require exhaustive scanning at nearby positions as in the case of the Cys127 in ScDGAT2. The sidedness of this introduced cysteine was deduced in the ER lumen only after two cysteine residues at flanking positions were proved to be in the ER lumen as well. A possible strategy to circumvent this hurdle is to use more complicated combinations of sulfhydryl reagents that differ in size and hydrophilicity, as demonstrated in the investigation of topological organization of ACAT1 expressed in mammalian cells (38). With the further development of this strategy in yeast cells, the proposed four-TMD model could be confirmed and/or refined by verifying the orientation of regions in the model established only by computational constraints.
In conclusion, we have provided new insights into the functional and structural characteristics of DGAT2 from S. cerevisiae using a combination of experimental and computational analysis. An updated topology model for ScDGAT2 has been proposed, which can serve as a basis for further structure-function studies on this enzyme.
Acknowledgments
We thank Dr. Sten Stymne and Ulf Ståhl for the yeast strains. We also thank Dr. Zhanyun Guo for useful discussion.
This work was supported by Natural Sciences and Engineering Research Council of Canada, the Canada Research Chairs Program, the Canada Foundation for Innovation, and the University of Alberta.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4 and Table S1.
Q. Liu, R. M. P. Siloto, and R. J. Weselake, unpublished data.
- TG
- triacylglycerol
- DGAT
- acyl-CoA:diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- ORF
- open reading frame
- TMD
- transmembrane domain
- NEM
- N-ethylmaleimide
- PEG-mal
- mPEG5000-maleimide.
REFERENCES
- 1. Lung S. C., Weselake R. J. (2006) Lipids 41, 1073–1088 [DOI] [PubMed] [Google Scholar]
- 2. Yen C. L., Stone S. J., Koliwad S., Harris C., Farese R. V., Jr. (2008) J. Lipid Res. 49, 2283–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hofmann K. (2000) Trends Biochem. Sci. 25, 111–112 [DOI] [PubMed] [Google Scholar]
- 4. Turkish A. R., Henneberry A. L., Cromley D., Padamsee M., Oelkers P., Bazzi H., Christiano A. M., Billheimer J. T., Sturley S. L. (2005) J. Biol. Chem. 280, 14755–14764 [DOI] [PubMed] [Google Scholar]
- 5. Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V., Jr. (2004) J. Biol. Chem. 279, 11767–11776 [DOI] [PubMed] [Google Scholar]
- 6. Yu X. X., Murray S. F., Pandey S. K., Booten S. L., Bao D., Song X. Z., Kelly S., Chen S., McKay R., Monia B. P., Bhanot S. (2005) Hepatology 42, 362–371 [DOI] [PubMed] [Google Scholar]
- 7. Choi C. S., Savage D. B., Kulkarni A., Yu X. X., Liu Z. X., Morino K., Kim S., Distefano A., Samuel V. T., Neschen S., Zhang D., Wang A., Zhang X. M., Kahn M., Cline G. W., Pandey S. K., Geisler J. G., Bhanot S., Monia B. P., Shulman G. I. (2007) J. Biol. Chem. 282, 22678–22688 [DOI] [PubMed] [Google Scholar]
- 8. Liu Y., Millar J. S., Cromley D. A., Graham M., Crooke R., Billheimer J. T., Rader D. J. (2008) Biochim. Biophys. Acta 1781, 97–104 [DOI] [PubMed] [Google Scholar]
- 9. Wakimoto K., Chiba H., Michibata H., Seishima M., Kawasaki S., Okubo K., Mitsui H., Torii H., Imai Y. (2003) Biochem. Biophys. Res. Commun. 310, 296–302 [DOI] [PubMed] [Google Scholar]
- 10. Shockey J. M., Gidda S. K., Chapital D. C., Kuan J. C., Dhanoa P. K., Bland J. M., Rothstein S. J., Mullen R. T., Dyer J. M. (2006) Plant Cell 18, 2294–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kroon J. T., Wei W., Simon W. J., Slabas A. R. (2006) Phytochemistry 67, 2541–2549 [DOI] [PubMed] [Google Scholar]
- 12. Snyder C. L., Yurchenko O. P., Siloto R. M., Chen X., Liu Q., Mietkiewska E., Weselake R. J. (2009) New Biotechnol. 26, 11–16 [DOI] [PubMed] [Google Scholar]
- 13. Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002) J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 14. Little D., Weselake R., Pomeroy K., Furukawa-Stoffer T., Bagu J. (1994) Biochem. J. 304, 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stone S. J., Levin M. C., Farese R. V., Jr. (2006) J. Biol. Chem. 281, 40273–40282 [DOI] [PubMed] [Google Scholar]
- 16. Liu Q., Siloto R. M., Weselake R. J. (2010) Biochemistry 49, 3237–3245 [DOI] [PubMed] [Google Scholar]
- 17. Heckman K. L., Pease L. R. (2007) Nat. Protoc. 2, 924–932 [DOI] [PubMed] [Google Scholar]
- 18. Gietz R. D., Schiestl R. H. (2007) Nat. Protoc. 2, 31–34 [DOI] [PubMed] [Google Scholar]
- 19. Gilstring C. F., Ljungdahl P. O. (2000) J. Biol. Chem. 275, 31488–31495 [DOI] [PubMed] [Google Scholar]
- 20. Siloto R. M., Truksa M., Brownfield D., Good A. G., Weselake R. J. (2009) Plant Physiol. Biochem. 47, 456–461 [DOI] [PubMed] [Google Scholar]
- 21. Siloto R. M., Truksa M., He X., McKeon T., Weselake R. J. (2009) Lipids 44, 963–973 [DOI] [PubMed] [Google Scholar]
- 22. Wang Y., Toei M., Forgac M. (2008) J. Biol. Chem. 283, 20696–20702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tusnády G. E., Simon I. (2001) Bioinformatics 17, 849–850 [DOI] [PubMed] [Google Scholar]
- 24. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 25. Claros M. G., Von Heijne G. (1994) Comput. Appl. Biosci. 10, 685–686 [DOI] [PubMed] [Google Scholar]
- 26. Yuan Z., Mattick J. S., Teasdale R. D. (2004) J. Comput. Chem. 25, 632–636 [DOI] [PubMed] [Google Scholar]
- 27. Hirokawa T., Boon-Chieng S., Mitaku S. (1998) Bioinformatics 14, 378–379 [DOI] [PubMed] [Google Scholar]
- 28. Arai M., Mitsuke H., Ikeda M., Xia J. X., Kikuchi T., Satake M., Shimizu T. (2004) Nucleic Acids Res. 32, W390–W393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hofmann K., Stoffel W. (1993) Biol. Chem. Hoppe-Seyler 347, 166. [DOI] [PubMed] [Google Scholar]
- 30. Kyte J., Doolittle R. F. (1982) J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 31. Normington K., Kohno K., Kozutsumi Y., Gething M. J., Sambrook J. (1989) Cell 57, 1223–1236 [DOI] [PubMed] [Google Scholar]
- 32. Tamura N., Konishi S., Iwaki S., Kimura-Someya T., Nada S., Yamaguchi A. (2001) J. Biol. Chem. 276, 20330–20339 [DOI] [PubMed] [Google Scholar]
- 33. Bogdanov M., Zhang W., Xie J., Dowhan W. (2005) Methods 36, 148–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu J., Deutsch C. (2001) Biochemistry 40, 13288–13301 [DOI] [PubMed] [Google Scholar]
- 35. Persson B. (2006) in Structural Genomics of Membrane Proteins (Lundstrom K. H. ed) pp. 5–20, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 36. Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., Farese R. V., Jr. (2009) J. Biol. Chem. 284, 5352–5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McFie P. J., Stone S. L., Banman S. L., Stone S. J. (2010) J. Biol. Chem. 285, 37377–37387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo Z. Y., Lin S., Heinen J. A., Chang C. C., Chang T. Y. (2005) J. Biol. Chem. 280, 37814–37826 [DOI] [PubMed] [Google Scholar]
- 39. Han G., Gable K., Yan L., Natarajan M., Krishnamurthy J., Gupta S. D., Borovitskaya A., Harmon J. M., Dunn T. M. (2004) J. Biol. Chem. 279, 53707–53716 [DOI] [PubMed] [Google Scholar]
- 40. Van Horn W. D., Kim H. J., Ellis C. D., Hadziselimovic A., Sulistijo E. S., Karra M. D., Tian C., Sönnichsen F. D., Sanders C. R. (2009) Science 324, 1726–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Geest M., Lolkema J. S. (2000) Microbiol. Mol. Biol. Rev. 64, 13–33 [DOI] [PMC free article] [PubMed] [Google Scholar]