Abstract
A single mutation (C73R) in the enzyme uroporphyrinogen III synthase (UROIIIS) is responsible for more than one-third of all of the reported cases of the rare autosomal disease congenital erythropoietic porphyria (CEP). CEP patients carrying this hotspot mutation develop a severe phenotype of the disease, including reduced life expectancy. Here, we have investigated the molecular basis for the functional deficit in the mutant enzyme both in vitro and in cellular systems. We show that a Cys in position 73 is not essential for the catalytic activity of the enzyme but its mutation to Arg speeds up the process of irreversible unfolding and aggregation. In the mammalian cell milieu, the mutant protein levels decrease to below the detection limit, whereas wild type UROIIIS can be detected easily. The disparate response is not produced by differences at the level of transcription, and the results with cultured cells and in vitro are consistent with a model where the protein becomes very unstable upon mutation and triggers a degradation mechanism via the proteasome. Mutant protein levels can be restored upon cell treatment with the proteasome inhibitor MG132. The intracellularly recovered C73R-UROIIIS protein shows enzymatic activity, paving the way for a new line of therapeutic intervention in CEP patients.
Keywords: Enzyme Catalysis, Enzyme Degradation, Enzyme Mutation, Protein Degradation, Protein Stability, Congenital Erithropoietic Porphyria, Proteasome Inhibition, Uroporphyrinogen III Synthase
Introduction
Congenital erythropoietic porphyria (CEP),4 or Gunther disease, is a rare autosomal disease caused by mutations in the gene that encodes for the enzyme uroporphyrinogen III synthase (UROIIIS) (1, 2). UROIIIS catalyzes the cyclization of the linear tetrapyrrole hydroxymethylbilane to produce uroporphyrin III (3, 4). Such condensation competes with the spontaneous degradation of the substrate to produce uroporphyrin I, which does not catabolize well (4). In CEP patients, lowered UROIIIS activity causes the accumulation of type I porphyrins in the bone marrow, teeth, stool, and urine as well as skin photosensitivity (2). CEP patients also suffer from hemolytic anemia and splenomegaly, and they have red-colored urine. Because porphyrins mostly accumulate in the marrow, current treatment has focused on marrow suppression and bone marrow transplantation with an uneven degree of success (5, 6).
At the molecular level, the severity of the disease is dependent ultimately on the nature of the inborn error (7–9). Genetic analyses of >110 CEP patients have identified ∼25 missense mutations as well as four defects in the promoter region (10, 11). As shown in supplemental Fig. S1, the distinct mutations are repeated only occasionally within patients. One striking exception is C73R, present in more than one-third of all reported cases (10). This hotspot mutation has a drastic effect on UROIIIS enzyme functionality and results in the most severe phenotype in CEP patients (11).
As a first step to therapy, it is crucial to understand the mechanism by which the mutation induces the loss of functionality. Structural studies and modeled complexes indicate that Cys-73 is far from the enzyme-substrate interaction site, suggesting that it does not play a critical role in catalysis (12–14). Consistent with this idea, we have shown recently that, in vitro, purified recombinant human C73R-UROIIIS partially retains its catalytic activity (15). Mutations can also reduce thermodynamic stability, and proteins that are mis- or unfolded are usually processed by cellular degradation pathways (16). Here, we have explored the relationship between enzyme activity and the stability of C73R-UROIIIS both in vitro and in cell cultures. Our results with pure protein show that the mutant protein is folded but undergoes a conformational change. The apparent enzyme activity decreases over time at a much faster rate than in wild type UROIIIS due to the premature unfolding of the molecule. In transfected cells, C73R-UROIIIS protein levels are depleted, but intracellular mRNA levels are not. Upon proteasome inhibition, mutant protein levels are restored, and the protein is proven to be functionally active.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
All media and reagents for tissue culture were purchased from Invitrogen. Human fibroblastoid M1 cells were grown in complete DMEM medium (Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 0.1 mg/ml streptomycin, and 100 units/ml penicillin). The murine hepatic MLP29 cell line was grown under the same culture conditions. When indicated, cells were cultured during 12 h in the presence of 10 μm MG132, 100 μm choloroquine, or 15 mm ammonium chloride, which were all from Sigma-Aldrich. Monoclonal antibodies were purchased from the following vendors: anti-Myc tag (clone 4A6) from Upstate (Lake Placid, NY), anti-neomycin phosphotransferase II (clone 4B4D1) from Abcam (Cambridge, UK), anti-Hsp90 (clone 68) from BD Biosciences, and anti-Hsp70 (clone BRM-22) and anti-alpha-tubulin (clone DM1A) from Sigma-Aldrich. HRP-conjugated secondary antibody was from GE Healthcare. All other reagents were of analytical grade and primarily acquired from Sigma-Aldrich.
DNA Constructs
To generate mammalian expression plasmids coding human UROIIIS (wild type and C73R) fused to the Myc epitope, the complete open reading frames of UROIIIS were amplified by PCR from the bacterial plasmid pHTUROIIIS expressing wild type or C73R versions of human UROIIIS. The products were purified and used as templates for nested PCR, using primers designed to append an EcoRI site upstream of the Kozak sequence and to replace the translation termination site with a SalI site, followed by Myc-coding sequence, a stop codon, and a NotI site. The nested PCR product was gel-purified and cloned in the EcoRI-SalI sites of the pEGFP-C2 and pEGFP-N3 vectors (Takara Bio, Clontech, Mountain View, CA) for expression of GFP-tagged proteins, or in the EcoRI-NotI of the pCR3.1 vector (Invitrogen) for expression of Myc-tagged proteins. All constructs were roughly verified by DNA sequencing.
The human UROIIIS sequence fused to the Myc epitope also was obtained by PCR of plasmid DNA (pHTUROIIIS) containing wild type human UROIIIS, using the forward and reverse primers 5′-cttccatatgatggaacaaaagcttatttc-3′ and 5′-cgcggatcctcagcagcaaccatg-3′ and cloned into the pET16b bacterial vector using the BamH1 and Nde1 restriction sites.
Generation of Stably Transfected Mammalian Cell Lines
For stable transfection, both M1 and MLP29 cell lines were grown to 60–70% confluency. M1 cells were transfected with C- or N-terminal Myc-tagged UROIIIS-WT or UROIIIS-C73R-encoding plasmids and MLP29 cells with C- or N-terminal GFP-tagged UROIIIS-WT or UROIIIS-C73R-encoding plasmids by using FuGENE 6 (Roche Diagnostics, Manheim, Germany) and OptiMEM medium (Invitrogen) as described by the manufacturer's instructions. The medium was changed to complete DMEM 5 h post-transfection and to complete DMEM containing 2 g/liter G418 24 h later. About 2 weeks later, clones of stably transfected cells were isolated and maintained in complete DMEM containing 2 mg/ml G418. The same stable cell line from each of the different WT and C73R versions of the UROIIIS protein was selected to perform this study.
Confocal Microscopy
MLP29 stable cell lines carrying GFP-tagged proteins were grown on glass coverslips, fixed with 2% (w/v) formaldehyde, washed twice with PBS, and mounted on glass slides with Fluoromount-G (Southern Biotech, Birmingham, AL) containing 0.7 mg/liter DAPI. Immunofluorescence staining was performed as described previously (27). Fluorescence was examined on a Leica TCS SP multiphoton confocal microscope, and images from different channels were acquired using the Leica Confocal Software.
Flow Cytometry Analysis
MLP29 cell lines stably expressing GFP-tagged UROIIIS-WT or UROIIIS-C73R proteins were cultured in 150-mm dishes, trypsinized using TRIPLE reagent (Invitrogen), washed twice with ice-cold PBS, and analyzed on a FACS Canto II cytometer (BD Biosciences). GFP fluorescence of a minimum of 20,000 cells was acquired using BD FACSDiva flow cytometry software (version 5.0).
UROIIIS Enzymatic Assay
The determination of UROIIIS specific activity was based on the method developed by Jordan and modified by Tsai et al. and Hart and Battersby (28–30). Purified protein samples were obtained as described previously (15). When testing the activity in eukaryotic cells, an aliquot of ten million untreated or MG132-treated cells were used, and the protein concentration was obtained by the Bradford protein assay (Bio-Rad) using BSA as the standard and corrected for the number of cells that contained the GFP tag that was determined by flow cytometry.
Circular Dichroism and Fluorescence Spectroscopy
CD experiments were collected in a JASCO J-810 spectropolarimeter and analyzed as described previously (31). CD experiments were performed in a quartz cuvette with a 0.2-cm path length, whereas fluorescence data were recorded in a JASCO FP-6500 fluorometer. Sample concentration was set to 350 nm and 40 nm for the CD and fluorescence experiments, respectively. The experiments were run at 25 °C with samples that had been purified and stored at 4 °C.
Computational Modeling
The C73R, C73A, C73D, C73S, C73N, C73L, and C73Y mutants of UROIIIS were modeled computationally using the Swiss Model Workspace (17) and the Protein Homology/analogY Recognition Engine (Phyre) (18). Solvent accessibility calculations were carried out using the MOLMOL program with a probe radius of 1.4 Å and using the 1JR2 Protein Data Bank structure (12) for wild type and the modeled structure for C73R. PyMOL software was used for data representation and analysis of interatomic distances.
Transcript Expression Analysis (qRT-PCR Assay)
Total RNA was isolated using the TRIzol Reagent (Invitrogen, Carlsbad, CA) as per the manufacturer's instructions, and quantitative real-time PCR (qRT-PCR) was performed in two steps. Single-stranded cDNA was synthesized from 1 μg of mRNA, treated previously with RNase-free DNase, using random hexamers and SuperScript II in presence of RNase OUT. (All reagents were from Invitrogen.) The real-time step was carried out with 5 μl of 1:20 cDNA dilution and SYBR Green (Invitrogen) for 30 cycles (for 30 s at 94 °C, for 30 s at 60 °C, and for 30 s at 72 °C) on a Bio-Rad iCycler thermocycler. The samples were examined twice in triplicate for expression of neomycin phosphotransferase II (NPTII), UROIIIS, and acidic ribosomal protein (ARP). For UROIIIS, both N- and C-terminal Myc-tagged versions of the wild type and C73R mutant proteins were analyzed. The specific primers (and expected product sizes, in base pairs) are shown in supplemental Table S1. The relative level of each transcript was calculated on the basis of the ΔΔCt method and normalized to ARP as the reference gene. Amplicon size and reaction specificity was also confirmed by 2.5% (w/v) agarose gel electrophoresis.
Western Blot Analysis
To prepare cell lysates, 106 trypsinized cells were lysed for 15 min on ice in the presence of 100 μl of lysis buffer (300 mm NaCl, 50 mm Tris, pH 7.4, 0.5% Triton X-100, and protease inhibitors). After clarification of the samples by centrifugation at 20,000 × g, the supernatant was transferred to a fresh Eppendorf tube. The protein concentration of the cell lysates was determined by means of the Bradford protein assay (Bio-Rad) using BSA as the standard. SDS sample buffer was added, and samples were incubated for 5 min at 37 °C, 65 °C, and 95 °C and separated on 4–12% precast acrylamide gels (Invitrogen). After transferring to PVDF membranes (Millipore, Bedford, MA) and blocking overnight (5% milk and 0.05% Tween 20 in PBS), the primary antibody was added for 1 h, followed by a PBS wash, and application of the secondary HRP-conjugated antibody. Chemiluminescent detection of bands was performed with ECL Plus reagent (GE Healthcare).
RESULTS
Reduced Stability of C73R-UROIIIS in Vitro
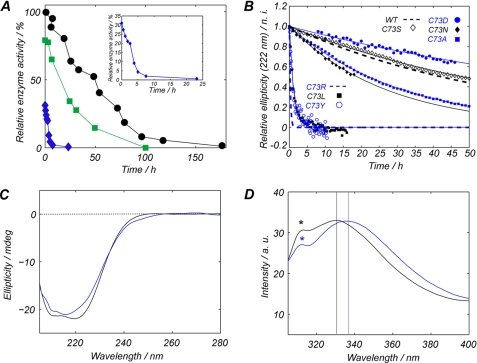
The catalytic activity of UROIIIS is time-dependent because the enzyme is not thermodynamically stable and undergoes irreversible denaturation, followed by aggregation (11). Freshly purified UROIIIS-C73R retained one-third of the activity of the wild type enzyme, but at physiological temperature, the activity of the mutant protein decayed >10 times faster than the wild type protein (Fig. 1A). The decrease in the folded conformation over time was determined using CD, monitored at 37 °C and 222 nm (this wavelength reports on the α-helical content of the protein), and is shown in Fig. 1B. Wild type UROIIIS had a half-life of 61 h as compared with 15 min observed with C73R-UROIIIS, which was in very good agreement with the time dependence of the catalytic activity of the enzyme.
FIGURE 1.
A, changes in enzyme activity over time at 37 °C and relative to wild type UROIIIS at 4 °C. Black circles, wild type UROIIIS; blue diamonds, C73R-UROIIIS; green squares, UROIIIS-Myc. The inset expands the information regarding C73R-UROIIIS. B, decay over time of the CD ellipticity at 222 nm (sensitive to the α-helical content of the protein) for WT and seven different Cys-73 mutants of UROIIIS. C, far-UV region of the CD spectra for wild type (black line) or C73R (blue line) UROIIIS. D, tryptophan emission fluorescence spectra for wild type (black line) or C73R (blue line) UROIIIS. The position of the maximum in each spectrum is highlighted by the vertical lines. The peaks labeled with an asterisk correspond to the Raman spectrum of water. a.u., arbitrary units; mdeg, millidegrees; n.i., normalized intensity.
Fig. 1C shows the CD spectra for the purified wild type (black line) and C73R (blue line) UROIIIS, which match significantly, consistent with the idea that both proteins share the same folding pattern. The lower intensity at 222 nm found in the C73R spectra is consistent with a slightly lower helical content for the mutant. The effect of the mutation was also investigated by fluorescence spectroscopy, as UROIIIS has two tryptophan residues, both of which are found at the N-terminal domain, and Trp-83 is only 10Å away from CYS-73. Fig. 1D shows the emission spectrum for wild type UROIIIS (black line) and C73R (blue line). The shift in the maximum of the emission spectra indicates that the tryptophan of the mutant protein is more exposed to a polar environment.
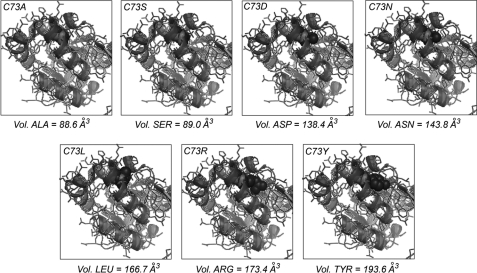
Using homology modeling programs (17, 18), we generated a structural model of C73R-UROIIIS. Fig. 2 shows the overlay of the N-terminal domains of wild type (Protein Data Bank code 1JR2) (12) and the C73R (model), where Cys-73 and Arg-73 side chains have been highlighted by red and blue spheres, respectively. According to the model, the arginine side chain penetrates into the domain core. To further investigate the origin of UROIIIS destabilization upon mutation, a set of Cys-73 mutations that differ in the side chain volume and charge has been generated: C73A, C73D, C73S, C73N, C73L, and C73Y. Structural models for the mutants are also shown in Fig. 2, whereas the loss of stability over time, analyzed by CD, is shown in Fig. 1B. A certain correlation between the side chain volume and the acceleration of the unfolding process is observed (Fig. 2), consistent with the increase in the number contacts that large side chains have with the hydrophobic surrounding (supplemental Table S2). However, C73D shows higher stability than wild type UROIIIS and retained 100% of its enzymatic activity, demonstrating that Cys-73 does not play a significant role in catalysis, consistent with the proposed reaction mechanism (14). This result suggests that the destabilization mechanism is complex, and the introduction of a negative charge in this position may also play a favorable role in increasing the free energy of the protein. By introducing a positive charge, C73R probably exerts the opposite effect, consistent with the very low stability found for this mutation.
FIGURE 2.
Structural models for the mutations in position 73. Overlay of wild type UROIIIS (Protein Data Bank code 1JR2, light ribbon) and the modeled structure for each Cys-73 mutant (dark ribbon). The side chains for Cys-73 and the specific mutation are represented by red and blue spheres, respectively. Vol., volume.
Reduced Expression Levels of C73R-UROIIIS in Mammalian Cells
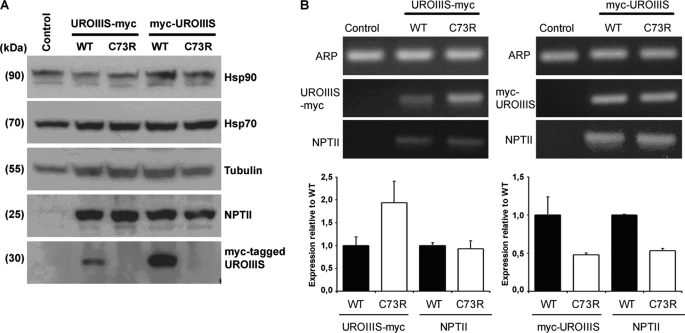
To investigate whether the loss of activity in vitro also is observed in the cell environment, several tagged versions of wild type and C73R-UROIIIS were analyzed in two different cell-types: a human fibroblast-derived cell line (named M1) and a murine hepatocyte-derived cell line (MLP29). M1 cell lines stably expressing N-terminal or C-terminal Myc-tagged versions of wild type and C73R mutant enzymes were generated, and the levels of the ectopically expressed proteins were analyzed by Western blotting. As expected, no signal was observed in the M1 parental cells (Fig. 3A). A clear specific band was observed in stable cell lines expressing wild type UROIIIS. When expressed in vitro and purified, this protein showed >80% of the activity of untagged UROIIIS (Fig. 1A), indicating that tagged version of this enzyme is functional. For the mutant, no specific bands were detected in stable cell lines expressing the C73R mutant protein. This result could not be assigned to additional stress induced by the ectopic expression of the different versions of UROIIIS protein because no significant differences were detected when two endogenous stress-inducible chaperones were analyzed (Hsp70 and Hsp90, Fig. 3A) (19). The mammalian vector pCR3.1 utilized in this study also carried the gene encoding NPTII that was detected at similar levels in all the stable cell lines, indicating that all of cell lines contained functional vectors.
FIGURE 3.
A, Western blot analysis of the unmodified M1 cell line (control) or M1 cells stably expressing WT or C73R versions of Myc-tagged UROIIIS proteins. Hsp70, Hsp90, and tubulin proteins serve as protein loading controls. Specific antibodies against NPTII and the Myc epitope were used to detect proteins from transfected vectors. Molecular masses of the detected proteins are indicated. B, quantitative real-time PCR analysis. Upper panel, transcripts of the housekeeping gene ARP, NPTII, and Myc-tagged UROIIIS mRNA were amplified in the different stable cell lines and examined on a 2.5% agarose gel. Lower panel, fold change of the indicated transcripts expressed relative to cell lines stably expressing WT versions of Myc-tagged proteins (lower panel). Triplicates for each stable cell line were evaluated to obtain the error bars.
qRT-PCR with the stable cell lines was used to discard the idea that the lack of mutant protein expression was due to a failure at the transcriptional level. Specific pair primers were designed to amplify the coding sequence for the N-terminal Myc-tagged UROIIIS (UROIIIS-Myc), C-terminal Myc-tagged UROIIIS (UROIIIS-Myc), vector-carried NPTII, and the housekeeping gene ARP. As an internal control, we certified that the corresponding primers amplified Myc-tagged UROIIIS and NPTII transcripts only in the stably transfected cell lines (Fig. 3B, upper panel). In the qRT-PCR assay, similar expression levels of NPTII were observed in stable cell lines transfected with the plasmids encoding the C-terminal Myc-tagged UROIIIS (wild type or C73R), whereas the expression levels of C73R UROIIIS-Myc were 2-fold higher than wild type UROIIIS-Myc (Fig. 3B, lower left panel). In the Myc-UROIIIS construct, a reduction in the transcript levels was observed upon mutation (Fig. 3B, lower right panel), but this was independent of the mutation because the transcript levels of NPTII and Myc-UROIIIS were similar. Together, the results obtained from the Western blot analysis and from qRT-PCR assay indicated that a defect in the transcription of the Myc-tagged UROIIIS mutant versions was not the origin of the lack of expression generated by the C73R mutation, supporting the idea that the protein mutant is unstable and quickly degraded in the cell, in agreement with the results obtained by the structural analysis.
Intracellular C73R Degradation Is Precluded by Proteasomal Inhibition
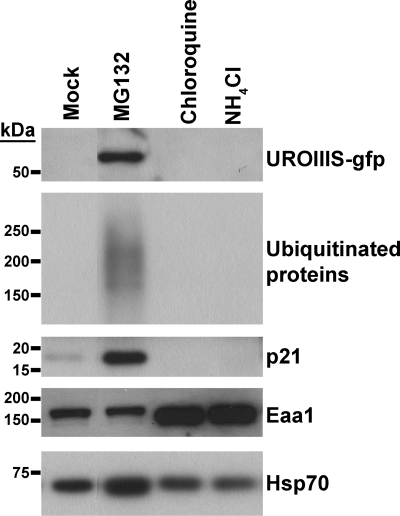
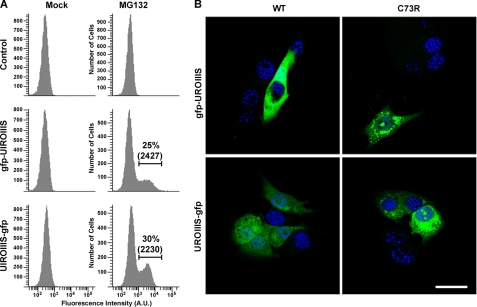
In this experiment, we analyzed the intracellular degradation of the C73R mutant. MLP29 cells stably expressing C73R-UROIIIS fused to GFP were incubated in the presence of proteasome (MG132) or lysosome (chloroquine, NH4Cl) inhibitors (Fig. 4). Treatment with chloroquine or NH4Cl affected the endosomal-lysosomal pathway (20) as could be observed by the accumulation of the early endosomal marker EAA1, but no effect was observed in the C73R mutant cells as compared with the mock treatment. On the contrary, treatment with MG132 resulted in the accumulation of C73R along with p21 and ubiquitinated proteins (Fig. 4). The protein accumulation induced by MG132 was independent of the tag position as shown by flow cytometry analysis (Fig. 5A); the GFP-fused proteins of the C73R mutant were absent under regular conditions but could be detected at similar levels of expression when proteasome activity was inhibited with MG132. This treatment also caused an increase in the level of WT protein (supplemental Fig. S2). However, the intracellular localization of WT and C73R were different; cytosolic staining was observed for WT UROIIIS, whereas a partially aggregated pattern was detected for C73R-UROIIIS (Fig. 5B). This aggregation was not observed in cells expressing GFP-tagged wild type version of UROIIIS even at the highest expression levels, indicating that the aggregation was caused by C73R mutation and not by the accumulation of GFP moeties. A fraction of these cells were disrupted and tested for enzyme activity. Wild type and C73R-UROIIIS were both catalytically active and, remarkably, the mutant retained 50% of the activity when normalized with respect to wild type. Altogether, these results demonstrate the implication of the proteasome degradation pathway in the intracellular clearance of the wild type enzyme and the misfolded C73R mutant.
FIGURE 4.
MLP29 cells stably expressing C73R UROIIIS-GFP protein cultured in the presence of dimethyl sulfoxide (mock), or inhibitors of degradative pathways, proteasome (MG132), and lysosome (chloroquine and NH4Cl), were analyzed by Western blotting. The C73R UROIIIS-GFP protein was detected by using an antibody against the GFP moiety. Note that Hsp70 serves as a protein loading control, and the accumulation of early endosomal antigen 1 (Eaa1), p21, and ubiquitinated proteins confirms that the inhibitors were working properly. Molecular mass size markers are indicated.
FIGURE 5.
A, GFP fluorescence of unmodified MLP29 cells (control) or cells stably expressing GFP-tagged versions of UROIIIS-C73R in the presence of dimethyl sulfoxide (mock) or MG132 was analyzed by flow cytometry. A threshold for GFP fluorescence was established based on the fluorescence observed for control. Percentage and median fluorescence intensity of GFP-positive cells are indicated. B, representative confocal images of formaldehyde-fixed MLP29 cells stably expressing GFP-tagged versions of WT and C73R proteins. GFP fluorescence (green) and DAPI nuclear staining (blue) of different cells are shown. Scale bar, 45 μm.
DISCUSSION
Congenital erythropoietic porphyria is a rare disease produced by the loss of activity in the enzyme uroporphyrinogen III synthase. Clinically speaking, C73R is, by far, the most relevant mutation in CEP (10). Here, we have studied the molecular basis of this hotspot mutation. Our results show that the mutation does not involve a residue important for catalysis. Instead, the C73R mutation, due to a combination of factors, produces a conformational change ultimately responsible for decreased kinetic stability of the molecule, because in vitro, the unfolding half-life dropped from 2.5 days (WT) to 15 min (C73R) at 37 °C. In two distinct mammalian cell types, the mutant was undetectable as compared with WT UROIIIS that could be overexpressed in the cell. From these data, a model emerges where the active conformation of WT UROIIIS is unstable thermodynamically but remains folded long enough to exert its function. This would not to be the case for the C73R-UROIIIS, which is expressed in the cell but rapidly unfolds and is degraded quickly, resulting in undetectable protein levels in the cell.
Numerous diseases have been related to mutations that result in decreased stability of an enzyme. For instance, the ΔPhe-508 mutation in the cystic fibrosis transmembrane receptor is responsible for >70% of the cases of cystic fibrosis (21), and destabilizing mutations in G protein-coupled receptors are responsible for diseases such as retinitis pigmentosa, nephrogenic diabetes insipidus, and hypogonadotropic hypogonadism (22). For some of these pathologies, it has been shown that certain substances can act as “chemical chaperones” to slow down or inhibit their tendency to aggregate, resulting in detectable enzyme levels in the cell (23). Our results suggest that some of these co-solutes could equally act on C73R-UROIIIS, and we are actively pursuing such studies.
The proteasome complex plays a critical role in signal transduction pathways for cell growth and survival, cell-cycle control, and in protein turnover, which is crucial to maintain cellular homeostasis. Here, we have shown that degradation of UROIIIS depends on the activity of the proteasome instead of the lysosome pathway, providing a molecular explanation for the failure of chloroquine treatment in CEP patients (24). Remarkably, the degradation process can be reverted by inhibiting proteasome activity, and even though the accumulation of the mutant protein in the cell results in partial aggregation, we show that the enzyme retains partial catalytic activity. The sensitivity of transformed cells to proteasome inhibitors along with the successful design of treatment protocols with tolerable therapeutic indices has made proteasome inhibition a viable strategy for cancer treatment. Clinical validation of the proteasome as a molecular target was achieved with the approval of bortezomib, a boronic acid proteasome inhibitor, for the treatment of multiple myeloma and mantle cell lymphoma (25). Several “next-generation” proteasome inhibitors (carfilzomib and PR-047, NPI-0052, and CEP-18770 represent the distinct structural classes of peptidyl epoxyketones, β-lactones, and peptidyl boronic acids, respectively) with different mechanisms of action, pharmacological and pharmacodynamic activity profiles, and therapeutic indices recently have entered clinical development (reviewed in Ref. 26). Our results argue positively for the possibility to expand the clinical utility of proteasome inhibitors for the treatment of specific non-oncological purposes. Although further studies are required, the partial inhibition of the proteasome may constitute a new therapeutic modality for CEP patients.
This work was supported by the Department of Industry, Tourism, and Trade of the Basque government (Etortek Research Programs 2005/2007), the Innovation Technology Department of Bizkaia County, the Ministerio de Ciencia y Tecnología (CTQ2006-09101/BQU and CTQ2009-10353/BQU), The Fundación Ramon Areces, and the Ramon y Cajal program (to J. M. F.-P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- CEP
- congenital erythropoietic porphyria
- UROIIIS
- uroporphyrinogen III synthase
- qRT-PCR
- quantitative real-time PCR
- NPTII
- neomycin phosphotransferase II
- ARP
- acidic ribosomal protein.
REFERENCES
- 1. Ged C., Moreau-Gaudry F., Richard E., Robert-Richard E., de Verneuil H. (2009) Cell Mol. Biol. 55, 53–60 [PubMed] [Google Scholar]
- 2. Desnick R. J., Astrin K. H. (2002) Br. J. Haematol. 117, 779–795 [DOI] [PubMed] [Google Scholar]
- 3. Battersby A. R. (2000) Nat. Prod. Rep. 17, 507–526 [DOI] [PubMed] [Google Scholar]
- 4. Schubert H. L., Raux E., Matthews M. A., Phillips J. D., Wilson K. S., Hill C. P., Warren M. J. (2002) Biochem. Soc. Trans. 30, 595–600 [DOI] [PubMed] [Google Scholar]
- 5. Dupuis-Girod S., Akkari V., Ged C., Galambrun C., Kebaïli K., Deybach J. C., Claudy A., Geburher L., Philippe N., de Verneuil H., Bertrand Y. (2005) Eur. J. Pediatr. 164, 104–107 [DOI] [PubMed] [Google Scholar]
- 6. Tezcan I., Xu W., Gurgey A., Tuncer M., Cetin M., Oner C., Yetgin S., Ersoy F., Aizencang G., Astrin K. H., Desnick R. J. (1998) Blood 92, 4053–4058 [PubMed] [Google Scholar]
- 7. Fontanellas A., Bensidhoum M., Enriquez de Salamanca R., Moruno Tirado A., de Verneuil H., Ged C. (1996) Eur. J. Hum. Genet. 4, 274–282 [DOI] [PubMed] [Google Scholar]
- 8. Xu W., Warner C. A., Desnick R. J. (1995) J. Clin. Invest. 95, 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. To-Figueras J., Badenas C., Mascaró J. M., Madrigal I., Merino A., Bastida P., Lecha M., Herrero C. (2007) Blood Cells Mol. Dis. 38, 242–246 [DOI] [PubMed] [Google Scholar]
- 10. Frank J., Wang X., Lam H. M., Aita V. M., Jugert F. K., Goerz G., Merk H. F., Poh-Fitzpatrick M. B., Christiano A. M. (1998) Ann. Hum. Genet. 62, 225–230 [DOI] [PubMed] [Google Scholar]
- 11. Bishop D. F., Johansson A., Phelps R., Shady A. A., Ramirez M. C., Yasuda M., Caro A., Desnick R. J. (2006) Am. J. Hum. Genet. 78, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mathews M. A., Schubert H. L., Whitby F. G., Alexander K. J., Schadick K., Bergonia H. A., Phillips J. D., Hill C. P. (2001) EMBO J. 20, 5832–5839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cunha L., Kuti M., Bishop D. F., Mezei M., Zeng L., Zhou M. M., Desnick R. J. (2008) Proteins 71, 855–873 [DOI] [PubMed] [Google Scholar]
- 14. Schubert H. L., Phillips J. D., Heroux A., Hill C. P. (2008) Biochemistry 47, 8648–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fortian A., Castaño D., Ortega G., Laín A., Pons M., Millet O. (2009) Biochemistry 48, 454–461 [DOI] [PubMed] [Google Scholar]
- 16. Knecht E., Aguado C., Cárcel J., Esteban I., Esteve J. M., Ghislat G., Moruno J. F., Vidal J. M., Sáez R. (2009) Cell Mol. Life Sci. 66, 2427–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 18. Bennett-Lovsey R. M., Herbert A. D., Sternberg M. J., Kelley L. A. (2008) Proteins 70, 611–625 [DOI] [PubMed] [Google Scholar]
- 19. Duncan R. F. (2005) FEBS J. 272, 5244–5256 [DOI] [PubMed] [Google Scholar]
- 20. Puertollano R., Alonso M. A. (1999) Mol. Biol. Cell 10, 3435–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kerem B., Rommens J. M., Buchanan J. A., Markiewicz D., Cox T. K., Chakravarti A., Buchwald M., Tsui L. C. (1989) Science 245, 1073–1080 [DOI] [PubMed] [Google Scholar]
- 22. Conn P. M., Ulloa-Aguirre A., Ito J., Janovick J. A. (2007) Pharmacol. Rev. 59, 225–250 [DOI] [PubMed] [Google Scholar]
- 23. Brown C. R., Hong-Brown L. Q., Biwersi J., Verkman A. S., Welch W. J. (1996) Cell Stress Chaperones 1, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lagarde C., Hamel-Teillac D., De Prost Y., Blanche S., Thomas C., Fischer A., Nordmann Y., Ged C., De Verneuil H. (1998) Ann. Dermatol. Venereol. 125, 114–117 [PubMed] [Google Scholar]
- 25. Schenkein D. (2002) Clin. Lymphoma 3, 49–55 [DOI] [PubMed] [Google Scholar]
- 26. Ruggeri B., Miknyoczki S., Dorsey B., Hui A. M. (2009) Adv. Pharmacol. 57, 91–135 [DOI] [PubMed] [Google Scholar]
- 27. Falcón-Pérez J. M., Dell'Angelica E. C. (2007) Exp. Cell Res. 313, 1473–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shoolingin-Jordan P. M., Leadbeater R. (1997) Methods Enzymol. 281, 327–336 [DOI] [PubMed] [Google Scholar]
- 29. Tsai S. F., Bishop D. F., Desnick R. J. (1987) Anal. Biochem. 166, 120–133 [DOI] [PubMed] [Google Scholar]
- 30. Hart G. J., Battersby A. R. (1985) Biochem. J. 232, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tadeo X., López-Méndez B., Trigueros T., Laín A., Castaño D., Millet O. (2009) PLoS Biol. 7, e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]