Abstract
The etiology of a variety of chronic inflammatory disorders has been attributed to the interaction of genetic and environmental factors. Herein, we identified a link between epigenetic regulation and IL-13-driven eotaxin-3 in the pathogenesis of chronic allergic inflammation. We first demonstrated that the cAMP-responsive element (CRE) site in the eotaxin-3 promoter affects IL-13-induced eotaxin-3 promoter activity. Furthermore, the CRE-binding protein-binding protein (CBP), a histone acetyltransferase, induced base-line and IL-13-induced eotaxin-3 promoter activity. Additionally, IL-13 treatment promoted global histone 3 acetylation as well as the formation of a complex containing CBP and STAT6 and the subsequent acetylation of histone 3 at the eotaxin-3 promoter. CBP gene silencing decreased IL-13-induced transcription of eotaxin-3. Conversely, inhibition of histone deacetylation increased IL-13-induced eotaxin-3 production. Clinical studies demonstrated markedly increased global acetylation of histone 3 in the inflamed tissue of patients with allergic inflammation. Collectively, these results identify an epigenetic mechanism involving CBP and chromatin remodeling in regulating IL-13-induced chemokine transcription.
Keywords: CBP, Chemokines, Chromatin Histone Modification, Epigenetics, Inflammation
Introduction
Epigenetic regulation of gene expression contributes to the risk of inflammatory diseases, but the mechanisms remain largely unexplored (1–4). Chromatin remodeling, via histone modification, is one of the key epigenetic mechanisms known to regulate development (5), cancer (6), and more recently inflammatory diseases (7, 8). Histone acetyltransferases such as cAMP-responsive element (CRE)3-binding protein-binding protein (CBP)/p300 (9, 10) and histone deacetylases (11) promote or inhibit transcription of select promoters, respectively (12). For example, post-translational acetylation of histone 3 typically activates transcription (13–15). Despite these molecular studies, there has been a paucity of data concerning the mechanisms of epigenetic contribution to chronic inflammatory diseases. Herein, we aimed to examine the role of histone 3 modifications in chronic allergic inflammation.
We focused our attention on an increasingly recognized disease, eosinophilic esophagitis (EE) (16) as we have developed evidence that the disease is mediated by gene-environment interaction and is associated with a marked alteration in a conserved mRNA transcriptome in the diseased tissue (esophagus) (17–19). Additionally, because direct analysis of the inflammatory tissue via endoscopic biopsy is routine, the study of EE has advantages compared with other inflammatory diseases where only surrogate tissue or markers can be consistently analyzed for epigenetic modification. Notably, a significant fraction of the EE-associated transcriptome is induced by IL-13 in esophageal epithelial cells. Of the entire EE-associated transcriptome, the gene with the greatest overexpression in esophageal biopsy samples and in IL-13-stimulated esophageal epithelial cells is eotaxin-3 (17, 20). As a well known potent eosinophil chemoattractant and activating factor, evidence is emerging that IL-13-induced eotaxin-3 has a key role in the pathogenesis of a number of allergic disorders (1, 18, 21–27).
Herein, we tested the hypothesis that histone 3 modification contributes to IL-13-induced eotaxin-3 responses, and we aimed to identify the mechanisms involved. In particular, we first demonstrated that the CRE site in the eotaxin-3 promoter affected IL-13-induced eotaxin-3 promoter activity. Additionally, we demonstrated that CBP promoted base-line and IL-13-induced eotaxin-3 promoter activity. Conversely, a CBP inhibitor (adenovirus 5 early region 1A (E1A) protein) repressed eotaxin-3 promoter activity in IL-13-stimulated cells. Furthermore, IL-13-induced eotaxin-3 production was enhanced by histone deacetylase inhibition. Chromatin immunoprecipitation (ChIP) revealed that stimulation of esophageal epithelial cells with IL-13 increased recruitment of CBP and the amount of acetylated histone 3 to the eotaxin-3 promoter. Moreover, transcriptional activation of eotaxin-3 and acetylation of histone 3 mediated by IL-13 were markedly reduced by shRNA-mediated gene silencing of CBP. Additionally, clinical studies demonstrated an increased level of histone 3 acetylation in esophageal tissue from EE patients compared with controls. Collectively, these results identify an epigenetic mechanism involving CBP and histone 3 modifications in regulating the IL-13-induced transcription of eotaxin-3. As such, we have identified a plausible mechanism for the epigenetic regulation of chronic allergic inflammation.
EXPERIMENTAL PROCEDURES
Culture Medium and Reagents
For human primary esophageal epithelial cell culture, the culture conditions and cytokine treatment have been described previously (17). Human esophageal epithelial cell lines (TE-1 and TE-7) were cultured in RPMI 1640 medium supplemented with 5% fetal bovine serum and 1% penicillin and streptomycin (28, 29). Recombinant human IL-13 was obtained from PeproTech, Inc. (Rocky Hill, NJ). Antibodies against STAT6, CBP, p300, and histone 3 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody against acetylated histone 3 was obtained from Millipore Corp. (Billerica, MA). Antibodies against phospho-STAT6 (Tyr-641) and phospho-ATF-2 (Thr-69/71) were obtained from Cell Signaling Technology (Beverly, MA).
Reporter Plasmids and Expression Constructs
The luciferase reporter constructs containing the human eotaxin-3 gene promoter (Eotaxin-3/Luc) and the eotaxin-3 promoter with the STAT6 binding site mutation were described previously (17, 30). The CRE and C/EBPβ sites were mutated by site-directed mutagenesis with a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutagenesis primers were as follows: CRE: forward primer, 5′-CCAGGCTGATCTTGAACTCCTagttcCAAGGGATCCGCCCACCTCGG-3′; reverse primer, 5′-CCGAGGTGGGCGGATCCCTTGgaactAGGAGTTCAAGATCAGCCTGG-3′; and C/EBPβ: forward primer, 5′CCAATTGTGCTATAAAGTAACTTAGTTCAGTTCACTGGAATATTACTcctcgTCCTCTGCTTTTTCTTTTTAAACAAG-3′; reverse primer, 5′-CTTGTTTAAAAAGAAAAAGCAGAGGAcgaggAGTAATATTCCAGTGAACTGAACTAAGTTACTTTATAGCACAATTGG-3′. Lowercase letters represent the mutated nucleotides, and the underlined sequences are the consensus binding sequences for CRE and C/EBPβ. The pRc/RSV-CBP expression plasmid was kindly provided by Edward M. Chan (Indiana University) (31). The adenovirus 5 E1A and CBP binding mutant were kindly provided by E. Pfitzner (Georg-Speyer-Haus, Frankfurt, Germany) (32, 33).
Transient Transfection and Luciferase Activity Assay
TE-7 cells were transiently co-transfected with 500 ng of pGL3-basic firefly reporter plasmid, 5 ng of reference Renilla luciferase reporter plasmid pHRL-TK (Promega, Madison, WI), and pRc/RSV expression plasmid by using Lipofectamine (Invitrogen) according to the manufacturer's instructions. The total amount of plasmid transfected per well was the same. After 48 h, cells were treated with 0 or 100 ng/ml of IL-13 for 24 h. Firefly and Renilla luciferase activities were assayed using the Promega Dual-Luciferase assay kit and a Synergy Mx Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT). All firefly measurements were normalized to Renilla luciferase activity.
CBP Gene Silencing
Two methods were utilized for knockdown of CBP. For transient knockdown of CBP, plasmids expressing shRNAs (SABiosciences Corp., Frederick, MD) were transfected into esophageal epithelial cells via Lipofectamine and screened for efficient knockdown. The hairpin targeting 5′-GAGCCATCTAGTGCATAAACT-3′ produced the most efficient CBP knockdown and was used in subsequent experiments. The vector has a GFP selection marker, and GFP-positive cells were sorted using FACS analysis. After transfection, we sorted GFP-positive cell populations for both constructs with the GFP marker (the control shRNA and CBP shRNA) as indicated by the presence of ∼100% GFP-positive cells (data not shown). Sorted cells showing significant knockdown of CBP expression by RT-PCR or Western blot were used. For generating cells in which CBP was stably knocked down, the sequence above was cloned into the pGreenPuro shRNA lentivector, which contains the puromycin selection marker (System Biosciences, Mountain, CA). The viral core at Cincinnati Children's Hospital Medical Center manufactured pseudoviral particles of CBP and non-targeting pGreenPuro shRNA for delivery. After transductions for 48 h, cells were selected by puromycin treatment (3 μg/ml).
RT-PCR
Total RNA was isolated from esophageal epithelial cells using the RNeasy Mini kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. Total RNA (100 ng) was reversed transcribed using the iScriptTM cDNA Synthesis kit (Bio-Rad). The levels of mRNA expression were determined using an iQ5 real time PCR detection system (Bio-Rad) with iQTM SYBR Green Supermix. Expression of the gene of interest was normalized to GAPDH. Relative expression was calculated using the comparative Ct method as described previously (34). cDNAs were amplified using the following primers: Eotaxin-3: forward primer, 5′-AACTCCGAAACAATTGTACTCAGCTG-3′; reverse primer, 5′-GTAACTCTGGGAGGAAACACCCTCTCC-3′; GAPDH: forward primer, 5′-TGGAAATCCCATCACCATCT-3′; reverse primer, 5′-GTCTTCTGGGTGGCAGTGAT-3′; CBP: forward primer, 5′-GAATCAGCTCTTCCGACTTC-3′; reverse primer, 5′-TGCCAGCCTTTCCTTACA-3′; and p300: forward primer, 5′-TGCACCTTCTGCTGACCA-3′; reverse primer, 5′-TTACAGCTGGCTGGGAAAG-3′.
Preparation of Total Cell Lysates and Nuclear Extracts
Cells were incubated with 100 ng/ml IL-13 for 0–120 min. Total cell lysates were prepared as described previously (35). Cells were rapidly washed with phosphate-buffered saline (PBS) and lysed in ice-cold lysis buffer (50 mm Tris/HCl, pH 8.0, 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P-40, 0.1 mm PMSF, 1× aprotinin, 1× leupeptin, and 1× pepstatin). Nuclear extracts were isolated as described previously (36). Briefly, cells (∼2 × 106) were grown on 10-mm dishes to 80% confluence, washed twice with ice-cold PBS, and harvested by scraping into 1 ml of ice-cold PBS. Cells were pelleted by centrifuging at 3,000 rpm for 10 min at 4 °C. The cell pellet was resuspended in 400 μl of buffer A (10 mm HEPES, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.5 mm PMSF). After 15 min of incubation on ice, 25 μl of 10% Nonidet P-40 was added. The cell homogenate was centrifuged in a microcentrifuge at 14,000 rpm for 1 min. The nuclear pellet was resuspended in 25–50 μl of buffer B (20 mm HEPES, pH 7.9, 0.4 m NaCl, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF). The tube was vigorously shaken by vortexing at 4 °C for 5 min. The clear supernatant was collected, and aliquots were flash frozen in liquid nitrogen and stored at −80 °C. All steps were performed at 4 °C or on ice.
Co-immunoprecipitation and Western Blot Analysis
Immunoprecipitations and Western blot analyses were performed as described (37). Briefly, the cleared lysates containing 500 μg of protein were incubated with 2 μg of anti-CBP or anti-STAT6 antibody for 16 h at 4 °C and then mixed together with 30 ml of 50% protein A/G-agarose beads for 1 h at 4 °C. The immunoprecipitates were washed three times with lysis buffer, then Laemmli buffer was added, and the samples were subjected to electrophoresis on 4–12% SDS-polyacrylamide gels. Western blot analysis was performed and visualized using an enhanced chemiluminescence reagent (Amersham Biosciences). Densitometry measurements were performed using Multi Gauge V3.0 (Fujifilm).
Chromatin Immunoprecipitation Assay
Esophageal epithelial (TE-1) cells were plated in 15-cm dishes and grown to ∼80–90% confluence. Cells were treated with 100 ng/ml IL-13 in medium for 0–30 min. Cells were cross-linked on the plates with 1% formaldehyde, and chromatin was prepared essentially as described (38) with some modifications. Sonication was performed using a Fisher Dismembrator Model 100 for eight cycles for 20-s pulses, and chromatin fragments were determined to be less than 500 bp in size. For ATF-2 (Santa Cruz Biotechnology, sc-6233), CBP (Santa Cruz Biotechnology, sc-369), acetyl-histone 3 (Millipore Corp.), and STAT6 (Santa Cruz Biotechnology, sc-621) immunoprecipitation, rabbit polyclonal antibodies against the respective antigens were used. In parallel reactions, an equivalent concentration of rabbit IgG was used as a negative control. After reversing cross-links, DNA was extracted using phenol-chloroform and then ethanol-precipitated. The purified pellet was resuspended in H2O and subjected to PCR. To amplify the regions containing the CRE and STAT6 elements in the eotaxin-3 promoter, PCR was performed with the following pairs of primers: forward primer, 5′-TTCTCCATGTTGGCCAGG-3′; and reverse primer, 5′-GATGCTGCAAATCAGGCC-3′. To amplify the promoter of β-actin, which was previously described (39), PCR was performed with the following pairs: forward primer, 5′-TCCTCCTCTTCCTCAATCTCG-3′; and reverse primer, 5′-AAGGCAACTTTCGGAACGG. PCR was carried out using Platinum Taq Supermix (Invitrogen) at 94 °C for 5 min followed by 34 cycles of 94 °C for 20 s, 58 °C for 30 s, and 72 °C for 30 s. PCR products were then resolved by 2% agarose gel electrophoresis with ethidium bromide and visualized by UV light.
Microarray Analysis
RNA extraction and microarray analysis were performed as described previously (40). The genome-wide Affymetrix Human Gene 1.0ST gene chip was used, and gene transcript levels were determined by using GeneSpring software (Agilent Technologies, Santa Clara, CA). A base of probe sets in transcriptome analyses was generated by requiring a minimum raw expression on the microarray in the 20th percentile. To identify CBP-regulated genes, those transcripts differentially regulated at least 2-fold between control shRNAs and expressed CBP-specific shRNAs were analyzed statistically using the t test (p < 0.05) with Benjamini-Hochberg false discovery rate correction.
Genome-wide Promoter Scan for STAT6 Binding Sites
The STAT6 binding site was searched from the 4-kb region upstream of the first exon of all genes in the genome using the TraFaC program, which analyzes non-coding genomic sequences that are evolutionarily conserved between mouse and human (41). The resulting list of genes was intersected with the CBP-regulated genes from the microarray analysis.
Esophageal Samples and Immunofluorescence Microscopy
Diagnostic criteria for normal and EE were defined as described previously (18). Patient biopsies, collected from the distal esophagus less than 5 cm from the lower esophageal sphincter, were submerged in formalin for routine pathological analysis with H&E staining. Diagnosis was established based on the maximum eosinophil count per high power field (400×) (18). Slide-mounted cryosections were air-dried and acetone-fixed, washed in PBS, incubated in a blocking solution containing 4% goat serum for 2 h, and then incubated with diluted anti-acetyl-histone 3 primary antibody. Sections were then washed with PBS and incubated for 2 h with Alexa Fluor 594-conjugated secondary antibodies at a 1:500 dilution in PBS containing 4% BSA. Slides were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI)/Supermount G solution (Fluoromount-G). Images were captured using an Olympus BX51 microscope fitted with UPlanApo lenses (200× magnification) and a MagnaFire camera and analyzed with MagnaFire 2.1c software (Olympus, Center Valley, PA). Postacquisition processing (brightness, opacity, contrast, and color balance) was applied to the entire image and accurately reflects that of the original. The number of acetylated histone 3-positive cells and DAPI-positive cells was quantified by Image-Pro Plus 4.1 (Media Cybernetics, Silver Spring, MD) (42). The number of acetylated histone 3-positive cells was normalized to the number of DAPI-positive cells per high power field (200×) for each biopsy.
Statistical Analysis
Values are reported as means ± S.E. Comparisons between two groups were determined by using a t test. Comparisons between more than two groups were determined by using one- or two-way analysis of variance followed by the Tukey post hoc test with Prism 5.0 software. A statistical probability of p < 0.05 was considered significant.
RESULTS
IL-13-induced Transcription of Eotaxin-3 Gene is CRE-dependent
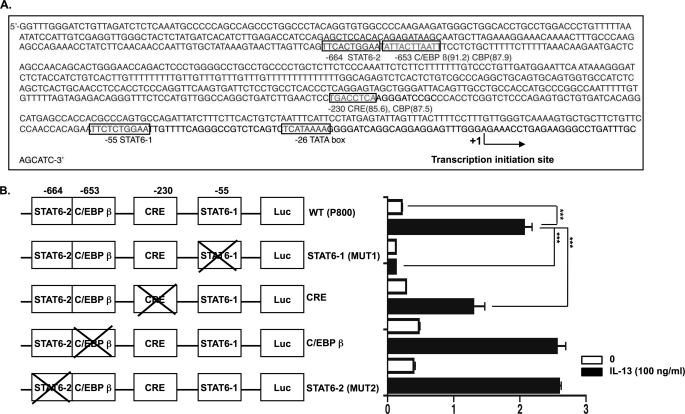
Previous studies have shown that a STAT6 consensus binding site located upstream of the eotaxin-3 transcription initiation site is required for IL-13-induced eotaxin-3 promoter activity in esophageal epithelial cells (17). Because CBP has been shown to function as a transcriptional coactivator for STAT6 (32), we examined whether CBP could mediate basal and IL-13-induced transcription of the eotaxin-3 gene. To monitor eotaxin-3 promoter activity, we used a construct containing the region of human eotaxin-3 from −929 to +35 base pairs (bp) relative to the transcription initiation site directly upstream of a firefly luciferase reporter gene. In addition to the STAT6 sites (−55 to −64 and −664 to −674), we identified two potential CBP-interacting protein binding sites, namely one CRE site (−230 to −237) and one C/EBPβ site (−653 to −662), in the promoter region of the eotaxin-3 gene (Fig. 1A). To determine the requirement of specific regulatory elements for IL-13-mediated eotaxin-3 promoter activity, we mutated the STAT6, CRE, and C/EBPβ consensus sequences within the reporter vector. The basal and IL-13-induced eotaxin-3 promoter activities of the resulting constructs were analyzed in esophageal epithelial cells (TE-7). As shown in Fig. 1B, mutation of the proximal STAT6 (Mut1) site abolished the response to IL-13. Interestingly, mutation of the CRE site significantly decreased the IL-13-induced eotaxin-3 promoter activity. In contrast, mutation of the distal STAT6 (Mut2) or C/EBPβ site had no effect. Taken together, these results indicate that the CRE site contributes to IL-13-mediated induction of the eotaxin-3 gene.
FIGURE 1.
CRE site in promoter region of eotaxin-3 gene affects IL-13-induced eotaxin-3 promoter activity. A, schematic representation of the nucleotide sequence of the promoter of the human eotaxin-3 gene. The TATA box and the transcription start site are located at nucleotides −26 and +1, respectively. Putative consensus binding sites for STAT6, CBP, C/EBPβ, and CRE are defined by boxes. B, site-directed mutagenesis studies of the eotaxin-3 promoter. Diagrams of constructs containing the eotaxin-3 promoter are shown with each of the transcription factor binding sites represented by white boxes (Luc, firefly luciferase). The crossed boxes denote the presence of mutated sites for the indicated transcription factors. These constructs were transfected into esophageal epithelial cells followed by treatment with or without IL-13 (100 ng/ml) for 24 h. For each sample, firefly luciferase activity was normalized to Renilla luciferase activity. Data are shown as mean ± S.E. (***, p < 0.001; n = 3 per group; data representative of three independent experiments).
Overexpression of CBP Enhances Both Basal and IL-13-induced Eotaxin-3 Promoter Activity, whereas CBP Inhibitor E1A Inhibits IL-13-induced Eotaxin-3 Promoter Activity
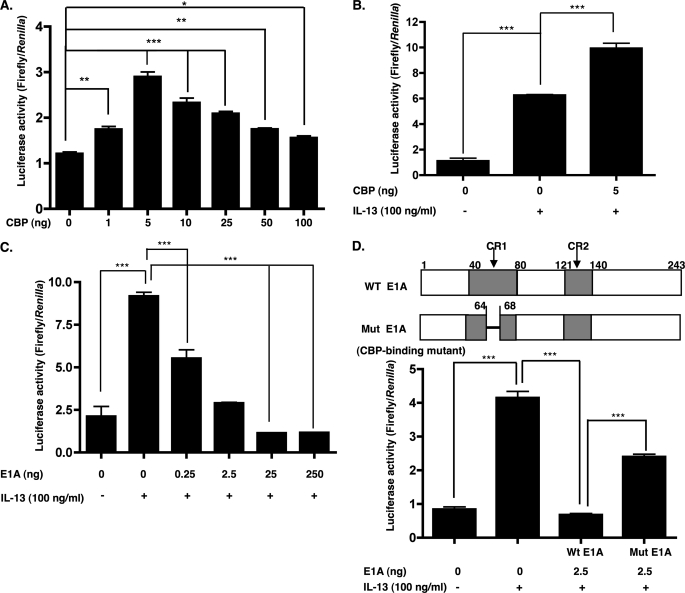
To provide evidence for the involvement of CBP in the transcription of the eotaxin-3 gene, we examined the effect of overexpression of the CBP gene, which possesses intrinsic histone acetyltransferase activity. Esophageal epithelial cells were co-transfected with the eotaxin-3 promoter construct in the presence of an expression vector containing CBP under the control of the SV40 promoter or the control vector. Overexpression of CBP enhanced basal eotaxin-3 promoter activity (Fig. 2A). CBP induced a dose-dependent increase in eotaxin-3 promoter activity with a maximum activity at 5 ng of CBP plasmid, whereas a decline was observed at higher doses. Overexpression of CBP also stimulated IL-13-induced eotaxin-3 mRNA production (Fig. 2B). These data indicate that CBP promotes transcriptional induction of eotaxin-3 both under basal and in IL-13-stimulated conditions.
FIGURE 2.
Regulation of eotaxin-3 transcriptional activation by CBP. A, esophageal epithelial cells were transiently co-transfected for 48 h with the eotaxin-3 luciferase reporter construct (Eotaxin-3/Luc; 500 ng) and either empty vector or the CBP expression plasmid (pRc/RSV-CBP; 1, 5, 10, 25, 50, and 100 ng). B, cells were transiently co-transfected with Eotaxin-3/Luc and either empty vector or the CBP expression plasmid (5 ng). After 48 h, the cells were treated with either 0 or 100 ng/ml IL-13 for 24 h. C, esophageal epithelial cells were transiently transfected with Eotaxin-3/Luc and either empty vector or the E1A expression plasmid (0.25, 2.5, 25, and 250 ng). Forty-eight hours after transfection, the cells were treated with either 0 or 100 ng/ml IL-13 for 24 h prior to measurement of luciferase activity. D, top panel, a schematic representation of E1A (WT E1A) showing the position of conserved regions 1 and 2 (CR1 and CR2) and the mutation in the CBP binding site (Mut E1A). D, bottom panel, cells were transiently co-transfected with Eotaxin-3/Luc and either WT E1A (2.5 ng) or mutant E1A (Mut E1A) (2.5 ng) for 48 h and then treated with IL-13 (100 ng/ml) for 24 h prior to measurement of luciferase activity. Data are shown as mean ± S.E. *, p < 0.05; **, p < 0.01; and ***, p < 0.001; n = 3 per group. Data are representative of three independent experiments.
To determine the effect of the CBP inhibitor adenovirus E1A protein (32, 33) on the transcriptional activity of the eotaxin-3 promoter, increasing amounts of an E1A expression vector were co-transfected with the eotaxin-3 promoter construct into esophageal epithelial cells. The eotaxin-3 promoter activity in the presence of 0.25, 2.5, 25, and 250 ng of the E1A expression vector was inhibited 40, 68, 88, and 87% compared with control IL-13-treated cells, respectively (Fig. 2C). To determine whether the inhibitory effect of E1A was due to competition for a limited amount of CBP, we examined the effect of mutating the CBP binding site in E1A (Fig. 2D, top panel) (32). Mutated E1A had reduced CBP-inhibitory activity in the context of IL-13-induced eotaxin-3 promoter activity (Fig. 2D, bottom panel). These data indicate that the repression of eotaxin-3 promoter activity by E1A correlated with its ability to bind CBP.
CBP Gene Silencing Significantly Reduces IL-13-induced Eotaxin-3 Gene Expression
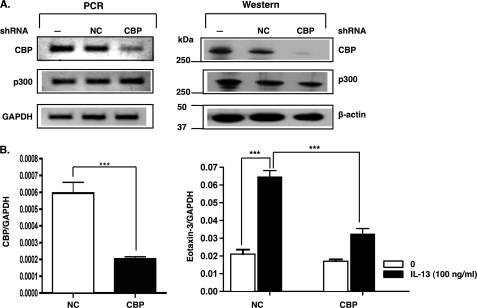
To further investigate the role of CBP in esophageal epithelial cells, we transfected cells for 24 h with either control shRNA vector or a vector expressing shRNA that targets CBP. The CBP shRNA reduced CBP transcripts by about 85% in comparison with the control shRNA (Fig. 3A, left panel), and the protein expression was similarly decreased (Fig. 3A, right panel). As controls, p300 and housekeeping gene mRNA and protein levels were unchanged in CBP shRNA-transfected cells, indicating the specificity of the CBP knockdown. We measured IL-13-induced eotaxin-3 gene expression in esophageal epithelial cells in which CBP expression was gene-silenced. CBP gene silencing significantly decreased IL-13-induced eotaxin-3 gene expression in esophageal epithelial cells (Fig. 3B). As such, these data demonstrate that CBP is required for IL-13-induced eotaxin-3 gene expression in esophageal epithelial cells.
FIGURE 3.
CBP gene silencing inhibits IL-13-induced eotaxin-3 gene expression. A, esophageal epithelial cells were either not transfected (lane 1; −), transfected with control shRNA (lane 2; NC), or CBP shRNA (lane 3; CBP). Twenty-four hours after transfection, the GFP-positive cells (control shRNA/CBP shRNA) were sorted using FACS analysis. The levels of CBP, actin, and p300 were analyzed by RT-PCR (left panel) and by Western blotting using anti-CBP, anti-actin, and anti-p300 antibodies (right panel). B, GFP-positive cells transfected with CBP shRNA plasmid and control plasmid were incubated overnight and then stimulated with or without IL-13 (100 ng/ml) for 24 h. Total RNA (100 ng) from these cells was reverse transcribed. Relative CBP (left) and eotaxin-3 (right) mRNA levels were measured by quantitative real time PCR and normalized to GAPDH. Data are shown as mean ± S.E. ***, p < 0.001; n = 5 per group. Data are representative of three independent experiments.
Histone Deacetylase Inhibition Enhances IL-13-induced Eotaxin-3 Expression
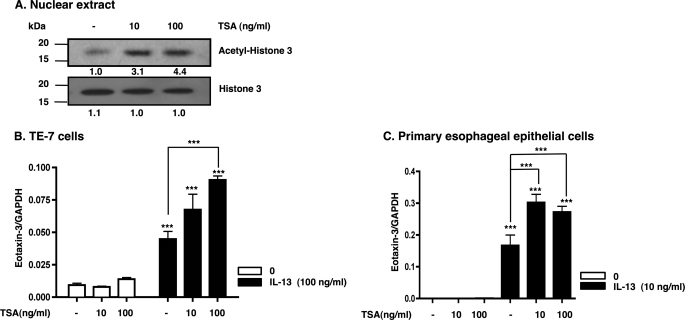
Histone deacetylases have an important role in the epigenetic regulation of gene expression by catalyzing the removal of acetyl groups, leading to chromatin condensation and transcriptional repression (11, 43, 44). We hypothesized that histone deacetylase inhibition (using trichostatin A (TSA)) would increase IL-13-induced eotaxin-3 gene expression. Primary esophageal epithelial cells were treated with TSA (10 and 100 ng/ml) or vehicle (ethanol) for 25 h, and nuclear extracts were used for Western blot analysis. As expected, TSA increased levels of acetylated histone 3 compared with vehicle-treated samples (Fig. 4A), confirming that TSA treatment was effective in our experimental system. Accordingly, cells were preincubated with TSA and subsequently treated with IL-13 for 24 h. TSA increased IL-13-induced eotaxin-3 expression in both TE-7 cells (Fig. 4B) and primary esophageal epithelial cells (Fig. 4C). In contrast, incubation of esophageal epithelial cells with TSA (10 and 100 ng/ml) alone did not induce eotaxin-3 gene expression. These data support the theory that histone acetylation promotes eotaxin-3 expression in IL-13-stimulated esophageal epithelial cells.
FIGURE 4.
TSA increases IL-13-induced eotaxin-3 gene expression in esophageal epithelial cells. A, Western blot analysis for acetylated and total histone 3 using esophageal epithelial cells. Esophageal epithelial cells were treated with either vehicle (−; ethanol) or TSA at a concentration of 10 or 100 ng/ml for 25 h. Proteins were extracted, electrophoresed, and blotted for acetylated histone 3 and total histone 3 levels. The protein levels were quantified by densitometric analysis. TE-7 cells (B) and primary esophageal epithelial cells (C) were pretreated with TSA (10 or 100 ng/ml) for 1 h and then incubated with IL-13 for 24 h. Relative eotaxin-3 mRNA levels were measured by quantitative real time PCR and normalized to GAPDH. Data are shown as mean ± S.E. ***, p < 0.001; n = 3 per group. Data are representative of three independent experiments.
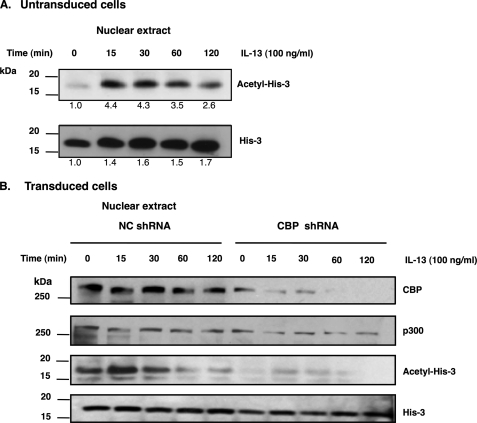
CBP Gene Silencing Reduces IL-13-induced Acetylation of Histone 3
Our results suggested that IL-13 induces a signaling pathway that promotes histone acetylation. Indeed, IL-13-stimulated acetylation of histone 3 within 15 min in esophageal epithelial cells was observed by Western blot analysis (Fig. 5A). This effect was relatively specific because IL-13 did not induce changes in acetylation of histone 4 (supplemental Fig. 1). We next aimed to determine whether CBP was responsible for IL-13-induced histone 3 acetylation. Using a lentivirus system that expressed CBP-specific shRNAs, knockdown of CBP markedly reduced the level of acetylated histone 3 following IL-13 stimulation (Fig. 5B). These data suggest that IL-13-induced histone 3 acetylation is indeed mediated by CBP.
FIGURE 5.
CBP gene silencing leads to reduction of acetylation of histone 3. A, human esophageal epithelial cells were harvested at the indicated time after treatment with IL-13 (100 ng/ml). Sixty micrograms of nuclear extracts were analyzed by immunoblotting with anti-histone 3 and anti-acetylated histone 3. B, esophageal epithelial cells were transduced with either CBP shRNA or control shRNA (NC). The levels of CBP, p300, histone 3, and acetylated histone 3 were analyzed by Western blotting using anti-CBP, anti-p300, anti-histone 3 (His-3), and anti-acetylated histone 3 (Acetyl-His-3) antibodies. Human esophageal epithelial cells were harvested at the indicated time after treatment with IL-13 (100 ng/ml). Data are representative of three independent experiments.
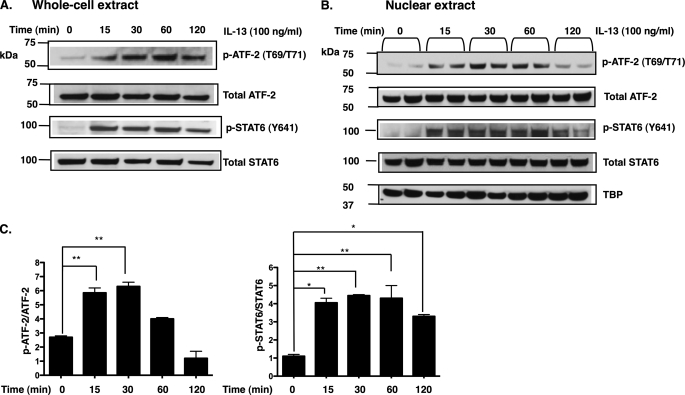
IL-13 Stimulation Increases ATF-2 Activation
Having established that the CRE site affects eotaxin-3 gene expression, we tested whether specific CRE-binding protein family members (CREB, ATF-1, and ATF-2) were activated by IL-13. We assessed the phosphorylation status of CREB, ATF-1, and ATF-2 by immunoblot analysis of isolated total cell lysates and nuclear extracts. Of the proteins tested, IL-13 induced the phosphorylation of threonine residues 69 and 71 of ATF-2 in both total lysates (Fig. 6A) and nuclear extracts 30 min following stimulation (Fig. 6, B and C); however, no phosphorylation of CREB (Ser-133) or ATF-1 (Ser-63) was observed (data not shown). As a positive control, in the same experiment, IL-13 induced the phosphorylation of STAT6. These data indicate that IL-13 induces phosphorylation of ATF-2 at Thr-69 and Thr-71, which correlate with activation of ATF-2 (45).
FIGURE 6.
IL-13 induces endogenous ATF-2 phosphorylation in esophageal epithelial cells. Human esophageal epithelial cells were harvested at the indicated time after treatment with IL-13 (100 ng/ml). Sixty micrograms of whole cell lysate (A) and nuclear extracts (B) was analyzed by immunoblotting with anti-ATF-2, anti-STAT6, anti-phospho-ATF-2 (p-ATF-2), and anti-phospho-STAT6 (p-STAT6) antibodies. TATA box-binding protein (TBP) was used as a nuclear loading control. C, densitometry results from B represent mean ± S.E. *, p < 0.05; **, p < 0.01; n = 2 per group. Data are representative of three independent experiments.
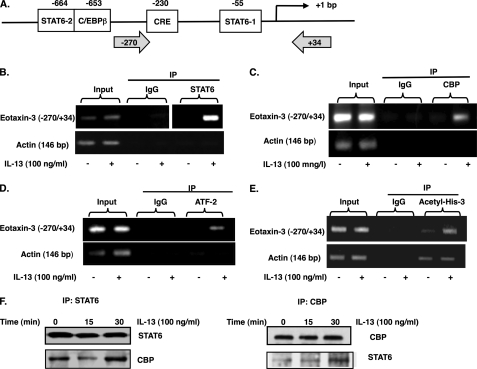
IL-13 Stimulation Promotes CBP, ATF-2, STAT6, and Acetylated Histone 3 Association with Eotaxin-3 Promoter
To determine the effect of IL-13 on STAT6, CBP, ATF-2, and acetyl-histone 3 on the eotaxin-3 promoter, ChIP assay was performed. Cross-linked and sheared chromatin from IL-13-treated esophageal epithelial cells was immunoprecipitated using anti-STAT6, anti-CBP, anti-ATF-2, or anti-acetyl-histone 3 antibody, and recovered DNA was subjected to PCR using primers spanning from −270 to +34 of the eotaxin-3 promoter sequence, which contains the CRE and STAT6–1 sites (Fig. 7A). As shown in Fig. 7, B–E, a 304-bp PCR product specific for the eotaxin-3 promoter was amplified from the immunoprecipitated samples for all four factors. In contrast, little or no product was observed when control IgG was used for immunoprecipitation. Interestingly, the amount of association with STAT6, CBP, ATF-2, and acetyl-histone 3 with the eotaxin-3 promoter was increased with IL-13 treatment compared with untreated cells (Fig. 7, B–E). As a control, the actin promoter was not amplified following ChIP analysis of STAT6, CBP, or ATF-2 (Fig. 7, B–D), and there was no increase of actin promoter association with acetyl-histone 3 after IL-13 treatment (Fig. 7E). These data indicate that IL-13 treatment promoted an increase in STAT6, CBP, ATF-2, and acetyl-histone 3 interaction with the eotaxin-3 promoter. We aimed to demonstrate an interaction between CBP and STAT6 in esophageal epithelial cells. As shown in Fig. 7F, IL-13 increased the formation of a complex that included CBP and STAT6 after 30 min in esophageal epithelial cells.
FIGURE 7.
Recruitment of STAT6, CBP, ATF-2, and acetylated histone 3 to eotaxin-3 promoter in IL-13-stimulated esophageal epithelial cells. A, diagram of the 5′-flanking region of the eotaxin-3 gene showing regulatory elements and putative binding sites of transcription factors. Gray arrows indicate PCR primer binding sites. B–E, chromatin immunoprecipitation (IP) was performed on esophageal epithelial cells cultured in medium alone or with IL-13 (100 ng/ml) for 30 min using control antibody and anti-STAT6 (B), anti-CBP (C), anti-ATF-2 (D), or anti-acetylated histone 3 (Acetyl-His-3) (E). PCR was used to detect the precipitated eotaxin-3 promoter regions (−270 to +34) or control (β-actin) promoter. One percent of the chromatin was amplified to verify equal loading (Input). F, esophageal epithelial cells were treated with IL-13 (100 ng/ml) for the indicated time. Total lysates were extracted and subjected to immunoprecipitation with anti-STAT6 antibody (F) followed by Western blot for CBP. Next, an inverse immunoprecipitation strategy was applied. Data are representative of three independent experiments.
Identification of CBP-regulated Genes in Esophageal Epithelial Cells
We identified genes regulated by CBP using genome-wide expression analysis after CBP silencing (supplemental Table 1). A subset of the top differentially regulated genes is listed in Table 1. Forty-five transcripts exhibited differential expression after CBP silencing compared with control. There are 29 genes that exhibited decreased expression and 16 genes that showed increased expression. We then determined the set of genes that had predicted STAT6 binding sites in the 4-kb region upstream of the first exon using the TraFaC program (41). Only two genes are regulated by CBP and also have predicted STAT6 binding sites (BCL11a and TGM1). Interestingly, one down-regulated gene (WNT5A) is a previously reported target for IL-13, but it does not have the STAT6 binding site (41), indicating that IL-13 may modulate histone acetylation independently of STAT6.
TABLE 1.
List of top down-regulated genes after CBP gene silencing compared with controls
MMTV, murine mammary tumor virus; CLL, chronic lymphocytic leukemia. Bolding indicates a previously reported target of IL-13.
| Gene | Description | GenBank accession no. | -Fold change |
|---|---|---|---|
| KRT10 | Keratin 10 | NM_000421 | 0.10 |
| CXCL14 | Chemokine (CXC motif) ligand 14 | NM_004887 | 0.10 |
| 15-PGDH | Hydroxyprostaglandin dehydrogenase 15 (NAD) | NM_000860 | 0.11 |
| KRT1 | Keratin 1 | NM_006121 | 0.16 |
| PTPRZ1 | Protein-tyrosine phosphatase, receptor-type, Z polypeptide 1 | NM_002851 | 0.17 |
| RARB | Retinoic acid receptor, β | NM_000965 | 0.18 |
| SEMA6D | Sema domain, transmembrane domain, and cytoplasmic domain, (semaphorin) 6D | NM_020858 | 0.18 |
| SERPINB10 | Serpin peptidase inhibitor, clade B (ovalbumin), member 10 | NM_005024 | 0.20 |
| SESN3 | Sestrin 3 | NM_144665 | 0.24 |
| WNT5A | Wingless-type MMTV integration site family, member 5A | NM_003392 | 0.26 |
| BCL11A | B-cell CLL/lymphoma 11A (zinc finger protein) | NM_022893 | 0.30 |
| DEFB1 | Defensin, β1 | NM_005218 | 0.33 |
| TGM1 | Transglutaminase 1 | NM_000359 | 0.33 |
| GJA1 | Gap junction protein, α1, 43 kDa | NM_000165 | 0.34 |
| KRT4 | Keratin 4 | NM_002272 | 0.35 |
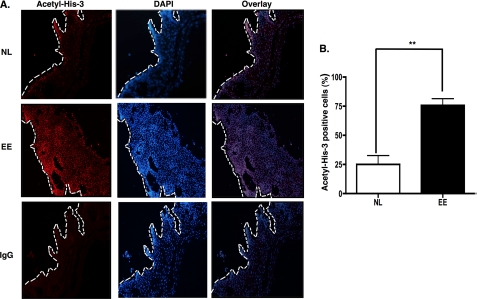
Increased Histone 3 Acetylation in Esophageal Tissue Is Associated with EE
We examined the level of histone 3 acetylation in esophageal biopsy specimens of healthy and EE patients. Biopsy specimens were immunostained with antibody specific for acetyl-histone 3, and nuclei were stained with DAPI. In biopsy specimens, the basal epithelial layer of the esophageal tissue is delineated by the white dashed line when compared with the hematoxylin and eosin-stained biopsy specimen (data not shown) (Fig. 8A). The acetyl-histone 3 staining was observed in the nuclei of the epithelial cells. Staining with control antibody confirmed that the observed signal was specific for the acetylated histone 3 antibody. The number of acetylated histone 3-positive cells was normalized to the number of DAPI-positive cells per high power field (200×) for each biopsy. In esophageal biopsies from patients with EE, there was a significant increase in acetylated histone 3-positive cells compared with normal individuals (Fig. 8B). These data imply that epigenetic regulation via chromatin remodeling is an important event in broadly regulating the EE-associated transcriptome.
FIGURE 8.
Level of acetylation of histone 3 in allergic inflammatory tissue from patients. The esophageal biopsies from normal (NL) individuals or patients with EE were subjected to immunofluorescence analysis with anti-acetyl-histone 3 (Acetyl-His-3) (red), and DAPI was used to visualize nuclei (blue). IgG control was a stained biopsy from a normal patient. The magnification is 200× (A). B, the immunofluorescence staining of anti-acetyl-histone 3 in esophageal biopsy specimens from patients with EE was quantified by using Image-Pro Plus. The number of acetylated histone 3-positive cells was normalized to DAPI-positive cells per high power field (200×) for each biopsy. Image-Pro Plus results from A represent mean ± S.E. **, p < 0.01; n = 3 or 4 per group. Data are representative of three independent experiments.
DISCUSSION
Herein, we have demonstrated that CBP and epigenetic modification of histone 3 are essential for eotaxin-3 gene expression mediated by IL-13 in esophageal epithelial cells. These conclusions are based on the following observations: 1) IL-13-induced eotaxin-3 promoter activity was decreased by mutation of the CRE site located in the eotaxin-3 promoter. 2) Eotaxin-3 promoter activity was induced by CBP and repressed by the E1A protein, a CBP inhibitor. 3) CBP gene silencing significantly decreased IL-13-induced eotaxin-3 gene expression and IL-13-induced acetylated histone 3. 4) Histone deacetylase inhibition enhanced IL-13-stimulated eotaxin-3 production. 5) IL-13-induced expression of eotaxin-3 was controlled by histone modification. 6) Stimulation with IL-13 resulted in recruitment of STAT6, CBP, and ATF-2 to the promoter region of eotaxin-3 with a corresponding increase of acetylated histone 3. 7) The esophageal epithelium from patients with EE had a high level of acetylated histone 3 compared with control individuals. Epithelial cells have been shown to be critically involved in linking innate and adaptive immunity in allergic responses in the respiratory and gastrointestinal mucosa. Herein, we have identified an epigenetic mechanism that regulates their activation by IL-13, a key cytokine involved in allergic inflammation (46–49). To the best of our knowledge, this is the first time that IL-13 has been shown to operate by modulating histone acetylation, although IL-13 induction itself has been shown to be increased by histone deacetylase inhibition in a preliminary report (50).
We observed that the promoter of eotaxin-3 contains the CRE site consensus binding sequence TGACCTCA (51). The CRE site in the eotaxin-3 promoter region contributed to IL-13-induced transcription as shown by site-directed mutagenesis. One group reported that the deletion of two CRE sites in the C/EBPβ gene promoter abrogates its expression in LPS-stimulated macrophages and also is required for macrophages to promote muscle fiber generation in vivo (52). We speculated that CBP could bind the CRE and the proximal STAT6 sites that were crucial for the eotaxin-3 promoter activity through interaction with transcription factors. We provided evidence that CBP activated basal and IL-13-induced eotaxin-3 promoter activity by showing that CBP overexpression increased eotaxin-3 promoter activity. Conversely, transfection of adenovirus protein E1A, but not mutated E1A, significantly decreased IL-13-stimulated eotaxin-3 promoter activity. The E1A protein acts by inhibiting the intrinsic histone acetyltransferase activity of CBP and/or by inhibiting interaction of CBP with the basal transcription machinery (53–55). The importance of CBP function in eotaxin-3 gene regulation was further demonstrated by CBP knockdown, which attenuated IL-13-induced eotaxin-3 expression.
In a previous study, Woisetschläger and co-worker (22) showed that two cis-acting elements (positions −233 to −105 and −928 to −783) increased the activity of the eotaxin-3 promoter (970 bp) following IL-13 stimulation and required the proximal STAT6 site in human dermal fibroblasts. In addition, the promoter sequence between −783 and −233 had repressor activity. We indeed found that the proximal STAT6 site is required for eotaxin-3 promoter activity and that the CRE binding sites (−230 to −237) acts in concert with the proximal STAT6 site and functions as a coactivator in esophageal epithelial cells; we did not find any repressor elements upstream of the CRE site. These results may be different due to distinct cell types or the use of different constructs in the two studies. Although Woisetschläger and co-worker (22) used deletion constructs, we used site-directed mutagenesis and only screened a limited number of sites between positions −783 and −233. It is likely that we did not mutate the putative repressor site in our constructs.
CBP acetylates histones at sites of transcription initiation, promoting an open chromatin structure and facilitating transcription (56–58). Inhibition of histone deacetylase activity in T cells leads to enhanced allergic airway inflammation and increased production of Th2 cytokines including IL-13 (50). Consistent with a proallergic effect, (59), we observed TSA-enhanced IL-13-induced eotaxin-3 gene expression in esophageal epithelial cells. TSA has been proven to be a remarkably specific antagonist that specifically contacts an active site conserved across the histone deacetylase family (60). IL-13 induced increased global acetylation of histone 3 (including at the eotaxin-3 gene) but not histone 4 (supplemental Fig. 1). Moreover, global acetylation of histone 3 mediated by IL-13 was markedly reduced by shRNA knockdown of CBP. Consistent with in vitro data, we also observed high levels of acetylated histone 3 directly in the esophageal tissue from patients with EE compared with control individuals. Although we showed that IL-13 could induce global acetylation of histone 3, it is possible that other factors are also involved in increasing acetyl-histone 3 in the esophageal tissue from EE patients. Our data demonstrated that CBP histone acetyltransferase activity contributes to chromatin remodeling required for IL-13-induced eotaxin-3 gene expression, and this process is likely applicable to a large subset of IL-13-induced genes.
CBP has been shown to interact with STAT6 to regulate target gene expression (32, 61, 62). Our results indeed demonstrate that CBP is located in the same complex with STAT6 in IL-13-stimulated cells, and ChIP analysis showed that these same transcription factors bind to the eotaxin-3 promoter in IL-13-treated esophageal epithelial cells. This combination of findings suggests that STAT6 and CBP bind to the eotaxin-3 promoter following IL-13 treatment. We speculate that STAT6 binding to the eotaxin-3 promoter facilitates recruitment of transcriptional coactivators such as CBP that enhance efficient transcription of eotaxin-3. These data suggest that STAT6 recruits CBP through either direct or indirect interactions, and CBP subsequently facilitates the recruitment of the basal transcriptional machinery to the eotaxin-3 promoter.
A number of transcription factors of the CREB/ATF family bind to CRE sites (63). Phosphorylation of the CREB/ATF family members promotes their binding to CRE sites. We tested whether candidate members of this family were activated following IL-13 treatment. Phosphorylation of CREB (Ser-133) or ATF-1 (Ser-63) was not observed after IL-13 treatment (data not shown). However, we observed that IL-13 increased phosphorylation of ATF-2 in esophageal epithelial cells. The specific residues phosphorylated (Thr-69/Thr-71) have been implicated previously in controlling CRE-dependent transcription (45). We found that ATF-2 binds in the vicinity of the CRE site on the eotaxin-3 promoter after IL-13 treatment using a ChIP assay. However, IL-13-induced eotaxin-3 gene expression was not reduced by shRNA knockdown of ATF-2 (supplemental Fig. 2), demonstrating specificity of CBP in regulating eotaxin-3.
In conclusion, CBP is involved in regulating the basal and IL-13-induced activity of the eotaxin-3 gene promoter. We provide direct evidence that CBP mediates eotaxin-3 gene regulation by showing that CBP gene silencing decreased IL-13-induced eotaxin-3 gene expression. CBP knockdown in esophageal epithelial cells documented that IL-13-induced acetylated histone 3 is CBP-dependent. Moreover, IL-13 promoted recruitment of CBP, STAT6, ATF-2, and acetylated histone 3 to the eotaxin-3 promoter.
To the best of our knowledge, this is the first report that IL-13 directly activates histone 3 acetylation of target genes via CBP. The finding that allergic inflammatory tissue has a markedly increased level of acetylated histone 3 highlights that epigenetic regulation of gene expression by chromatin modification is likely operational in Th2-associated human diseases. Indeed, maintaining proper CBP activity may be a potential therapeutic target for eotaxin-3-induced eosinophil infiltration in EE. Furthermore, the molecular insights obtained here should allow for therapeutic manipulation of this regulatory pathway to either promote or impair eosinophil infiltration mediated by eotaxin-3. The findings present a novel of mechanism for how environmental factors may contribute to Th2 responses as histone acetylation is modulated by a number of environmental factors such as tobacco smoke exposure, oxidative stress, particulate matter, allergen exposure, microbial exposure including endotoxin, diet, and nutritional factors (64). Collectively, these results identify an epigenetic mechanism involving CBP and modification of histone 3 in regulating the IL-13-induced transcription of eotaxin-3. As such, we have identified a plausible mechanism for the epigenetic regulation of chronic allergic inflammation.
Acknowledgments
We thank Drs. Nives Zimmermann and Julie M. Caldwell for review of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 DK76893 and U19 AI070235. This work was also supported by the Food Allergy Project, Food and Anaphylaxis Network Grant, the Campaign Urging Research for Eosinophilic Disorders, and the Buckeye Foundation. M. E. R. has an equity interest in reslizumab, a drug being developed by Cephalon, Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Table 1.
- CRE
- cAMP-responsive element
- CBP
- CRE-binding protein-binding protein
- CREB
- CRE-binding protein
- EE
- eosinophilic esophagitis
- E1A
- early region 1A
- ChIP
- chromatin immunoprecipitation
- C/EBP
- CCAAT/enhancer-binding protein
- ATF
- activating transcription factor
- TSA
- trichostatin A.
REFERENCES
- 1. Wierda R. J., Geutskens S. B., Jukema J. W., Quax P. H., van den Elsen P. J. (2010) J. Cell. Mol. Med. 14, 1225–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes P. J. (2009) Proc. Am. Thorac. Soc. 6, 693–696 [DOI] [PubMed] [Google Scholar]
- 3. Pearce E. L., Shen H. (2006) Immunol. Rev. 211, 197–202 [DOI] [PubMed] [Google Scholar]
- 4. Adcock I. M., Tsaprouni L., Bhavsar P., Ito K. (2007) Curr. Opin. Immunol. 19, 694–700 [DOI] [PubMed] [Google Scholar]
- 5. Reik W. (2007) Nature 447, 425–432 [DOI] [PubMed] [Google Scholar]
- 6. Jones P. A., Baylin S. B. (2007) Cell 128, 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumar R. K., Hitchins M. P., Foster P. S. (2009) Dis. Model. Mech. 2, 549–553 [DOI] [PubMed] [Google Scholar]
- 8. Richardson B. (2007) Nat. Clin. Pract. Rheumatol. 3, 521–527 [DOI] [PubMed] [Google Scholar]
- 9. Ito K., Hanazawa T., Tomita K., Barnes P. J., Adcock I. M. (2004) Biochem. Biophys. Res. Commun. 315, 240–245 [DOI] [PubMed] [Google Scholar]
- 10. Marmorstein R., Roth S. Y. (2001) Curr. Opin. Genet. Dev. 11, 155–161 [DOI] [PubMed] [Google Scholar]
- 11. de Ruijter A. J., van Gennip A. H., Caron H. N., Kemp S., van Kuilenburg A. B. (2003) Biochem. J. 370, 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nishihira T., Hashimoto Y., Katayama M., Mori S., Kuroki T. (1993) J. Cancer Res. Clin. Oncol. 119, 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han S., Lu J., Zhang Y., Cheng C., Li L., Han L., Huang B. (2007) Immunol. Lett. 108, 143–150 [DOI] [PubMed] [Google Scholar]
- 14. Konikoff M. R., Noel R. J., Blanchard C., Kirby C., Jameson S. C., Buckmeier B. K., Akers R., Cohen M. B., Collins M. H., Assa'ad A. H., Aceves S. S., Putnam P. E., Rothenberg M. E. (2006) Gastroenterology 131, 1381–1391 [DOI] [PubMed] [Google Scholar]
- 15. Schmeck B., Lorenz J., N'guessan P. D., Opitz B., van Laak V., Zahlten J., Slevogt H., Witzenrath M., Flieger A., Suttorp N., Hippenstiel S. (2008) J. Immunol. 181, 940–947 [DOI] [PubMed] [Google Scholar]
- 16. Furuta G. T., Schmidt-Choudhury A., Wang M. Y., Wang Z. S., Lu L., Furlano R. I., Wershil B. K. (1997) Gastroenterology 113, 1560–1569 [DOI] [PubMed] [Google Scholar]
- 17. Blanchard C., Mingler M. K., Vicario M., Abonia J. P., Wu Y. Y., Lu T. X., Collins M. H., Putnam P. E., Wells S. I., Rothenberg M. E. (2007) J. Allergy Clin. Immunol. 120, 1292–1300 [DOI] [PubMed] [Google Scholar]
- 18. Blanchard C., Wang N., Stringer K. F., Mishra A., Fulkerson P. C., Abonia J. P., Jameson S. C., Kirby C., Konikoff M. R., Collins M. H., Cohen M. B., Akers R., Hogan S. P., Assa'ad A. H., Putnam P. E., Aronow B. J., Rothenberg M. E. (2006) J. Clin. Investig. 116, 536–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rothenberg M. E., Spergel J. M., Sherrill J. D., Annaiah K., Martin L. J., Cianferoni A., Gober L., Kim C., Glessner J., Frackelton E., Thomas K., Blanchard C., Liacouras C., Verma R., Aceves S., Collins M. H., Brown-Whitehorn T., Putnam P. E., Franciosi J. P., Chiavacci R. M., Grant S. F., Abonia J. P., Sleiman P. M., Hakonarson H. (2010) Nat. Genet. 42, 289–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanchard C., Mishra A., Saito-Akei H., Monk P., Anderson I., Rothenberg M. E. (2005) Clin. Exp. Allergy 35, 1096–1103 [DOI] [PubMed] [Google Scholar]
- 21. Shinkai A., Yoshisue H., Koike M., Shoji E., Nakagawa S., Saito A., Takeda T., Imabeppu S., Kato Y., Hanai N., Anazawa H., Kuga T., Nishi T. (1999) J. Immunol. 163, 1602–1610 [PubMed] [Google Scholar]
- 22. Hoeck J., Woisetschläger M. (2001) J. Immunol. 167, 3216–3222 [DOI] [PubMed] [Google Scholar]
- 23. Rothenberg M. E. (2004) J. Allergy Clin. Immunol. 113, 11–28; quiz 29 [DOI] [PubMed] [Google Scholar]
- 24. Yang M., Hogan S. P., Mahalingam S., Pope S. M., Zimmermann N., Fulkerson P., Dent L. A., Young I. G., Matthaei K. I., Rothenberg M. E., Foster P. S. (2003) J. Allergy Clin. Immunol. 112, 935–943 [DOI] [PubMed] [Google Scholar]
- 25. Wills-Karp M. (2004) Immunol. Rev. 202, 175–190 [DOI] [PubMed] [Google Scholar]
- 26. Kitaura M., Suzuki N., Imai T., Takagi S., Suzuki R., Nakajima T., Hirai K., Nomiyama H., Yoshie O. (1999) J. Biol. Chem. 274, 27975–27980 [DOI] [PubMed] [Google Scholar]
- 27. van Wetering S., Zuyderduyn S., Ninaber D. K., van Sterkenburg M. A., Rabe K. F., Hiemstra P. S. (2007) Mol. Immunol. 44, 803–811 [DOI] [PubMed] [Google Scholar]
- 28. Blanchard C., Wang N., Rothenberg M. E. (2006) J. Allergy Clin. Immunol. 118, 1054–1059 [DOI] [PubMed] [Google Scholar]
- 29. Blanchard C., Rothenberg M. E. (2008) Gastrointest. Endosc. Clin. N. Am. 18, 133–143; x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Blanchard C., Durual S., Estienne M., Emami S., Vasseur S., Cuber J. C. (2005) Int. J. Biochem. Cell Biol. 37, 2559–2573 [DOI] [PubMed] [Google Scholar]
- 31. Chan E. M., Chan R. J., Comer E. M., Goulet R. J., 3rd, Crean C. D., Brown Z. D., Fruehwald A. M., Yang Z., Boswell H. S., Nakshatri H., Gabig T. G. (2007) Exp. Hematol. 35, 1782–1792 [DOI] [PubMed] [Google Scholar]
- 32. Gingras S., Simard J., Groner B., Pfitzner E. (1999) Nucleic Acids Res. 27, 2722–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reid J. L., Bannister A. J., Zegerman P., Martínez-Balbás M. A., Kouzarides T. (1998) EMBO J. 17, 4469–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 35. Bannister A. J., Kouzarides T. (1996) Nature 384, 641–643 [DOI] [PubMed] [Google Scholar]
- 36. Lim E. J., Kim C. W. (2007) Gene 386, 183–190 [DOI] [PubMed] [Google Scholar]
- 37. Lim E. J., Smart E. J., Toborek M., Hennig B. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H3340–H3347 [DOI] [PubMed] [Google Scholar]
- 38. Ravnskjaer K., Kester H., Liu Y., Zhang X., Lee D., Yates J. R., 3rd, Montminy M. (2007) EMBO J. 26, 2880–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shang Y., Myers M., Brown M. (2002) Mol. Cell 9, 601–610 [DOI] [PubMed] [Google Scholar]
- 40. Zimmermann N., King N. E., Laporte J., Yang M., Mishra A., Pope S. M., Muntel E. E., Witte D. P., Pegg A. A., Foster P. S., Hamid Q., Rothenberg M. E. (2003) J. Clin. Investig. 111, 1863–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jegga A. G., Sherwood S. P., Carman J. W., Pinski A. T., Phillips J. L., Pestian J. P., Aronow B. J. (2002) Genome Res. 12, 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fukasawa M., Korc M. (2004) Clin. Cancer Res. 10, 3327–3332 [DOI] [PubMed] [Google Scholar]
- 43. Barnes P. J., Adcock I. M., Ito K. (2005) Eur. Respir. J. 25, 552–563 [DOI] [PubMed] [Google Scholar]
- 44. Rosato R. R., Grant S. (2003) Cancer Biol. Ther. 2, 30–37 [DOI] [PubMed] [Google Scholar]
- 45. Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y., Yokoyama K. K. (2000) Nature 405, 195–200 [DOI] [PubMed] [Google Scholar]
- 46. Grünig G., Warnock M., Wakil A. E., Venkayya R., Brombacher F., Rennick D. M., Sheppard D., Mohrs M., Donaldson D. D., Locksley R. M., Corry D. B. (1998) Science 282, 2261–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wills-Karp M., Luyimbazi J., Xu X., Schofield B., Neben T. Y., Karp C. L., Donaldson D. D. (1998) Science 282, 2258–2261 [DOI] [PubMed] [Google Scholar]
- 48. Kuperman D. A., Huang X., Koth L. L., Chang G. H., Dolganov G. M., Zhu Z., Elias J. A., Sheppard D., Erle D. J. (2002) Nat. Med. 8, 885–889 [DOI] [PubMed] [Google Scholar]
- 49. Foster P. S., Martinez-Moczygemba M., Huston D. P., Corry D. B. (2002) Pharmacol. Ther. 94, 253–264 [DOI] [PubMed] [Google Scholar]
- 50. Su R. C., Becker A. B., Kozyrskyj A. L., Hayglass K. T. (2008) J. Allergy Clin. Immunol. 121, 57–63.e3 [DOI] [PubMed] [Google Scholar]
- 51. Walton K. M., Rehfuss R. P. (1990) Mol. Neurobiol. 4, 197–210 [DOI] [PubMed] [Google Scholar]
- 52. Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R. G., Rosenthal N., Nerlov C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17475–17480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ait-Si-Ali S., Ramirez S., Barre F. X., Dkhissi F., Magnaghi-Jaulin L., Girault J. A., Robin P., Knibiehler M., Pritchard L. L., Ducommun B., Trouche D., Harel-Bellan A. (1998) Nature 396, 184–186 [DOI] [PubMed] [Google Scholar]
- 54. Perissi V., Dasen J. S., Kurokawa R., Wang Z., Korzus E., Rose D. W., Glass C. K., Rosenfeld M. G. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3652–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Arany Z., Newsome D., Oldread E., Livingston D. M., Eckner R. (1995) Nature 374, 81–84 [DOI] [PubMed] [Google Scholar]
- 56. Karamouzis M. V., Konstantinopoulos P. A., Papavassiliou A. G. (2007) Cell Res. 17, 324–332 [DOI] [PubMed] [Google Scholar]
- 57. Goodman R. H., Smolik S. (2000) Genes Dev. 14, 1553–1577 [PubMed] [Google Scholar]
- 58. Ito K., Charron C. E., Adcock I. M. (2007) Pharmacol. Ther. 116, 249–265 [DOI] [PubMed] [Google Scholar]
- 59. Grausenburger R., Bilic I., Boucheron N., Zupkovitz G., El-Housseiny L., Tschismarov R., Zhang Y., Rembold M., Gaisberger M., Hartl A., Epstein M. M., Matthias P., Seiser C., Ellmeier W. (2010) J. Immunol. 185, 3489–3497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Finnin M. S., Donigian J. R., Cohen A., Richon V. M., Rifkind R. A., Marks P. A., Breslow R., Pavletich N. P. (1999) Nature 401, 188–193 [DOI] [PubMed] [Google Scholar]
- 61. Shankaranarayanan P., Chaitidis P., Kühn H., Nigam S. (2001) J. Biol. Chem. 276, 42753–42760 [DOI] [PubMed] [Google Scholar]
- 62. McDonald C., Reich N. C. (1999) J. Interferon Cytokine Res. 19, 711–722 [DOI] [PubMed] [Google Scholar]
- 63. Hai T., Hartman M. G. (2001) Gene 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 64. Ho S. M. (2010) J. Allergy Clin. Immunol. 126, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]