Abstract
Chronic exposure to elevated levels of glucose and fatty acids leads to dysfunction of pancreatic β-cells by mechanisms that are only partly understood. The transcription factor peroxisome proliferator-activated receptor α (PPARα) is an important regulator of genes involved in fatty acid metabolism and has been shown to protect against lipid-induced β-cell dysfunction. We and others have previously shown that expression of the PPARα gene in β-cells is rapidly repressed by glucose. Here we show that the PPARα gene is transcribed from five alternative transcription start sites, resulting in three alternative first exons that are spliced to exon 2. Expression of all PPARα transcripts is repressed by glucose both in insulinoma cells and in isolated pancreatic islets. The observation that the dynamics of glucose repression of PPARα transcription are very similar to those of glucose activation of target genes by the carbohydrate response element-binding protein (ChREBP) prompted us to investigate the potential role of ChREBP in the regulation of PPARα expression. We show that a constitutively active ChREBP lacking the N-terminal domain efficiently represses PPARα expression in insulinoma cells and in rodent and human islets. In addition, we demonstrate that siRNA-mediated knockdown of ChREBP abrogates glucose repression of PPARα expression as well as induction of well established ChREBP target genes in insulinoma cells. In conclusion, this work shows that ChREBP is a critical and direct mediator of glucose repression of PPARα gene expression in pancreatic β-cells, suggesting that ChREBP may be important for glucose suppression of the fatty acid oxidation capacity of β-cells.
Keywords: Chromatin Immunoprecipitation (ChIP), Fatty Acid Oxidation, Gene Regulation, Glucose, PPAR, ChREBP, Mlx, Pancreatic β-Cells
Introduction
The primary function of pancreatic β-cells is to continuously secrete an appropriate amount of insulin in response to the concentration of glucose and other nutrients in the blood. This process requires constant metabolic adjustment within the β-cell, which not only involves rapid changes in the post-translational state of key metabolic enzymes but also includes regulation of metabolic gene expression at the transcriptional level. Accordingly, the expression levels of a large number of active genes are regulated by glucose in β-cells (1, 2).
The peroxisome proliferator-activator receptor α (PPARα)4 is a lipid-activated nuclear transcription factor that controls glucose and fatty acid metabolism at the cellular and whole body levels (3, 4). It is highly expressed in metabolically active tissues such as liver, brown adipose tissue, and heart and plays a key role in the adaptive response to fasting (5–7). Several studies suggest that PPARα plays an important role in the regulation of fatty acid metabolism in pancreatic β-cells (8–10). Notably, the function of pancreatic islets is impaired in PPARα-deficient mice with an obese (ob/ob) background (11, 12), and PPARα activation improves β-cell function when challenged with elevated levels of free fatty acids by mechanisms that are related to increased glucose and fatty acid metabolism (9, 13). Furthermore, it has been suggested that PPARα protects against lipid-induced β-cell dysfunction by binding to a PPAR response element in the pancreas/duodenum homeobox protein 1 promoter, thereby counteracting lipid-induced repression of pancreatic/ duodenum homeobox protein 1 expression (14). Importantly, sustained exposure to elevated glucose levels, which are detrimental to the function of β-cells (15), down-regulates the expression of PPARα at the transcriptional level (16). This down-regulation may aggravate the harmful effects of elevated levels of free fatty acids on β-cells by compromising the fatty acid oxidative capacity of the β-cells. Thus, characterization of the molecular mechanisms involved in repression of PPARα expression by glucose is central to understanding nutrient-induced β-cell dysfunction and the pathophysiology of type 2 diabetes.
The basic helix-loop-helix leucine zipper transcription factor carbohydrate response element-binding protein (ChREBP) is a key mediator of glucose induction of glycolytic and lipogenic gene expression in both hepatocytes (17–21) and β-cells (22–25). ChREBP exerts its actions by binding to DNA on carbohydrate response elements together with Max-like protein X (Mlx) another basic helix-loop-helix leucine zipper protein (26–28). ChREBP is mainly found in the cytoplasm but shuttles in and out of the nuclear compartment by mechanisms that are regulated by association with 14-3-3 and importin proteins (29–31). The activity of ChREBP is linked to increased glucose metabolism (32), and glucose-sensing was recently found to be mediated by a glucose-sensing module located in the N-terminal part of ChREBP (33). Furthermore, glucose seems to activate ChREBP by increasing its nuclear import as well as by relieving repression of transcriptional activity (29, 34).
In hepatocytes, activation of ChREBP by glucose appears to be mediated by activation of protein phosphatase 2A (PP2A) via generation of xylulose-5-phosphate, an intermediate of the pentose phosphate pathway (35–37). Furthermore, dephosphorylation of ChREBP by PP2A was found to relieve inhibitory phosphorylations by protein kinase A and AMP-activated protein kinase (AMPK), which have been shown to interfere with nuclear localization and DNA binding, respectively (35, 36, 38, 39).
The mechanism of ChREBP-mediated activation of gene expression by glucose has been extensively studied in the past few years. Thus, with regards to the liver type pyruvate kinase (L-PK), it was recently shown that glucose facilitates recruitment of a complex containing ChREBP, hepatocyte nuclear factor 4α, and the histone acetyl transferase cAMP-responsive element-binding protein-binding protein to the proximal promoter (22). Similarly, ChREBP and the histone acetyl-transferase p300 are recruited to the TxNIP promoter in a glucose-dependent manner (23), and ChREBP is required for glucose-induced acetylation of histones H3 and H4, which is a characteristic of transcriptionally active genes (40). Interestingly, another basic helix-loop-helix leucine zipper transcription factor, c-Myc, has recently been shown to cooperate with ChREBP in glucose induction of L-PK and other ChREBP target genes (41).
Although glucose-induced gene expression has been thoroughly studied, the mechanism of glucose-mediated gene repression is not well understood. We have shown that the repressive effect of glucose on PPARα expression is not linked to β-cell excitation or insulin secretion but rather involves activation of PP2A and inactivation of AMPK (42). Furthermore, it was recently demonstrated that repression of PPARα expression by glucose does not involve the p38 mitogen-activated protein kinase (43). Thus, the nature of the downstream events and the transcription factors involved in the negative effect of glucose on PPARα expression have not been identified. Here we show that transcription of the rat PPARα gene is initiated from several alternative transcription start sites, and we identify two PPARα transcripts containing a novel promoter proximal exon that are spliced to exon 2. All of the transcripts are repressed by glucose in pancreatic β-cells, indicating that glucose represses the entire PPARα locus and not a particular promoter. Interestingly, by employing gain-of-function as well as loss-of-function experiments, we demonstrate that activation of ChREBP is involved and required for glucose repression of PPARα expression in pancreatic β-cells.
EXPERIMENTAL PROCEDURES
Cell Cultures
The INS-1E cell line was cultured as described previously (44) and used between passage numbers 50 and 80. Medium and supplements were from Invitrogen, and cycloheximide and metformin were from Sigma-Aldrich. For isolation of islets, rat pancreata from male Wister rats (250–300 g) were subjected to retrograde infusion from the common bile duct with 10 ml of ice-cold Hanks' balanced salt solution containing 0.5 mg/ml collagenase. Isolated pancreata were digested by incubation at 37 °C for ∼10 min, and the islets were isolated by centrifugation in Histopaque® (Sigma) and handpicked under the microscope in RPMI 1640 medium. Freshly isolated human islets were obtained from D. Bosco and T. Berney (Cell Isolation and Transplantation Center, Department of Surgery, Geneva University Hospitals). Donors had provided written informed consent. Human islets were maintained in CMRL-1066 medium at 5.6 mm glucose supplemented with 10% FCS and antibiotics for 2–4 days before experiments. Human and rat islets were cultured free floating in their respective media supplemented as for INS-1E cells.
5′-RNA Ligase-mediated Rapid Amplification of cDNA Ends (5′-RLM-RACE)
Transcriptional start site determination by 5′-RLM-RACE was performed on 5 μg of total RNA from INS-1E cells using the GeneRacer kit (Invitrogen). First strand cDNA was synthesized using a gene-specific primer PPARα-942 (5′-CTTTCAGATCGTGTTCACAGGTAA) and Superscript III reverse transcriptase (Invitrogen). Full-length 5′ cDNA ends were amplified with PPARα-515 primer (5′-GGAACTCTCCTCTCCGAGGGACTGA) followed by a nested PCR with PPARα-423 primer (5′-CTTCAAGTGGGGAGAGAGGACAGAT) using Pfu polymerase (Promega). PCR cycling parameters were as described previously (45). The PCR products were subcloned in the pGL3-basic vector (Promega) and sequenced.
Molecular Cloning
The construct pcDNA3-MycEGFP-mChREBPζ was kindly provided by Giuseppe Merla (30). The construct was cut by NaeI and XhoI to obtain a 1.9-kb fragment encoding the region 240–864 of mouse ChREBPζ (GenBankTM accession number AF245475). This fragment was cloned into pEGFP-C3 (Clontech) using the BglII/XhoI sites. The generated construct was cut by AgeI and XhoI, and the 2.6-kb fragment obtained was cloned into pShuttle-CMV (Stratagene) using the NotI/XhoI sites to create the construct pShuttle-CMV-GFP-mChREBPζ(240–864). All of the restriction sites except XhoI/NaeI sites were Klenow filled during the cloning procedures described above. To obtain the construct pShuttle-CMV-GFP-mChREBPζ, pcDNA3-MycEGFP-mChREBPζ was cut by XhoI, Klenow filled, and cut by HindIII. The 3.4-kb GFP-mChREBPζ cassette obtained was cloned into pShuttle-CMV using the HindIII/EcoRI sites. Correct insertions of fragments into vectors were confirmed by DNA sequencing of the ligation points.
Adenovirus Generation and Transduction
Recombinant adenoviruses were generated using the AdEasy cloning system (Stratagene). The CMV-GFP-mChREBPζ(240–864) cassette and the CMV-GFP-mChREBPζ cassette were transferred from the pShuttle vectors to the AdEasy-1 vector by homologous recombination in electrocompetent Escherichia coli cells BJ5183 generating the constructs pAd-CMV-GFP-mChREBPζ(240–864) and pAd-CMV-GFP-mChREBPζ, respectively. Following linearization, these constructs were transfected into the adenovirus trans-complementing cell line HEK293 (ATCC CRL-1573; ATCC, Manassas, VA) using METAFECTENE PRO (Biontex Laboratories GmbH). The produced adenoviruses Ad-CMV-GFP-mChREBPζ(240–864) and Ad-CMV-GFP-mChREBPζ were amplified and subsequently purified by ultracentrifugation using CsCl gradients. Equal viral concentrations in the different experiments were verified by determining expression of the adenoviral transcript AdE4 normalized to TFIIB expression by real time qPCR. After an overnight culture at 3 mm glucose, rat or human islets were treated with 1 ml of Hanks' balanced salt solution containing 1 mg/ml collagenase for 10 min at 37 °C before infection. INS-1E cells and islets were infected with recombinant adenovirus for 90 min at a multiplicity of infection of 20 when assuming 3000 cells/islet. They were then cultured for 20 h in new RPMI 1640 medium containing either low (3 or 5 mm) or high (25 mm) glucose before analysis.
RNA Analysis
Total RNA was isolated using TRIzol® reagent (Invitrogen) according to the manufacturer's instructions. 1 μg of RNA was treated with DNase I and reverse transcribed using MML-V reverse transcriptase (Invitrogen). Oligonucleotide primers were designed using the Primer3 software with amplicon lengths ranging from 75 to 150 bp and spanning introns. Primer specificity and efficacy was validated before use. Quantitative real time PCR was performed using a Stratagene MX3000 qPCR system and SYBR® Green Master Mix with passive reference (Sigma). All qPCRs were performed in duplicate. Expression levels in rat cells were normalized to TFIIB and human cells to an average of tubulin G and GAPDH.
Immunoblotting
For preparation of nuclear extracts INS-1E cells were washed twice with ice-cold PBS and incubated in buffer A (20 mm HEPES, pH 7.9, 1 mm EDTA, 1 mm MgCl2, 10 mm KCl, 1 mm dithioerythritol, 0.5 mm PMSF, 20% glycerol, 0.5% Triton X-100, and 1× complete protease inhibitor mixture) for 20 min on ice. The nuclei were scraped off, pelleted by centrifugation at 1000 × g for 5 min, and resuspended in buffer A containing 400 mm NaCl without Triton X-100. The samples were subjected to gentle shaking for 30 min at 4 °C and then centrifuged at 20,000 × g for 30 min before supernatant was used for subsequent analysis. For total protein extraction INS-1E cells were lysed in hypotonic lysis buffer containing 2.5% SDS. Primary antibodies anti-PPARγ (sc-7273), anti-TFIIB (sc-225), and anti-ChREBP (sc-21189) were from Santa Cruz Biotechnology Inc. sc-7273 (E-8) is raised against the C terminus of PPARγ, which is highly conserved between the PPAR subtypes. Using the sc-7273 antibody, we recently showed that PPARα and PPARδ but not PPARγ is detectable in INS-1E cells (46).
siRNA Transfections
INS-1E cells were reverse transfected with 50 nm of siRNA duplexes (Dharmacon) in OptiMEM using Dharmafect Reagent 1 (Dharmacon). Duplexes were targeted to 19-bp regions of the rat ChREBP cDNA sequence (GenBankTM accession number AB074517). The siRNA target sequences were as follows: siChREBP#1, AAGAGGCGTTTCAATATTA; siChREBP#2, GCAACTGAGGGATGAAATA. A siRNA duplex targeting the luciferase gene (siLuc, CGTACGCGGAATACTTCGA) with no known mammalian sequence homology or biological effect was used as a control. After a 4–6-h transfection period, the cells were cultured for 36 h in normal medium and subsequently preincubated in 5 mm glucose medium for 24 h before they were subjected to either 5 or 25 mm glucose medium for a further 24 h and harvested.
Chromatin Immunoprecipitation
INS-1E cells were cross-linked with 2 mm disuccinimidyl glutarate for 45 min at room temperature and subsequently cross-linked with 1% formaldehyde for another 10 min at room temperature. Cross-linking was stopped by adding glycine to a final concentration of 0.125 m (10 min), before the cells were washed twice in cold PBS and harvested in lysis buffer (1% Triton X-100, 0.1% SDS, 150 mm NaCl, 1 mm EDTA, 20 mm Tris, pH 8.0, and complete protease inhibitor mixture). DNA concentration was determined by absorption at 260 nm, and the samples were diluted to equal concentrations and sonicated 16 times (30 s on/off, maximum) at 4 °C using a Diagenode Bioruptor. After centrifugation at 10,000 × g for 1 min at 4 °C, the supernatants were collected and incubated rotating with ChREBP antibody (NB400-135; Novus Biologicals) for 3 h at 4 °C. Protein A beads (Amersham Biosciences) were washed three times in lysis buffer and diluted four times in lysis buffer containing 1 μg/μl BSA and 1× complete for 2 h at 4 °C rotating. Chromatin (400 μl) was incubated with prepared beads (100 μl) overnight at 4 °C rotating and then washed twice with buffer 1 (1% Triton X-100, 0.1% SDS, 0.1% NaDOC, 150 mm NaCl, 1 mm EDTA, 20 mm HEPES), once with buffer 2 (1% Triton X-100, 0.1% SDS, 0.1% NaDOC, 500 mm NaCl, 1 mm EDTA, 20 mm HEPES), once with buffer 3 (0.25 m LiCl, 0.5% Nonidet P-40, 0.5% NaDOC, 1 mm EDTA, 20 mm HEPES), and twice with buffer 4 (1 mm EDTA, 20 mm HEPES). Each wash was for 5 min at 4 °C rotation followed by centrifugation for 1 min at 700 × g. DNA-protein complexes were eluted with 400 μl of elution buffer (1% SDS and 0.1 m NaHCO3) and de-cross-linked by adding 0.2 m NaCl and shaking overnight at 65 °C. Enriched DNA was purified by phenol-chloroform-isoamyl alcohol extraction and precipitated using 10 μg of glycogen, volume of 3 m sodium acetate (pH 5.2), and 2.5 volumes of cold ethanol.
[1-14C]Oleic Acid Oxidation
INS-1E cells were cultured in RPMI 1640 medium containing 5 or 25 mm glucose for 24 h and then incubated for a further 48 h in 1% serum medium containing 5 or 25 mm glucose and Me2SO or GW7647 (100 nm). Fatty acid oxidation measurements were performed essentially as described (47) in serum-free medium containing 5 mm glucose and [1-14C]oleic acid complexed to BSA. Cultures were left for [1-14C]CO2 trapping overnight, and the data were normalized to total cellular protein.
RESULTS
Characterization of the Rat PPARα 5′-UTR
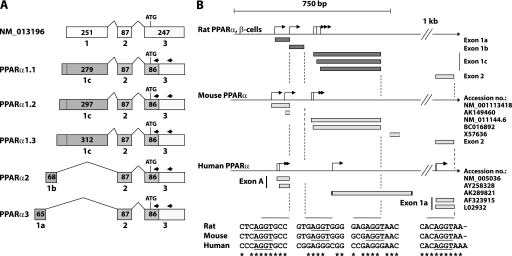
The 5′-UTR of human PPARα has been well characterized in hepatocytes, where it is subject to alternative splicing (45, 48). In contrast, little is known about the architecture of the rat PPARα 5′-flanking region. Thus, to study the regulation of PPARα expression in rat pancreatic β-cells, we characterized the 5′-UTR of the PPARα gene in the pancreatic β-cell line INS-1E using 5′-RNA ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACE). PCR amplification of full-length cDNAs was conducted using gene-specific primers that anneal to the coding sequence of exon 3 (Fig. 1A). By this approach, we obtained five RACE-PCR products of different sizes (data not shown). Sequence analysis revealed that all five RACE-PCR products share 100% sequence similarity with genomic regions of the rat PPARα core promoter region. Furthermore, the products aligned perfectly with the 5′-end of exon 3 as well as the entire exon 2 of the rat PPARα gene previously characterized and deposited in GenBankTM (accession number NM_013196) (49) (Fig. 1A). Three of the RACE-PCR products (PPARα1.1–1.3) contain the previously characterized exon 1 (designated exon 1c); however, none of the transcripts contain the start site indicated in NM_013960. In contrast, all three transcripts have an extended 5′-end compared with NM_013196 because of a more upstream initiation of transcription. Interestingly, we also identified two novel promoter proximal exons (exon 1a and 1b) that are alternatively spliced to exon 2 and thereby give rise to the two RACE-PCR products designated PPARα2 and PPARα3, respectively (Fig. 1A). The sequences of these five novel 5′-UTR variants of rat PPARα have been deposited in GenBankTM (accession numbers HM117636–HM117640).
FIGURE 1.
Characterization of the rat PPARα 5′-UTR. Transcriptional start sites of the rat PPARα gene were mapped by 5′-RNA ligase-mediated rapid amplification of cDNA ends (5′-RLM-RACE) performed on total RNA from INS-1E cells. A, schematic representation of the characterized PPARα 5′-UTR and its exon structure. Sequences identified by 5′-RLM-RACE are marked by dark shading. The number of nucleotides in each exon is indicated. Arrows indicate primers used for nested PCR amplification of cDNAs. B, alignment of the characterized rat 5′-UTR with published sequence data from the mouse and human (promoter A) PPARα genes. Identified transcription start sites are indicated by arrows. Exon-intron boundary sequences are depicted and consensus 5′-splice donor sites (AGGT) are underscored. The mouse and human illustrations are based on sequence data obtained from GenBankTM.
To compare our findings with data available from the mouse and human PPARα promoters, a local alignment was performed, and a schematic representation is depicted in Fig. 1B. The alignment shows that exon 2 of rat and mouse PPARα as well as exon 1a of human PPARα terminate in a conserved consensus 5′-splice donor site (AGGT). Furthermore, analysis of the sequence covering the exon-intron boundary of the novel exon 1a revealed that it terminates in a consensus 5′-splice donor site (AGGT) that is conserved between rat, mouse, and human PPARα, and this site is involved in the splicing of exon A and exon 1a of the human PPARα transcripts NM_005036 and AY258328 (Fig. 1B).
Expression of PPARα Transcripts Is Regulated by Glucose in Pancreatic β-Cells
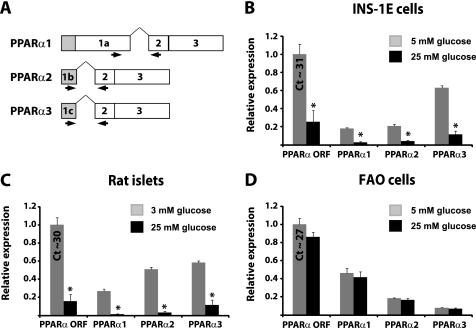
To confirm the existence of the two novel PPARα transcripts, designated PPARα2 and PPARα3, and to evaluate their respective regulation by glucose, we designed quantitative real time PCR primers able to distinguish between the three different PPARα transcripts (Fig. 2A). We found that PPARα2 and PPARα3 were expressed in both INS-1E cells, isolated rat islets, and FAO cells (Fig. 2, B–D). Interestingly, all three PPARα transcripts are down-regulated by glucose to a similar degree in INS-1E cells and isolated rat islets (Fig. 2, B and C). The degree of repression by glucose is similar to what we previously observed using primers that anneal to sequences within the open reading frame of PPARα (42). All PPARα transcripts are also expressed in the FAO rat hepatoma cell line; however, expression is independent of the glucose concentration in these cells (Fig. 2D). Taken together, these findings show that the entire PPARα promoter region is repressed by glucose in rat β-cells but not in hepatoma cells.
FIGURE 2.
PPARα transcripts are expressed in INS-1E cells and isolated rat pancreatic islets and are all down-regulated by glucose. A, schematic representation of the binding sites of qPCR primers used to differentiate between different PPARα transcripts. B–D, INS-1E cells (B), isolated rat pancreatic islets (C), and FAO hepatoma cells (D) were preincubated in low glucose concentrations (5, 3, and 5 mm, respectively) for 24 h and then incubated in either low glucose or high glucose (25 mm) for a further 24 h before analysis of PPARα mRNA expression as determined by qPCR. The bars represent the means ± S.D. of three independent experiments. *, p < 0.01 versus low glucose (t test).
PPARα Expression Is Rapidly Down-regulated by Glucose in INS-1E Cells
Studies from the Prentki group (16) and from our group (42) have clearly demonstrated a fast and direct effect of glucose on PPARα expression independent of de novo protein synthesis and corresponding to complete inhibition of transcription. Furthermore, we have also shown that transcriptional repression of PPARα expression by glucose in pancreatic β-cells involves activation of PP2A and inactivation of AMPK (42). However, the DNA binding factor(s) mediating the repressive effect on PPARα expression have not yet been identified. Interestingly, PP2A and AMPK have been implicated in the regulation of the glucose-responsive basic helix-loop-helix leucine zipper transcription factor ChREBP at the level of post-translational modifications (38), and we therefore considered the possibility that ChREBP could mediate the repression of PPARα in pancreatic β-cells. ChREBP is known to activate the glucose-induced lipogenic genes fatty acid synthase (FAS) and L-PK in β-cells (24). Furthermore, preliminary data from our laboratory indicated that another lipogenic gene, cytosolic glycerol-3-phosphate dehydrogenase (GPDH), which is a direct target of ChREBP in primary rat hepatocytes (20), is also a very sensitive ChREBP target gene in INS-1E cells (data not shown). This notion is in line with a recent study showing that ChREBP is recruited to the GPDH carbohydrate response element in a glucose-regulated manner in INS-1-derived 832/13 insulinoma cells (41).
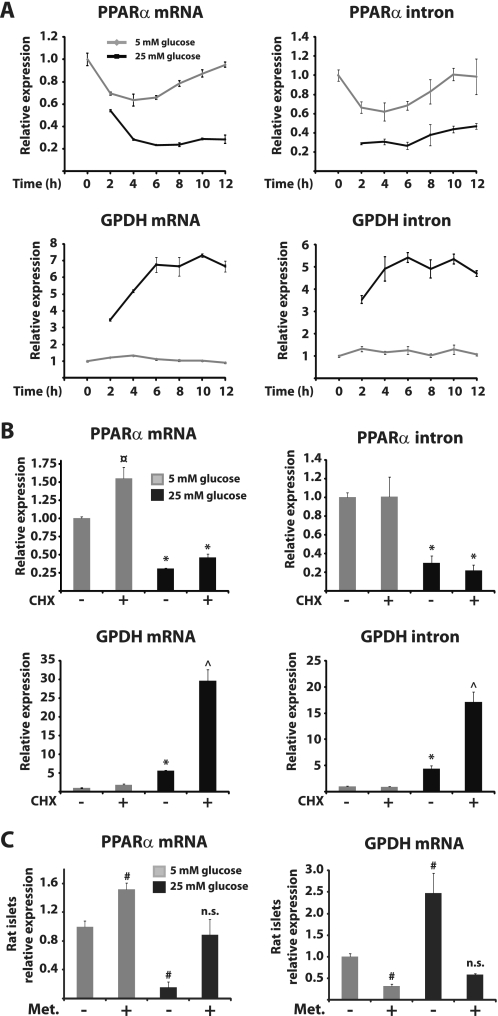
To compare the dynamics of repression of PPARα with those of glucose-induced gene activation mediated by ChREBP, we performed a time course experiment at low and high concentrations of glucose. INS-1E cells were preincubated at 5 mm glucose for 24 h and subsequently cultured at either 5 or 25 mm glucose for various time periods. The mRNA expression level of PPARα as well as that of GPDH was determined by real time PCR, and the data showed that PPARα mRNA expression is significantly repressed, whereas GPDH mRNA is significantly induced by 25 mm glucose after 2 h (Fig. 3A). The maximal repression of PPARα and the maximal induction of GPDH are reached after 6 h. To evaluate changes in the rate of transcription, we also measured the pre-mRNA expression levels of PPARα and GPDH. PPARα pre-mRNA levels are markedly repressed by 25 mm glucose within 2 h, and this repression is maintained throughout the 12-h time period (Fig. 3A). Similarly, GPDH pre-mRNA levels are induced by 25 mm glucose within 2 h but reach a maximum level after 6 h. This shows that transcription from the PPARα gene is rapidly and effectively repressed by glucose and that the dynamics of this regulation are very similar to glucose induction of the ChREBP target gene GPDH.
FIGURE 3.
Transcription from the PPARα gene is rapidly and effectively repressed by glucose in β-cells. A, INS-1E cells were preincubated in 5 mm glucose medium for 24 h and subsequently cultured at either 5 or 25 mm glucose for the indicated time periods. B, INS-1E cells were preincubated in 5 mm glucose medium for 24 h and subsequently cultured in 5 or 25 mm glucose medium with or without 10 μg/ml cycloheximide for 6 h. C, isolated rat pancreatic islets were preincubated in 3 mm glucose for 24 h before culture in either 3 or 25 mm glucose medium with or without 1 mm metformin (Met.). Expression levels of PPARα and GPDH were determined by quantitative PCR. The bars represent the means ± S.D. of three independent experiments. *, p < 0.05 versus 5 mm glucose ± cycloheximide (CHX); ¤, p < 0.01 versus 5 mm glucose and 25 mm glucose ± cycloheximide; ^, p < 0.05 versus 5 mm glucose ± cycloheximide and 25 mm glucose; #, p < 0.05 versus 5 mm glucose (one-way ANOVA).
The protein synthesis inhibitor cycloheximide has previously been used to show that glucose repression of PPARα mRNA expression after 24 h occurs in the absence of de novo protein synthesis in INS(832/13) cells (16). To evaluate the effect of blocking protein synthesis on glucose-regulated gene transcription, we measured both PPARα and GPDH mRNA and pre-mRNA levels in the presence of cycloheximide at high versus low glucose concentration in INS-1E cells (Fig. 3B). In line with previous data on PPARα mRNA levels, cycloheximide treatment does not affect glucose repression of PPARα mRNA or pre-mRNA expression after 6 h. In contrast, cycloheximide leads to super induction of GPDH expression by glucose (Fig. 3B), which is likely due to secondary effects of translational arrest as previously reported for several immediate early genes (50). This demonstrates that repression of PPARα transcription by glucose is independent of de novo protein synthesis.
Our hypothesis that ChREBP is involved in repression of PPARα expression by glucose was further supported by the finding that PPARα and GPDH expression in isolated rat islets is subject to reciprocal regulation not only by glucose but also by the AMPK activator metformin (Fig. 3C). These results suggest that glucose regulates GPDH and PPARα expression by a common signaling pathway involving AMPK and ChREBP. Furthermore, the results indicate that AMPK can counteract glucose-induced ChREBP activity in β-cells as previously reported in hepatocytes (39).
PPARα Expression Is Repressed by ChREBP in Pancreatic β-Cells
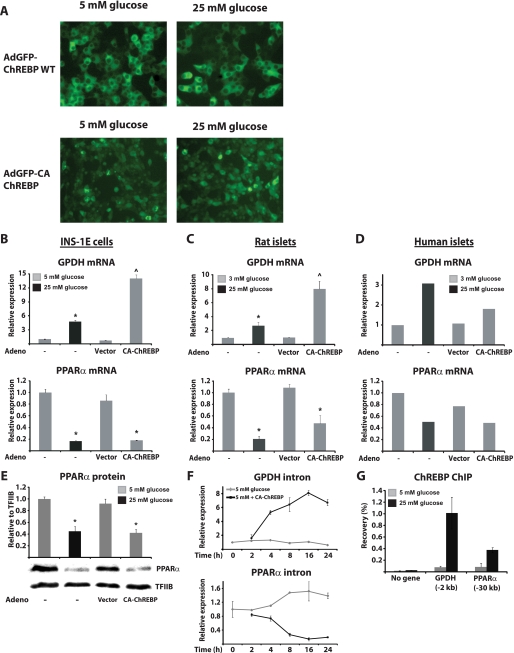
To further investigate the possibility that PPARα could be a direct target of ChREBP in pancreatic β-cells, we ectopically expressed WT GFP-tagged ChREBP together with its heterodimerization partner Mlx in INS-1E cells. However, despite high expression levels, we did not observe significant effects on known ChREBP target genes or PPARα, neither at 5 nor at 25 mm glucose (data not shown). The lack of effect following WT-ChREBP overexpression is in line with previous studies in INS(832/13) cells (33, 34). It may at least in part reflect the predominant nuclear exclusion of ectopically expressed WT-ChREBP at both 5 and 25 mm glucose in INS-1-derived cells (33, 34) (Fig. 4A). The N-terminal part of ChREBP (amino acids 1–192) has previously been shown to repress activity under low glucose conditions, and a mutant ChREBP lacking this region has been shown to be constitutively active at low glucose conditions (33, 51). In parallel with these investigations by others, we constructed an adenoviral vector expressing a ChREBP GFP fusion protein lacking the first 239 amino acid residues of ChREBP (Ad-CMV-GFP-mChREBPξ (240–864)). This N-terminally truncated ChREBP protein (denoted CA-ChREBP) displays both constitutive nuclear localization (Fig. 4A) and constitutive activity in INS-1E cells as assessed by expression of a number of ChREBP target genes under low glucose conditions including GPDH (Fig. 4B and data not shown). Likewise, induction of GPDH and other ChREBP target genes is also observed in isolated rat and human islets transduced with this adenovirus (Fig. 4, C and D, and data not shown). Most interestingly, PPARα mRNA expression is strongly suppressed by CA-ChREBP in both INS-1E cells and isolated rat islets cultured in low glucose conditions (Fig. 4, B–D). In human islets, we made the novel discovery that high glucose also represses PPARα expression (Fig. 4D) in line with data from experiments performed in rodents. Furthermore, although the effect is less dramatic than in rodent islets, CA-ChREBP also suppresses PPARα expression in isolated human islets (Fig. 4D). The suppression of PPARα mRNA expression by CA-ChREBP at 5 mm glucose in INS-1E cells was reflected in significantly decreased PPARα protein expression to levels similar to that observed in cells cultured in 25 mm glucose medium (Fig. 4E). Thus, acute ectopic expression of CA-ChREBP not only induces well known ChREBP target genes but also mimics the effect of high glucose on PPARα expression. To investigate the kinetics by which CA-ChREBP suppresses PPARα expression, we ectopically expressed CA-ChREBP in INS-1E cells and determined pre-mRNA expression levels of GPDH and PPARα at different time points. We found that induction of GPDH and repression of PPARα pre-mRNA occurred rapidly and simultaneously starting at 4 h and reaching a maximal effect after 16 h (Fig. 4F). Cumulatively, this strongly indicates that ChREBP is directly involved in repression of PPARα expression in pancreatic β-cells.
FIGURE 4.
ChREBP activation represses PPARα expression at low glucose levels in pancreatic β-cells and is recruited to the distal PPARα promoter in a glucose-regulated manner. A, INS-1E cells were co-transduced with adenovirus expressing Mlx and either GFP-mChREBP or GFP-CA-ChREBP for 1 h. Fluorescence pictures were acquired after subsequent incubation in RPMI medium containing either 5 or 25 mm glucose for 12 h. B–D, INS-1E cells and isolated rat and human islets were preincubated in low glucose medium (5, 3, and 3 mm glucose, respectively) for 24 h. The cells were subsequently cultured in either low (3 or 5 mm) or high glucose (25 mm) or transduced with an adenovirus expressing CA-ChREBP or a control adenovirus in low glucose medium and cultured for a further 20 h before analysis of GPDH and PPARα expression. E, INS-1E cells were preincubated in 5 mm glucose medium for 24 h and then transduced with an adenovirus expressing CA-ChREBP or a control adenovirus for 1 h and cultured for 20 h before nuclear extraction of proteins. F, INS-1E cells were preincubated 5 mm glucose medium and transduced with CA-ChREBP for 1 h prior to subsequent culture and harvest at the indicated time points. G, INS-1E cells were preincubated in 5 mm glucose for 24 h and then cultured in either 5 or 25 mm glucose medium before formaldehyde cross-linking and harvest of chromatin. Relative occupancy at the indicated sites was determined using an antibody against ChREBP and evaluated by ChIP-qPCR. INS-1E and rat islet bars represent the means ± S.D. of at least three independent experiments, whereas the data from human islets represent the values of one experiment. *, p < 0.01 versus 5 mm glucose ± vector; ^, p < 0.01 versus 5 mm glucose ± vector and 25 mm glucose (one-way ANOVA).
ChREBP Is Recruited to the Distal PPARα Promoter
In a parallel study, we have investigated the binding of GFP-tagged CA-ChREBP to the genome using ChIP combined with deep sequencing. Data from these analyses revealed binding of CA-ChREBP to several known carbohydrate response elements, including that of GPDH (20, 41), FAS, and L-PK (24) (data not shown). Interestingly, we also found CA-ChREBP to be recruited to a distal site ∼30 kb upstream of the PPARα promoter, whereas no other binding site closer to the PPARα locus was observed. Subsequent ChIP-PCR analysis revealed that endogenous ChREBP is recruited to this distal site in the PPARα promoter in a glucose-dependent manner (Fig. 4G). These data suggest that glucose may repress PPARα expression by modulating the recruitment of ChREBP to the distal binding site 30 kb upstream of the PPARα promoter region.
ChREBP Knockdown Blunts Glucose Repression of PPARα Expression in INS-1E Cells
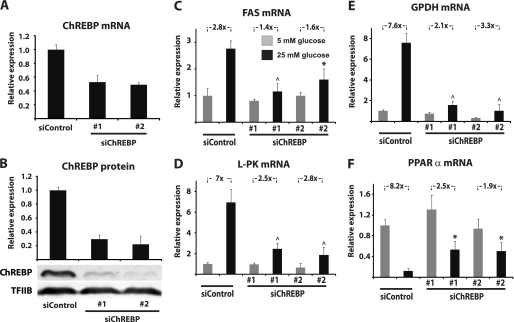
To directly evaluate the role of ChREBP in glucose repression of PPARα expression, INS-1E cells were transfected with siRNA duplexes targeting the rat ChREBP gene (siChREBP#1 and siChREBP#2) or with a control siRNA with no mammalian sequence homology (siControl). The siChREBP duplexes were able to reduce ChREBP mRNA levels by ∼50% (Fig. 5A) and ChREBP protein levels by 70–75% (Fig. 5B). This reduction in ChREBP expression severely blunts glucose-induced FAS and L-PK expression (Fig. 5, C and D) in line with data from a previous study performed in the mouse insulinoma cell line MIN6 (24). Similarly, glucose induction of GPDH expression is also drastically reduced (Fig. 5E). Importantly, knockdown of ChREBP significantly impairs glucose repression of PPARα mRNA expression (Fig. 5F), thereby demonstrating that ChREBP is required for repression of PPARα expression by glucose in β-cells.
FIGURE 5.
ChREBP knockdown blunts glucose repression of PPARα expression in INS-1E cells. A and B, INS-1E cells were transfected with siChREBP#1, siChREBP#2, or siControl and ChREBP knockdown was evaluated at the level of mRNA (A) and protein 90 h after transfection (B). C–F, the effects on mRNA levels of the ChREBP target genes FAS (C), L-PK (D), and GPDH (E), as well as PPARα (F), were assessed. The bars represent the means ± S.D. of three independent experiments. *, p < 0.05; ^, p < 0.01 versus siControl at 25 mm glucose (one-way ANOVA).
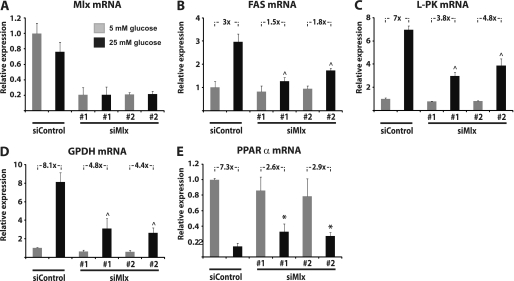
To further investigate the molecular mechanisms involved in repression of PPARα expression by glucose in β-cells, we decided to evaluate the role of Mlx, which is the functional heterodimeric partner of ChREBP when mediating glucose-induced gene activation (20, 27, 28). Knockdown of Mlx by siRNA reduces Mlx mRNA levels by 70–80% (Fig. 6A) and significantly reduces glucose induction of the ChREBP target genes FAS, L-PK, and GPDH (Fig. 6, B–D). Interestingly, knockdown of Mlx also significantly blunts the repression of PPARα mRNA expression by glucose (Fig. 6E). These findings demonstrate that glucose regulation of classical ChREBP target genes, as well as repression of PPARα, is dependent on Mlx in β-cells, indicating that Mlx is the functional heterodimeric partner of ChREBP not only when ChREBP mediates glucose-induced gene expression but also when it acts as a transcriptional repressor.
FIGURE 6.
Ablation of Mlx by siRNA blunts glucose regulation of classical ChREBP target genes and PPARα in INS-1E cells. A, INS-1E cells were transfected with siMlx#1, siMlx#2, or siControl, and Mlx knockdown was evaluated at the level of mRNA 90 h after transfection. B–E, the effects on mRNA levels of the ChREBP target genes FAS (B), L-PK (C), and GPDH (D), as well as PPARα (E), were assessed. The bars represent the means ± S.D. of three independent experiments. *, p < 0.05; ^, p < 0.01 versus siControl at 25 mm glucose (one-way ANOVA).
ChREBP Knockdown Blunts Glucose Repression of Fatty Acid Oxidation Capacity Induced by PPARα Activation
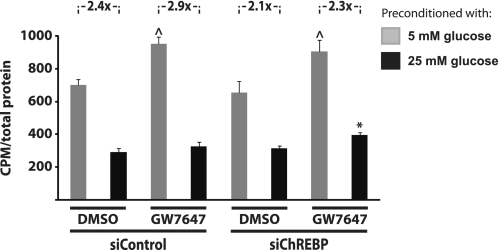
Sustained exposure to elevated levels of glucose and fatty acids is detrimental to the function of pancreatic β-cells in vitro (52–54) and in vivo (55). This may in part be due to a compromised fatty acid oxidation capacity as a result of glucose repression of PPARα expression mediated by ChREBP. We therefore examined the effect of glucose and PPARα activation on the fatty acid oxidation capacity of INS-1E cells with and without knockdown of ChREBP. Activation of PPARα by the specific agonist GW7647 significantly induces fatty acid oxidation capacity at 5 mm glucose, and this induction is not affected by ChREBP knockdown (Fig. 7). As previously reported using INS-1 β-cells (16), preincubation with high glucose for 72 h dramatically reduces fatty acid oxidation capacity at 5 mm glucose of INS-1E cells treated with a control siRNA (Fig. 7). This reduction (2.4-fold) is slightly diminished by knockdown of ChREBP (to 2,1-fold), although it does not reach statistical significance. Consistent with the suppression of PPARα expression by high glucose, preincubation with 25 mm glucose for 72 h completely blocks induction of fatty acid oxidation capacity by PPARα activation. Importantly, however, this inhibition by preincubation with high glucose is slightly but significantly reversed by siRNA-mediated ChREBP knockdown, indicating that ChREBP mediates the negative effects of chronic high glucose on PPARα-induced fatty acid oxidation capacity. Thus, these results support the functional significance of ChREBP as a glucose-induced regulator of PPARα expression and activity in β-cells.
FIGURE 7.
ChREBP knockdown blunts glucose repression of fatty acid oxidation capacity induced by the PPARα specific agonist GW7647 in INS-1E cells. The cells were incubated with 5 or 25 mm glucose for 24 h and then incubated for a further 48 h in 1% serum medium containing 5 or 25 mm glucose and Me2SO or GW7647 (100 nm). Fatty acid oxidation measurements were performed in triplicate in serum-free medium containing 5 mm glucose and [1-14C]oleic acid. ^, p < 0.01 versus Me2SO at 5 mm glucose; *, p < 0.05 versus siControl plus Me2SO or GW7647 at 25 mm glucose and siChREBP plus Me2SO at 25 mm glucose (one-way ANOVA).
DISCUSSION
PPARα is a regulator of fatty acid metabolism in pancreatic β-cells, and PPARα has been reported to protect against lipid-induced β-cell dysfunction (9, 12–14). Studies performed in rodents suggest that reduced activity of PPARα is implicated in nutrient-induced β-cell dysfunction. Thus, PPARα expression is down-regulated by glucose in rat β-cells (16, 42), and PPARα function is compromised in β-cells from rodent models of diabetes (8). Here we show for the first time that glucose also down-regulates PPARα expression in human islets, which indicates that observations regarding this regulation in rodent β-cells are also relevant to human health and disease.
Previous data from Prentki's group (16) and our group (42) have shown that glucose down-regulates PPARα expression independent of de novo protein synthesis by mechanisms that appear to involve transcriptional regulation. However, the DNA binding mediators governing this regulation have not yet been identified. In an attempt to identify the cis-acting elements on which these factors are acting, we used 5′-RLM-RACE to characterize the rat PPARα 5′-UTR and pinpoint the core promoters affected by glucose. We identified five PPARα transcripts expressed from five different start sites within just a few hundred base pairs. We also identified two novel promoter proximal exons that are alternatively spliced to exon 2. Notably, however, all identified PPARα transcripts are regulated in a similar manner by glucose in INS-1E cells and in isolated rat islets. Thus, glucose represses the entire PPARα promoter region without any specificity toward certain transcriptional start sites.
Based on the identified reciprocal and rapid regulation by glucose of PPARα and ChREBP target genes, we speculated that ChREBP might be directly mediating the negative effect of glucose on PPARα expression. So far ChREBP has mainly been found to act as a glucose-stimulated transcriptional activator of glycolytic and lipogenic genes in hepatocytes (17–21) and in pancreatic β-cells (22, 23). Recently, however, it was reported that ChREBP acts as a negative regulator of the ARNT/HIF-1α gene (56). We employed a combination of gain-of-function and loss-of-function experiments to investigate the possible role of ChREBP in glucose repression of PPARα expression in β-cells. The results showed that constitutively active ChREBP at low glucose concentrations acutely represses PPARα expression in both INS-1E cells and isolated rat and human islets, thereby mimicking the effect of high glucose on PPARα expression. Importantly, knockdown of ChREBP by siRNA impairs glucose repression of PPARα expression in INS-1E cells as well as the induction of ChREBP-activated genes. Interestingly, knockdown of the heterodimeric partner Mlx, known to be required for ChREBP activation of target genes, also resulted in impaired glucose repression of PPARα. This indicates that the repressive effect of ChREBP on PPARα gene expression is also mediated through the ChREBP-Mlx heterodimer. The Mlx dependence further suggests that ChREBP-mediated repression occurs by direct binding to DNA, rather than by tethering, although firm proof of that would require substantial additional work.
By combining overexpression of constitutively active ChREBP with ChIP sequencing, we identified a putative ChREBP-binding site 30 kb upstream of the PPARα promoter. Subsequent analysis revealed that endogenous ChREBP is recruited to this site in a glucose-dependent manner. It is conceivable that binding of ChREBP to this site is involved in the repression of PPARα; however, direct proof thereof requires very detailed analyses that are beyond the scope of this work. In conclusion, our data show that ChREBP is a critical and direct mediator of glucose repression of PPARα expression in pancreatic β-cells and provide further evidence that ChREBP can act as a transcriptional repressor.
ChREBP has previously been shown to activate the expression of the lipogenic genes L-PK and FAS in pancreatic β-cells (22, 24). In line with a recent study performed in INS(832/13) cells (41), we show here that the induction by glucose of another lipogenic gene, GPDH, is mediated by ChREBP in INS-1E cells. Cytosolic GPDH is a NADPH-dependent enzyme that catalyzes the conversion of dihydroxyacetone phosphate to glycerol-3-phosphate, and GPDH is therefore an important link between glycolysis and de novo lipogenesis. Furthermore, we show for the first time that cytosolic GPDH expression is also induced by glucose in rat and human islets.
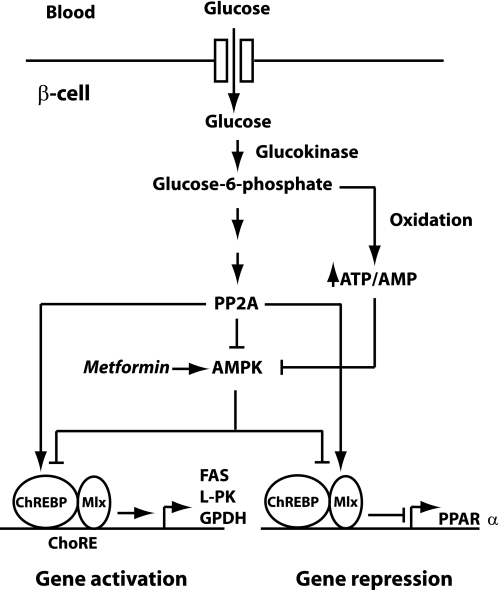
Our previous finding that PP2A and AMPK are involved in regulation of PPARα expression by glucose in pancreatic β-cells (42) supports our current model of ChREBP-mediated glucose repression of PPARα expression because both PP2A and AMPK have been implicated in the regulation of ChREBP activity in hepatocytes. Thus, some studies suggest that activation of ChREBP by glucose is mediated by xylulose-5-phosphate-induced PP2A activity, which leads to ChREBP dephosphorylation (35–37). However, the fact that the contribution of the pentose phosphate pathway to total glucose oxidation is very low in islets and purified islet β-cells (57, 58) suggests that it is not xylulose-5-phosphate but rather another glucose metabolite that activates PP2A in β-cells. Furthermore, AMPK has been reported to impair the transcriptional activity of ChREBP in primary mouse hepatocytes and to inhibit the DNA binding activity of mouse ChREBP by phosphorylating ChREBP on Ser566 in vitro (39). Moreover, the same site was found to be phosphorylated in primary rat hepatocytes in an independent study (59). Based on these findings, we propose a model for glucose-regulated gene expression in pancreatic β-cells as shown in Fig. 8. Our data indicate that AMPK, as well as ChREBP, is involved both in the induction and the repression of gene expression by glucose. Whether AMPK and PP2A regulate PPARα expression by directly or indirectly modulating the activity of ChREBP in β-cells has yet to be determined.
FIGURE 8.
Working model for molecular mechanisms involved in ChREBP-mediated regulation of gene expression by glucose in pancreatic β-cells. Glucose metabolism activates PP2A and inactivates AMPK, which in turn leads to activation of ChREBP by a so far undefined mechanism. ChREBP subsequently mediates glucose induction of FAS, L-PK, and GPDH gene expression by binding to carbohydrate response elements together with Mlx. In addition, glucose also induces binding of ChREBP to the distal PPARα promoter, which leads to repression of PPARα expression.
The molecular mechanism underlying ChREBP repression of PPARα expression also remains to be further investigated. We found that ChREBP binds to a site ∼30 kb upstream of the PPARα promoter, yet careful investigation of the sequence under the peak did not reveal any consensus carbohydrate response element, suggesting that ChREBP may bind to a single E-box motif present near this site as a heterodimer with Mlx. This would be in line with a previous study showing that ChREBP-Mlx heterodimers can bind to E-box motifs and repress transcription in vitro (26).
In conclusion, we have characterized the 5′-UTR of PPARα in insulinoma cells and identified five alternative promoters that give rise to three alternatively spliced first exons. Expression of all transcripts is significantly suppressed by glucose. Using gain-of-function as well as loss-of-function experiments, we have identified ChREBP as a critical and direct novel mediator of glucose repression of PPARα in rat pancreatic β-cells, and this repression may be mediated through a distal ChREBP-binding site located ∼30 kb upstream of the PPARα promoter region. Furthermore, we show that the ability of PPARα to induce fatty acid oxidation when the glucose concentration is low is compromised following exposure to chronic high glucose levels and that this is partly mitigated by knockdown of ChREBP. This suggests that ChREBP may be implicated in the molecular mechanisms of glucolipotoxicity by mediating glucose repression of PPARα expression and thereby compromising the ability of endogenous PPARα ligands to induce fatty acid oxidation. Thus, ChREBP, which has primarily been implicated in glucose-induced activation of gene expression, may play an important and hitherto unrecognized role in glucose repression of gene expression in pancreatic β-cells.
Acknowledgments
We thank Dr. Giuseppe Merla for providing the pcDNA3-MycEGFP-mChREBPζ construct. Fresh human islets were provided by the Cell Isolation and Transplantation Center of the Geneva School of Medicine, thanks to the European Consortium for Islet Transplantation “Islet for Research” distribution program sponsored by the Juvenile Diabetes Research Foundation (to Thierry Berney).
This work was supported by grants from the Danish Health Science Research Council, the Danish Diabetes Foundation, and NordForsk given to the Nordic Center of Excellence MitoHealth (to S. M.), as well as grants from the Swiss National Science Foundation (to P. M.).
- PPAR
- peroxisome proliferator-activated receptor
- AMPK
- AMP-activated protein kinase
- GPDH
- glycerol-3-phosphate dehydrogenase
- L-PK
- liver type pyruvate kinase
- PP2A
- protein phosphatase 2A
- 5′-RLM-RACE
- 5′-RNA ligase-mediated rapid amplification of cDNA ends
- qPCR
- quantitative PCR
- FAS
- fatty acid synthase
- ANOVA
- analysis of variance
- ChREBP
- carbohydrate response element-binding protein
- Mlx
- Max-like protein X.
REFERENCES
- 1. Shalev A., Pise-Masison C. A., Radonovich M., Hoffmann S. C., Hirshberg B., Brady J. N., Harlan D. M. (2002) Endocrinology 143, 3695–3698 [DOI] [PubMed] [Google Scholar]
- 2. Schuit F., Flamez D., De Vos A., Pipeleers D. (2002) Diabetes 51, (Suppl. 3) S326–S332 [DOI] [PubMed] [Google Scholar]
- 3. Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. (1998) J. Biol. Chem. 273, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 4. Djouadi F., Weinheimer C. J., Saffitz J. E., Pitchford C., Bastin J., Gonzalez F. J., Kelly D. P. (1998) J. Clin. Invest. 102, 1083–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Desvergne B., Wahli W. (1999) Endocr. Rev. 20, 649–688 [DOI] [PubMed] [Google Scholar]
- 6. Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. (1999) J. Clin. Invest. 103, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leone T. C., Weinheimer C. J., Kelly D. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 7473–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou Y. T., Shimabukuro M., Wang M. Y., Lee Y., Higa M., Milburn J. L., Newgard C. B., Unger R. H. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 8898–8903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellemans K., Kerckhofs K., Hannaert J. C., Martens G., Van Veldhoven P., Pipeleers D. (2007) FEBS J. 274, 6094–6105 [DOI] [PubMed] [Google Scholar]
- 10. Gremlich S., Nolan C., Roduit R., Burcelin R., Peyot M. L., Delghingaro-Augusto V., Desvergne B., Michalik L., Prentki M., Wahli W. (2005) Endocrinology 146, 375–382 [DOI] [PubMed] [Google Scholar]
- 11. Holness M. J., Smith N. D., Greenwood G. K., Sugden M. C. (2007) Am. J. Physiol. Endocrinol. Metab. 292, E1087–E1094 [DOI] [PubMed] [Google Scholar]
- 12. Lalloyer F., Vandewalle B., Percevault F., Torpier G., Kerr-Conte J., Oosterveer M., Paumelle R., Fruchart J. C., Kuipers F., Pattou F., Fiévet C., Staels B. (2006) Diabetes 55, 1605–1613 [DOI] [PubMed] [Google Scholar]
- 13. Frigerio F., Brun T., Bartley C., Usardi A., Bosco D., Ravnskjaer K., Mandrup S., Maechler P. (2010) Diabetologia 53, 331–340 [DOI] [PubMed] [Google Scholar]
- 14. Sun Y., Zhang L., Gu H. F., Han W., Ren M., Wang F., Gong B., Wang L., Guo H., Xin W., Zhao J., Gao L. (2008) Endocrinology 149, 662–671 [DOI] [PubMed] [Google Scholar]
- 15. Robertson R. P., Harmon J., Tran P. O., Tanaka Y., Takahashi H. (2003) Diabetes 52, 581–587 [DOI] [PubMed] [Google Scholar]
- 16. Roduit R., Morin J., Massé F., Segall L., Roche E., Newgard C. B., Assimacopoulos-Jeannet F., Prentki M. (2000) J. Biol. Chem. 275, 35799–35806 [DOI] [PubMed] [Google Scholar]
- 17. Denechaud P. D., Bossard P., Lobaccaro J. M., Millatt L., Staels B., Girard J., Postic C. (2008) J. Clin. Invest. 118, 956–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iizuka K., Bruick R. K., Liang G., Horton J. D., Uyeda K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7281–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ishii S., Iizuka K., Miller B. C., Uyeda K. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15597–15602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ma L., Robinson L. N., Towle H. C. (2006) J. Biol. Chem. 281, 28721–28730 [DOI] [PubMed] [Google Scholar]
- 21. Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burke S. J., Collier J. J., Scott D. K. (2009) FASEB J. 23, 2855–2865 [DOI] [PubMed] [Google Scholar]
- 23. Cha-Molstad H., Saxena G., Chen J., Shalev A. (2009) J. Biol. Chem. 284, 16898–16905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. da Silva Xavier G., Rutter G. A., Diraison F., Andreolas C., Leclerc I. (2006) J. Lipid Res. 47, 2482–2491 [DOI] [PubMed] [Google Scholar]
- 25. Wang H., Wollheim C. B. (2002) J. Biol. Chem. 277, 32746–32752 [DOI] [PubMed] [Google Scholar]
- 26. Cairo S., Merla G., Urbinati F., Ballabio A., Reymond A. (2001) Hum. Mol. Genet. 10, 617–627 [DOI] [PubMed] [Google Scholar]
- 27. Ma L., Tsatsos N. G., Towle H. C. (2005) J. Biol. Chem. 280, 12019–12027 [DOI] [PubMed] [Google Scholar]
- 28. Stoeckman A. K., Ma L., Towle H. C. (2004) J. Biol. Chem. 279, 15662–15669 [DOI] [PubMed] [Google Scholar]
- 29. Li M. V., Chen W., Poungvarin N., Imamura M., Chan L. (2008) Mol. Endocrinol. 22, 1658–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Merla G., Howald C., Antonarakis S. E., Reymond A. (2004) Hum. Mol. Genet. 13, 1505–1514 [DOI] [PubMed] [Google Scholar]
- 31. Sakiyama H., Wynn R. M., Lee W. R., Fukasawa M., Mizuguchi H., Gardner K. H., Repa J. J., Uyeda K. (2008) J. Biol. Chem. 283, 24899–24908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li M. V., Chen W., Harmancey R. N., Nuotio-Antar A. M., Imamura M., Saha P., Taegtmeyer H., Chan L. (2010) Biochem. Biophys. Res. Commun. 395, 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li M. V., Chang B., Imamura M., Poungvarin N., Chan L. (2006) Diabetes 55, 1179–1189 [DOI] [PubMed] [Google Scholar]
- 34. Davies M. N., O'Callaghan B. L., Towle H. C. (2008) J. Biol. Chem. 283, 24029–24038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kabashima T., Kawaguchi T., Wadzinski B. E., Uyeda K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishimura M., Fedorov S., Uyeda K. (1994) J. Biol. Chem. 269, 26100–26106 [PubMed] [Google Scholar]
- 38. Uyeda K., Yamashita H., Kawaguchi T. (2002) Biochem. Pharmacol. 63, 2075–2080 [DOI] [PubMed] [Google Scholar]
- 39. Kawaguchi T., Osatomi K., Yamashita H., Kabashima T., Uyeda K. (2002) J. Biol. Chem. 277, 3829–3835 [DOI] [PubMed] [Google Scholar]
- 40. Burke S. J., Collier J. J., Scott D. K. (2009) J. Mol. Biol. 392, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang P., Metukuri M. R., Bindom S. M., Prochownik E. V., O'Doherty R. M., Scott D. K. (2010) Mol. Endocrinol. 24, 1274–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ravnskjaer K., Boergesen M., Dalgaard L. T., Mandrup S. (2006) J. Mol. Endocrinol. 36, 289–299 [DOI] [PubMed] [Google Scholar]
- 43. Joly E., Roduit R., Peyot M. L., Habinowski S. A., Ruderman N. B., Witters L. A., Prentki M. (2009) J. Diabetes 1, 263–272 [DOI] [PubMed] [Google Scholar]
- 44. Merglen A., Theander S., Rubi B., Chaffard G., Wollheim C. B., Maechler P. (2004) Endocrinology 145, 667–678 [DOI] [PubMed] [Google Scholar]
- 45. Chew C. H., Samian M. R., Najimudin N., Tengku-Muhammad T. S. (2003) Biochem. Biophys. Res. Commun. 305, 235–243 [DOI] [PubMed] [Google Scholar]
- 46. Ravnskjaer K., Frigerio F., Boergesen M., Nielsen T., Maechler P., Mandrup S. (2009) J. Lipid Res. 51, 1370–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Berge K., Tronstad K. J., Bohov P., Madsen L., Berge R. K. (2003) J. Lipid Res. 44, 118–127 [DOI] [PubMed] [Google Scholar]
- 48. Pineda Torra I., Jamshidi Y., Flavell D. M., Fruchart J. C., Staels B. (2002) Mol. Endocrinol. 16, 1013–1028 [DOI] [PubMed] [Google Scholar]
- 49. Göttlicher M., Widmark E., Li Q., Gustafsson J. A. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4653–4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Edwards D. R., Mahadevan L. C. (1992) EMBO J. 11, 2415–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Iizuka K., Horikawa Y. (2008) Biochem. Biophys. Res. Commun. 374, 95–100 [DOI] [PubMed] [Google Scholar]
- 52. Kelpe C. L., Moore P. C., Parazzoli S. D., Wicksteed B., Rhodes C. J., Poitout V. (2003) J. Biol. Chem. 278, 30015–30021 [DOI] [PubMed] [Google Scholar]
- 53. Olofsson C. S., Collins S., Bengtsson M., Eliasson L., Salehi A., Shimomura K., Tarasov A., Holm C., Ashcroft F., Rorsman P. (2007) Diabetes 56, 1888–1897 [DOI] [PubMed] [Google Scholar]
- 54. Gremlich S., Bonny C., Waeber G., Thorens B. (1997) J. Biol. Chem. 272, 30261–30269 [DOI] [PubMed] [Google Scholar]
- 55. Fontés G., Zarrouki B., Hagman D. K., Latour M. G., Semache M., Roskens V., Moore P. C., Prentki M., Rhodes C. J., Jetton T. L., Poitou V. (2010) Diabetologia 53, 2369–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Noordeen N. A., Khera T. K., Sun G., Longbottom E. R., Pullen T. J., da Silva Xavier G., Rutter G. A., Leclerc I. (2010) Diabetes 59, 153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hedeskov C. J., Capito K. (1975) Biochem. J. 152, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schuit F., De Vos A., Farfari S., Moens K., Pipeleers D., Brun T., Prentki M. (1997) J. Biol. Chem. 272, 18572–18579 [DOI] [PubMed] [Google Scholar]
- 59. Tsatsos N. G., Davies M. N., O'Callaghan B. L., Towle H. C. (2008) Biochem. J. 411, 261–270 [DOI] [PubMed] [Google Scholar]