Abstract
The biochemical basis of modified kernel texture in Quality Protein Maize (QPM) is poorly understood. Proteomic analysis of several QPM lines indicated increased levels of granule-bound starch synthase I in the soluble nonzein protein fraction of these genotypes. Increased extraction of this enzyme reflected a change in starch structure, which was manifested as shorter amylopectin branches and increased starch-granule swelling. In mature kernels, these alterations in starch structure were associated with interconnections between starch granules that resulted in a vitreous kernel phenotype. Understanding the molecular basis for this previously uncharacterized starch structure will accelerate the development of QPM.
Keywords: opaque2, high-lysine corn, modified opaque2, starch fine structure, endosperm texture
Maize is an important food and animal feed in many parts of the world, but the deficiency in essential amino acids, particularly lysine and tryptophan, limit its nutritional value. The discovery by Mertz et al. (1) that the opaque2 (o2) mutation creates a kernel with higher lysine content held great promise for improving maize protein quality, but excitement over its potential was tempered by the fact that the soft, starchy endosperm of o2 grain makes it prone to damage (2). To overcome these deficiencies, plant breeders selected o2 lines with hard, translucent (vitreous) kernels that retained a high lysine content (3). The resulting Quality Protein Maize (QPM) varieties were shown to be comparable with wild type but with an improved amino acid composition (4). In 2000, the World Food Prize was awarded to Villegas and Vasal for their efforts to develop QPM for worldwide use (5).
The mechanism by which the starchy phenotype of o2 endosperm is modified in QPM is not understood. Genetic mapping of o2 modifiers revealed linkage with the locus encoding the 27-kDa γ-zein storage protein and a region near the end of the long arm of chromosome 7 (6). A possible role for the 27-kDa γ-zein in the formation of vitreous endosperm was supported by the observation that this protein was increased 2- to 3-fold in QPM compared with soft o2 (7). Consequently, it was postulated that an increase in the number of zein protein bodies and their compaction between starch grains is at least partially responsible for endosperm modification in QPM (8).
Given the complexity of o2 modifier inheritance (4), it is believed that several unlinked loci control the creation of a vitreous kernel phenotype in QPM. To discover additional factors that influence endosperm modification in QPM, we performed a proteomic analysis of the nonzein proteins from several QPM lines. This study showed that one of the most prominent changes in modified o2 (mo2) endosperm was the amount of a starch-synthesis enzyme, and subsequent analysis of starch structure showed that amylopectin branching is altered in QPM. The consequence of this difference is that QPM starch swells more than normal, and the starch granules in mature endosperm associate with one another. These results suggest that suppression of the opaque endosperm phenotype in QPM could be related to the properties of starch grains.
Materials and Methods
Protein Extraction and Identification. Endosperm from degermed maize kernels was ground in a ball mill. Proteins were extracted from 50 mg of flour in 1 ml of borate extraction buffer (12.5 mM NaBO3/2% 2-mercaptoethanol/1% SDS) at 37°C with agitation for 2 h (7). The samples were centrifuged at room temperature in a microcentrifuge at 12,000 × g for 10 min. The supernatant was transferred to a fresh tube, ethanol was added to a final concentration of 70%, and the sample was incubated for 1 h at –20°C. The precipitated nonzein proteins were collected by centrifugation at 12,000 × g, the pellet was washed two times with 70% ethanol, and the ethanol-soluble zein fraction was dried under vacuum in a centrifugal concentrator.
All electrophoretic separations were carried out according to manufacturer instructions. Proteins were resuspended in the appropriate sample buffer for either 1D or 2D SDS/PAGE. The first dimension of 2D electrophoresis was performed by using immobilized pH gradient strips on the Multiphor II flatbed electrophoresis system (Amersham Biosciences). The second dimension was performed on either the Multiphor II system or the vertical Mini Protean II system (Bio-Rad).
Proteins were identified by peptide mass mapping with matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Protein spots were cut from SDS/PAGE gels and digested in-gel with trypsin (9). The peptides were purified on C-18 reversed-phase media fixed in pipette tips (Millipore). The purified peptides were spotted on MALDI target plates with α-cyano-4-hydroxycinnamic acid. Mass spectra were acquired on a Reflex III MALDI-TOF mass spectrometer (Bruker Daltonics, Billerica, MA). Peptide mass maps were searched against protein databases with the ProFound search engine (http://prowl.rockefeller.edu).
Starch Isolation. Starch granules were purified essentially as described by Gutierrez et al. (10). Mature kernels were soaked for 24 h in 0.5% Na2S2O5 at 50°C. The endosperm was dissected from the pericarp and germ and ground lightly in a mortar. The sample was blended with 50 mM NaCl for 30 s and filtered through two layers of Miracloth (Calbiochem). The filtered material was extracted and pelleted by centrifugation five times in 1:4 toluene/50 mM NaCl, followed by extraction two times with acetone. The starch was dried for 48 h before use.
Starch Sedimentation. A 20% slurry of the starchy pellet recovered after protein isolation was prepared in double deionized H2O with a small amount of bromophenol blue. The slurry was mixed by vortexing, and a sample was drawn into a hematocrit capillary tube (VWR Scientific). The samples settled overnight and were scanned on a flatbed scanner. The length of the column of settled starch and the total length occupied by the sample were measured by using NIH image software.
Amylose Quantification. A 10 mg/ml sample of starch was gelled in 1 M NaOH by heating to 65°C for 1 h, and it was neutralized in 9 vol of 0.11 M acetic acid. An aliquot of the sample was diluted to 0.1 mg/ml, and 100 μl was mixed with 100 μl of I2/KI solution (2 mg/ml I2/20 mg/ml KI) in a microtiter plate. The OD was measured on a microplate reader (Dynatech) using a 670-nm filter.
Wide-Angle X-Ray Scattering. Purified starch was placed in a glass sample holder and mounted in the beamline of a Scintag XDS 2000 diffractometer. The x-ray source was filtered for Kα radiation through a copper screen; the source was operated at 40 mA and 40 kV. The sample was scanned over the range of 5–35° 2θ with steps of 0.02°.
Amylopectin Branch Analysis. A 2 mg/ml starch sample was gelled in 50 mM sodium acetate, pH 4.5, by boiling for 1 h. Isoamylase (Sigma) was added to a final concentration of 1,000 units per mg of starch and incubated for 24 h at 37°C (11). The enzyme was inactivated by boiling for 5 min. Analysis of the branch length distribution for each sample was performed by MALDI-TOF (12). The sample was mixed with 4 vol of 0.1 M 2,5,6-trihydroacetophenone in 50% acetonitrile, spotted on a MALDI target plate, and dried in air. Mass spectra were acquired on a Reflex III MALDI-TOF mass spectrometer (Bruker Daltonics). The baseline was corrected, and the area under the peaks was measured with the polymer analysis tools in xmass (Bruker Daltonics).
Results
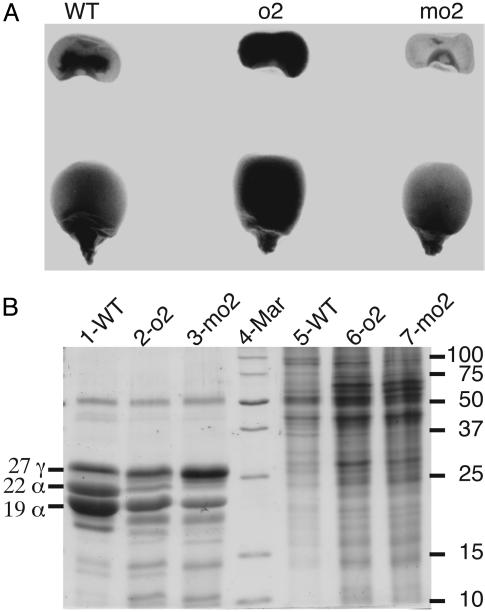
CM105mo2 was created by introgressing the o2 modifier genes from CIMMYT Pool 33 QPM into CM105o2, which has an opaque kernel phenotype at maturity. Progeny from each backcross generation were self-pollinated, and true-breeding vitreous kernels were used for the subsequent backcross to CM105o2. After four backcrosses, CM105+, o2, and mo2 plants were similar in stature, leaf, and floral architecture; their kernels were similar in size and shape but varied in texture and protein composition. CM105+ and CM105mo2 endosperms transmitted light more effectively than CM105o2. SDS/PAGE analysis of zein storage proteins revealed patterns characteristic of each genotype: there was a reduction of 19- and 22-kDa α-zeins in CM105o2 and CM105mo2 compared with wild type and a marked increase in the 27-kDa γ-zein in CM105mo2 (Fig. 1B). Based on equal amounts of endosperm flour, CM105o2 and CM105mo2 contained nearly twice as much nonzein protein as wild type (Fig. 1B, lane 5 compared with lanes 6 and 7), which is largely responsible for their higher lysine content (1).
Fig. 1.
Phenotypic comparison of CM105+, o2, and mo2 kernels. (A) Transmission of light by mature kernels. (B) SDS/PAGE analysis of zein and nonzein proteins, respectively, from equal amounts of endosperm flour of CM105+ (lanes 1 and 5), CM105o2 (lanes 2 and 6), and CM105mo2 (lanes 3 and 7). The identity of specific types of zeins is shown on the left, and molecular masses of the size markers in lane 4 are shown on the right.
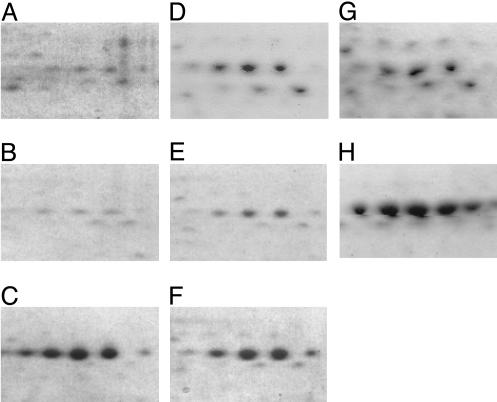
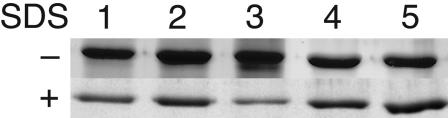
Analysis of the nonzein proteins from these inbreds by 2D isoelectric-focusing SDS/PAGE revealed a prominent difference in a cluster of 56-kDa polypeptides with pI values of 5.45–5.80 (Fig. 2). Relative to CM105+ (Fig. 2 A) and CM105o2 (Fig. 2B), these proteins were increased ≈10-fold in CM105mo2 (Fig. 2C). The polypeptides were identified by peptide mass mapping as granule-bound starch synthase I (GBSS I), the product of the Waxy1 gene (13). To determine whether increased GBSS I in the nonzein fraction is a common feature of the mo2 phenotype, a segregating F2 ear from a cross between a South African QPM, K0326Y (14), and W64Ao2 was used to make pools of opaque and vitreous kernels. Compared with W64Ao2 or the opaque F2 kernels (Fig. 2 D and E, respectively), vitreous F2 kernels had more soluble GBSS I in the nonzein fraction (Fig. 2F). Furthermore, analysis of kernels from recombinant inbred (RI) lines generated by crossing W22o2, a soft endosperm o2, with Pool 33 QPM (15) showed the same result. The opaque RI-5 (Fig. 2G) yielded less soluble GBSS I than the vitreous RI-32 (Fig. 2H). The total amount of granule-associated GBSS I was similar for CM105+ (Fig. 3 Upper, lane 1), CM105o2 (Fig. 3, lane 2), and CM105mo2 (Fig. 3, lane 3) when SDS was not present in the extraction buffer. However, when endosperm was extracted in the presence of SDS, there was a marked decrease in granule-associated GBSS I in CM105mo2 (lower lane 3) compared with CM105+ or CM105o2 (lanes 1 and 2, respectively), and the protein appeared in the soluble fraction (Fig. 2C). Immunoblot analysis of nonzein proteins with GBSS I antiserum (13) produced similar results (data not shown). Endosperm from vitreous and opaque kernels of a K0326Y × W64Ao2 cross (Fig. 3, lanes 4 and 5, respectively) behaved similarly, although the difference in the extraction of GBSS I in the presence of SDS was not as extreme. This result is consistent with the observation that less GBSS I was found in the nonzein fraction from the vitreous kernels (Fig. 2F). The difference between mo2 and wild-type or o2 lines was in the level of extractable versus GBSS I, which had an inverse relationship: Lines with high GBSS I in the nonzein fraction showed reduced granule-associated GBSS I, and the opposite was true of lines with low extractable GBSS I. Thus, GBSSI was more extractable from mo2 starch, which possibly indicated altered starch-granule structure.
Fig. 2.
Two-dimensional gel patterns of GBSS I proteins from wild-type, o2, and mo2 genotypes. Nonzein proteins were separated by isoelectric focusing with immobilized pH 4–7 gradient strips and 12.5% SDS/PAGE. Compared with CM105+ (A) and CM105o2 (B), nonzeins from CM105mo2 contained larger amounts of a cluster of 56-kDa polypeptides identified as GBSS I. Vitreous mo2 kernels (F) from a segregating F2 ear of a cross between K0326Y QPM and W64Ao2 contained more GBSS I than opaque kernels from the same ear (E) or the W64Ao2 parent (D). The opaque RI-5 (G) had smaller amounts of GBSS I than the vitreous RI-32 (H).
Fig. 3.
Differential extraction of GBSS I in the presence of SDS from wild-type, o2, and mo2 genotypes. Endosperm flour was extracted without (Upper) or with (Lower) SDS in the extraction buffer, and the resulting starch was solubilized by boiling in SDS/PAGE sample buffer. The total amount of granule-associated GBSS I was similar from all three CM105 lines (lane 1, CM105+; lane 2, CM105o2; lane 3, CM105mo2) and from vitreous or opaque kernels from a K0326Y × W64Ao2 cross (lane 4, vitreous kernels; lane 5, opaque kernels). In the presence of SDS, more GBSS I was extracted from starch granules of CM105mo2 (Lower, lane 3, compared with lanes 1 and 2) and vitreous kernels from the K0326Y × W64Ao2 cross (Lower, lane 4, compared with lane 5).
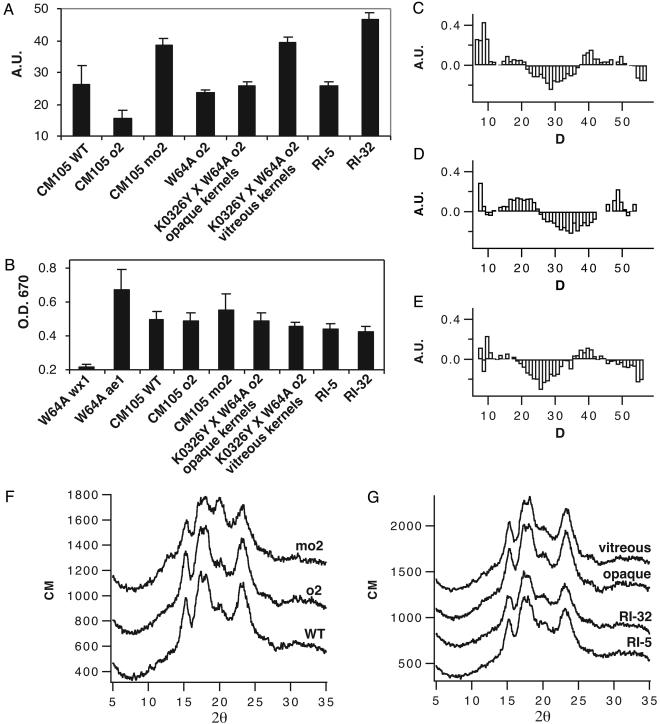
After protein extraction from endosperm of these inbred lines, we noticed that the starch pellet of mo2 genotypes was considerably larger than that of comparable wild-type and o2 genotypes. To quantify the increased swelling, a sedimentation assay was performed (Fig. 4A). Purified starch was hydrated in deionized water and allowed to settle overnight in capillary tubes, and the volume was recorded. All mo2 lines showed significantly greater starch swelling (t test, P < 0.05; n > 6), which was two to three times the volume measured for comparable wild-type and soft o2 lines. Because amylose is the product of GBSS I, we measured the amount of this polymer to determine whether variation in its level might explain the difference in starch-granule swelling (Fig. 4B). Samples of gelled starch were incubated with I2/KI solution, and the OD at 670 nm was measured. For comparison, starch from low-amylose [waxy1 (wx1)] and high-amylose [amylose extender1 (ae1)] mutants were analyzed. CM105mo2 had somewhat higher amylose content than CM105+ or CM105o2, but this was not true of other mo2 lines. None of the differences in amylose content between o2 and mo2 lines were significant (t test, P > 0.05; n > 4), indicating that amylose content was not related to swelling of mo2 starch.
Fig. 4.
Analysis of starch structure in wild-type, o2, and mo2 maize genotypes. (A) Sedimentation assay measuring swelling of starch granules in water at room temperature. (B) Measurement of amylose content by I2 binding. (C–E) Difference plots contrasting the lengths of α-1,4-linked glucose polymers of debranched amylopectin molecules from mo2 and o2 genotypes. (C) Compared with CM105o2, CM105mo2 has more very short α-1,4-linked glucose chains and a lower abundance of chains with a DP of 25–35. (D) Vitreous mo2 kernels from an F2 ear of K0326Y QPM by W64Ao2 have a greater proportion of α-1,4-glucose chains with a DP of 12–25 and a decreased proportion of chains with a DP of 27–42, compared with opaque kernels from the same ear. (E) The vitreous RI-32 had a slightly increased proportion of short α-1,4-linked glucose chains and a prominent reduction in chains with a DP of 20–35 compared with the opaque RI-5. (F) Compared with CM105+ and o2, wide-angle x-ray scattering of CM105mo2 starch granules showed an increase in the peak at 20.0° 2θ and a small new peak at 13.2° 2θ. (G) The other opaque and vitreous lines tested had x-ray scattering patterns similar to CM105+ and CM105o2.
Because amylose did not seem to be involved in the increased swelling of mo2 starch, the structure of the amylopectin in these lines was analyzed. To investigate amylopectin branching, the α-1,6 linkages were hydrolyzed with isoamylase, and the α-1,4 chain lengths were measured by MALDI-TOF (12). Compared with CM105+ and CM105o2, CM105mo2 contained more amylopectin chains with a degree of polymerization (DP) <10 and fewer intermediate-length chains with a DP of 20–35 (Fig. 4C). Likewise, amylopectin of mo2 kernels from a segregating F2 ear of a cross between K0326Y and W64Ao2 had reduced intermediate chains with a DP of 25–40 and had an additional increase in chains with a DP of <25 (Fig. 4D). The vitreous RI-32 had a prominent reduction in the level of chains with a DP of 20–35 compared with the opaque RI-5 (Fig. 4E). These results demonstrated that a common feature of the amylopectin in all mo2 lines was fewer intermediate-length α-1,4-linked glucose chains.
We investigated whether the variation in amylopectin branch length affected the crystallinity of the amylopectin with wide-angle x-ray scattering. The x-ray-scattering plots of CM105+ and CM105o2 starch were typical for A-type maize starch (16). CM105mo2 starch showed a prominent increase in the peak at 20.0° 2θ and a small new peak at 13.2° 2θ (Fig. 4F), indicating partial conversion to the V-type crystalline structure (17). In contrast, RI-32 and vitreous F2 kernels from a K0326Y × W64Ao2 cross had x-ray-scattering patterns indistinguishable from the A-type pattern of their opaque counterparts or CM105+ (Fig. 4G).
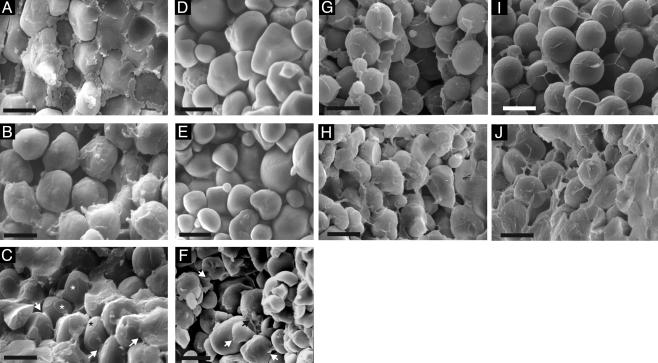
Scanning electron microscopy of mature maize kernels and the starch purified from them revealed marked differences in the gross structure and organization of mo2 starch granules (Fig. 5). Starch granules in CM105+ endosperm had a smooth surface and were embedded in a matrix of dried cytosol and aggregated protein bodies (Fig. 5A). Starch granules in CM105o2 endosperm were also smooth; however, there was relatively little proteinaceous material filling the spaces between them (Fig. 5B). In contrast, the surfaces of starch granules in CM105mo2 had depressions at points of contact between starch grains (Fig. 5C, asterisks). Additionally, there were interconnections between many adjacent starch granules (Fig. 5C, arrows). We noted similar starch interconnections in previously published micrographs of R802mo2 (18), but their significance was not described by the authors. The nature of the material between starch grains in CM105mo2 was not clear, but it had a morphology different from that observed in wild-type kernels (Fig. 5A). It completely filled the spaces between starch granules in vitreous regions of mo2 endosperm (e.g., the area under the scale bar in Fig. 5C). Scanning electron microscopy analysis was performed on purified starch granules to determine whether the contacts were starch. Starch granules purified from CM105+ and CM105o2 were smooth and did not adhere to one another (Fig. 5 D and E, respectively). In contrast, the surface of many CM105mo2 starch granules was irregular, and there were points at which adjacent granules formed contacts (Fig. 5F). After drying, the CM105mo2 starch granules adhered to one another, forming a hard pellet. These observations lead us to believe that the material connecting starch granules in CM105mo2 endosperm is carbohydrate.
Fig. 5.
Scanning electron microscopy analysis of starch granules in mature endosperms and purified starch. (A) The CM105+ kernel contained spherical starch grains embedded in a proteinaceous matrix containing protein bodies. (B) The CM105o2 kernel had smooth, spherical starch grains with very little matrix surrounding them. (C) The CM105mo2 kernel contained starch grains with circular surface depressions at which adjacent starch granules made contact (*). The arrows mark connections between adjacent starch granules. Purified starch from CM105+ (D) and CM105o2 (E) consisted of round, smooth granules, whereas that from CM105mo2 (F) consisted of irregularly shaped granules, and there were many points at which adjacent granules form contacts (arrows). Opaque (G) and vitreous (H) kernels from a segregating F2 ear of K0326Y QPM by W64Ao2 were similar to CM105o2 and CM105mo2, respectively. Likewise, opaque RI-5 (I) and vitreous RI-32 (J) kernels were similar to CM105o2 and CM105mo2, respectively.
Starch properties similar to these were observed in kernels of other o2 and mo2 genotypes. Starch granules in endosperm of an opaque kernel from a segregating F2 ear of a K0326Y × W64Ao2 cross showed somewhat more irregular surfaces than those in CM105o2, but the spaces between them were not filled and there were no extensive contacts between starch granules (Fig. 5G). In contrast, a vitreous kernel from the same ear had extensive starch granule contacts, and the spaces between them were filled with “starchy” material (Fig. 5H). Starch granules in the opaque RI-5 kernels were similar to CM105o2, although small depressions were observed where granules made contact (Fig. 5I). The arrangement of starch granules in endosperm of the vitreous RI-32 was similar to CM105mo2, with extensive contacts between adjacent granules (Fig. 5J), and the spaces between them were completely filled in the vitreous areas (Fig. 5J, right side of image).
Discussion
These studies demonstrated that altered starch structure, which leads to increased extractability of GBSS I, is a common feature of mo2 genotypes. Compared with wild-type and soft o2 genotypes, amylopectin in mo2 lines had reduced levels of intermediate-length α-1,4-linked glucose chains. This change in starch structure was associated with increased swelling in water and the formation of tight contacts between starch granules in mature endosperm; this was also observed in purified starch. The reduced level of intermediate-length chains in amylopectin of mo2 genotypes appeared to predominantly affect the structure of the amorphous regions of the starch granule. This is consistent with the observation that mo2 starch granules swelled more than those of wild type or o2, because water uptake into amorphous regions is responsible for granule swelling (19). Furthermore, CM105mo2 starch was significantly more sensitive to acid hydrolysis than that from CM105+ or CM105o2, indicating more accessible amorphous starch (data not shown).
It is interesting that the crystalline structure of the starch in mo2 genotypes was not identical. CM105mo2 had partial V-type crystallinity, whereas RI-32 and vitreous F2 kernels from a K0326Y × W64Ao2 cross had A-type crystallinity, which is typical of cereal starches. This suggests that the alterations to starch structure in mo2 genetic backgrounds may arise from changes in different enzymatic activities of the starch-synthesis pathway. Considerable progress has been made in recent years to understand the influence of individual isoforms of starch-synthesis enzymes on amylopectin structure (20, 21). Reduced expression of starch-synthesis genes in mutants and rice antisense RNA expression lines have been very useful to elucidate the role of individual enzymes in production of amylopectin. Perturbation of several genes in rice resulted in amylopectin branch differences similar to those described for maize mo2 genotypes. Consequently, these are candidate genes that could be responsible for the altered biochemical activities that lead to the unusual starch structure in mo2; they include starch synthase IIa (22, 23), branching enzyme I (21), and isoamylase debranching enzyme (24, 25).
Normally, maize starch granules do not form cohesive contacts with one another. Consequently, we hypothesize that physical connections between starch granules in mo2 could be an important component of the mechanism that restores the hard, vitreous phenotype to o2 mutants. Such a mechanism would be similar to that proposed for other cereals. In wheat, for example, puroindoline proteins influence grain hardness (26). It is thought that these proteins become bound to starch granules, preventing their association in the mature seed, which results in a softer endosperm texture. When little puroindoline is present, the starch granules coalesce during seed desiccation, leading to a harder endosperm.
The results from this study suggest that amorphous, noncrystalline amylopectin molecules at the surface of starch granules in mo2 endosperm can interact and form contacts that link starch granules together. These contacts could provide a mechanism that complements the one postulated for γ-zein-rich protein bodies, which are proposed to fill the spaces between starch granules and crosslink proteins, creating a vitreous kernel phenotype (8, 27). Understanding the mechanisms by which modifier genes create a vitreous kernel phenotype will accelerate the development QPM, which is of growing economic and nutritional importance in many parts of the developing world.
Acknowledgments
This work was supported by Department of Energy Grant 96ER20242 and National Science Foundation Grant DBI-0077676 (to B.A.L.).
Abbreviations: QPM, Quality Protein Maize; o2, opaque2; mo2, modified o2; MALDI, matrix-assisted laser desorption/ionization; TOF, time of flight; GBSS I, granule-bound starch synthase I; RI, recombinant inbred; DP, degree of polymerization.
References
- 1.Mertz, E. T., Bates, L. S. & Nelson, O. E. (1964) Science 145, 279–280. [DOI] [PubMed] [Google Scholar]
- 2.Glover, D. V. & Mertz, E. T. (1987) in Nutritional Quality of Cereal Grains: Genetic and Agronomic Improvement, eds. Olson, R. A. & Frey, K. J. (American Society for Agronomy, Crop Science Society of America, Soil Science Society of America, Madison, WI), pp. 183–336.
- 3.Villegas, E., Vasal, S. K. & Bjarnason, M. (1992) in Quality Protein Maize, ed. Mertz, E. T. (Purdue University, West Lafayette, IN), pp. 49–78.
- 4.Prasanna, B. M., Vasal, S. K., Kassahun, B. & Singh, N. N. (2001) Curr. Sci. 81, 1308–1319. [Google Scholar]
- 5.Vietmeyer, N. D. (2000) Diversity 16, 29–32. [Google Scholar]
- 6.Lopes, M. A., Takasaki, K., Bostwick, D. E., Helentjaris, T. & Larkins, B. A. (1995) Mol. Gen. Genet. 247, 603–613. [DOI] [PubMed] [Google Scholar]
- 7.Wallace, J. C., Lopes, M. A., Paiva, E. & Larkins, B. A. (1990) Plant Physiol. 92, 191–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dannenhoffer, J. M., Bostwick, D. E., Or, E. & Larkins, B. A. (1995) Proc. Natl. Acad. Sci. USA 92, 1931–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hellman, U., Wernstedt, C., Gonez, J. & Heldin, C. H. (1995) Anal. Biochem. 224, 451–455. [DOI] [PubMed] [Google Scholar]
- 10.Gutierrez, O. A., Campbell, M. R. & Glover, D. V. (2002) Crop Sci. 42, 355–359. [Google Scholar]
- 11.Nishi, A., Nakamura, Y., Tanaka, N. & Satoh, H. (2001) Plant Physiol. 127, 459–472. [PMC free article] [PubMed] [Google Scholar]
- 12.Broberg, S., Koch, K., Andersson, R. & Kenne, L. (2000) Carbohydr. Polym. 43, 285–289. [Google Scholar]
- 13.Shure, M., Wessler, S. & Fedoroff, N. (1983) Cell 35, 225–233. [DOI] [PubMed] [Google Scholar]
- 14.Gevers, H. O. & Lake, J. K. (1992) in Quality Protein Maize, ed. Mertz, E. T. (Purdue University, West Lafayette, IN), pp. 49–78.
- 15.Lopes, M. A. & Larkins, B. A. (1995) Theor. Appl. Genet. 91, 274–281. [DOI] [PubMed] [Google Scholar]
- 16.Jane, J. L., Wong, K. S. & McPherson, A. E. (1997) Carbohydr. Res. 300, 219–227. [Google Scholar]
- 17.Rundle, R. E. & Edwards, F. C. (1943) J. Am. Chem. Soc. 65, 2200–2203. [Google Scholar]
- 18.Paulis, J. W., Bietz, J. A., Felker, F. C. & Nelsen, T. C. (1992) in Quality Protein Maize, ed. Mertz, E. T. (Purdue University, West Lafayette, IN), pp. 122–140.
- 19.Donald, A. M., Kato, K. L., Perry, P. A. & Weigh, T. A. (2001) Starch 53, 504–512. [Google Scholar]
- 20.James, M. G., Denyer, K. & Myers, A. M. (2003) Curr. Opin. Plant Biol. 6, 215–222. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, Y. (2002) Plant Cell Physiol. 43, 718–725. [DOI] [PubMed] [Google Scholar]
- 22.Umemoto, T., Yano, M., Satoh, H., Shomura, A. & Nakamura, Y. (2002) Theor. Appl. Genet. 104, 1–8. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura, Y., Sakurai, A., Inaba, Y., Kimura, K., Iwasawa, N. & Nagamine, T. (2002) Starch 54, 117–131. [Google Scholar]
- 24.Fujita, N., Kubo, A., Suh, D. S., Wong, K. S., Jane, J. L., Ozawa, K., Takaiwa, F., Inaba, Y. & Nakamura, Y. (2003) Plant Cell Physiol. 44, 607–618. [DOI] [PubMed] [Google Scholar]
- 25.Wong, K. S., Kubo, A., Jane, J. L., Harada, K., Satoh, H. & Nakamura, Y. (2003) J. Cereal Sci. 37, 139–149. [Google Scholar]
- 26.Morris, C. F. (2002) Plant Mol. Biol. 48, 633–647. [DOI] [PubMed] [Google Scholar]
- 27.Chandrashekar, A. & Mazhar, H. (1999) J. Cereal Sci. 30, 193–207. [Google Scholar]