Abstract
A growing body of evidence indicates that G protein-coupled receptors (GPCRs) are involved in breast tumor progression and that targeting GPCRs may be a novel adjuvant strategy in cancer treatment. However, due to the redundant role of multiple GPCRs in tumor development, it may be necessary to target a common signaling component downstream of these receptors to achieve maximum efficacy. GPCRs transmit signals through heterotrimeric G proteins composed of Gα and Gβγ subunits. Here we evaluated the role of Gβγ in breast tumor growth and metastasis both in vitro and in vivo. Our data show that blocking Gβγ signaling with Gαt or small molecule inhibitors blocked serum-induced breast tumor cell proliferation as well as tumor cell migration induced by various GPCRs in vitro. Moreover, induced expression of Gαt in MDA-MB-231 cells inhibited primary tumor formation and retarded growth of existing breast tumors in nude mice. Blocking Gβγ signaling also dramatically reduced the incidence of spontaneous lung metastasis from primary tumors and decreased tumor formation in the experimental lung metastasis model. Additional studies indicate that Gβγ signaling may also play a role in the generation of a tumor microenvironment permissive for tumor progression, because the inhibition of Gβγ signaling attenuated leukocyte infiltration and angiogenesis in primary breast tumors. Taken together, our data demonstrate a critical role of Gβγ signaling in promoting breast tumor growth and metastasis and suggest that targeting Gβγ may represent a novel therapeutic approach for breast cancer.
Keywords: Breast Cancer, G Protein-coupled Receptors (GPCR), G Proteins, Mouse, Signal Transduction, Tumor Growth, Tumor Metastasis
Introduction
Breast cancer is the most frequently diagnosed cancer in women (1). Despite recent advances in its diagnosis and treatment with adjuvant targeted therapies, breast cancer remains the second most common cause of cancer death among women in the United States (2). As many as 90% of cancer deaths are caused by metastatic spread from primary tumors because the majority of currently marketed anticancer drugs have little effect on tumors at this stage. In light of this situation, efforts to better understand the mechanisms underlying tumor metastasis and to identify novel treatment strategies are warranted.
Emerging experimental and clinical data indicate that G protein-coupled receptors (GPCRs),2 a major family of cell surface proteins, play a critical role at multiple stages of breast tumor development (3). A variety of GPCRs, such as chemokine receptors CXCR4 and CCR7 (4–6), protease-activated receptors (PARs) (7), and receptors for bioactive lipids, such as lysophosphatidic acid (LPA) (8), are up-regulated in breast tumor cells and tissues. Moreover, breast tumor and stromal cells frequently produce higher levels of GPCR agonists (9). Furthermore, overexpressed GPCRs in tumor cells, such as CXCR4, may mediate the metastatic spread of tumor cells to distant organs that produce a high level of their agonists, such as SDF1α (stromal cell-derived factor-1α) (10). Finally, the increased production of some GPCR ligands by tumor and stromal cells has been associated with chronic leukocyte infiltration into tumors (9). The infiltrated leukocytes in turn produce a plethora of protumorigenic factors, including matrix metalloproteases, angiogenic factors, growth factors, and immunosuppressive cytokines and chemokines (9, 11, 12).
Given the critical involvement of GPCRs in breast tumor progression, targeting GPCRs and their ligands has been proposed as a novel adjuvant strategy in cancer treatment (3, 13–15). However, due to the involvement of diverse GPCRs in breast tumor progression, it is unlikely that targeting a single receptor and its ligand can produce full effects in therapy. Indeed, no drugs targeting a single GPCR have been approved for cancer treatment. Thus, there is a need for an alternative approach, preferably one that targets the common pathway downstream of the receptors.
GPCRs transmit extracellular signals through heterotrimeric G proteins, which comprise three subunits, Gα, Gβ, and Gγ (16). Gβ and Gγ form a dimer and are associated with Gα in the inactive state. There are many gene products encoding each G protein subunit (21 α, 6 β, and 12 γ gene products are known) (16). However, based on the function and sequence homology of the Gα subunits, four main classes of G proteins can be distinguished: Gs, which activates adenylyl cyclase; Gi, which inhibits adenylyl cyclase; Gq, which activates phospholipase C; and G12 and G13, which regulate RhoA via PDZ-RhoGEFs. G proteins are activated by receptor-catalyzed guanine nucleotide exchange, resulting in GTP binding to the α subunit (16). Once GTP-bound Gα dissociates from Gβγ, both free Gα and Gβγ can activate downstream effectors to transmit GPCR signals. Heterotrimeric G proteins therefore represent a point of signal convergence from multiple GPCRs and may be targeted to simultaneously block the various GPCRs involved in tumors. Indeed, small molecular inhibitors that non-selectively target all classes of Gα subunits have been shown to suppress tumor growth and invasion in vitro and enhance the activity of several classic anticancer agents in xenograft mouse models of human tumors (17, 18). Blocking Gβγ signaling by a Gβγ scavenger, the C-terminal tail of G protein-coupled receptor kinase 2 obliterates prostate tumor formation and progression (19). Recently, it has been shown that inhibition of Gβγ signaling also prevents SDF1α-induced and NIH-3T3 conditioned medium-induced breast tumor cell migration and invasion in vitro (20). However, many GPCRs that are known to overexpress in breast tumor cells, such as LPA receptors and PARs, couple to multiple G proteins, and they may mediate their effect through either Gα or Gβγ subunits or both (21–23). It remains unclear if Gβγ signaling is required for the function of these GPCRs in breast tumor migration. Moreover, it is unknown if blockage of Gβγ signaling alone is sufficient to limit tumorigenesis and metastasis of breast tumors in vivo, where tumor cells encounter a plethora of protumorigenic factors, including growth factors and GPCR agonists, within the tumor microenvironment.
Here, we investigated the effect of blocking Gβγ signaling on breast tumor growth and metastasis both in vitro and in vivo. Our data show that Gβγ signaling is critical for the growth of breast tumor cells but is not required for the growth of non-transformed breast epithelia. Blocking Gβγ signaling by a Gβγ scavenger, Gαt, and small molecule inhibitors selectively abrogated breast tumor cell migration induced by diverse GPCR agonists. Moreover, we demonstrated that inhibition of Gβγ signaling suppressed primary tumor formation and retarded the progression of preexisting tumors in xenograft mouse models of MDA-MB-231 cells. Notably, inhibiting Gβγ signaling dramatically reduced the formation of spontaneous lung metastases from primary tumors and suppressed the formation of tumor colonies in the lung in the experimental lung metastasis models. These findings thus establish Gβγ as a key mediator in breast tumor growth and metastasis and indicate that Gβγ may be a novel therapeutic target for breast cancer treatment.
EXPERIMENTAL PROCEDURES
Reagents
Human SDF-1α was from Pepro Tech. LPA, pertussis toxin (PTx), epidermal growth factor (EGF), and fibronectin were from Sigma. Matrigel (growth factor-reduced) was from BD Biosciences. d-Luciferin (potassium salt) was from Gold Bio Technology. Doxycycline was from MP Biomedicals. Fura-2/AM was from Invitrogen. Rabbit anti-Akt, mouse anti-phospho-Akt, rabbit anti-ERK, and mouse anti-phospho-ERK antibodies were from Cell Signaling Technology, Inc. Rat anti-CD31, F4/80, and CD45 antibodies were from eBioscience. Mouse anti-Gαt was kindly provided by Dr. Heidi Hamm (Vanderbilt University, Nashville, TN). M119 and M119B were obtained from NCI, National Institutes of Health. Gallein and fluorescein were from TCI America.
Lentiviral and Retroviral Constructs and Virus Generation
Human Gαt and EGFP cDNAs were amplified by PCR and ligated into the entry vector pEN_Tmcs containing a tetracycline-inducible TRE promoter (ATCC) between the restriction enzyme sites SpeI and NotI. Using the Gateway cloning technology (Invitrogen), these cDNAs were then transferred by LR reaction into a pSLIK lentiviral destination vector (ATCC) (24) containing a hygromycin resistance marker and a reverse tetracycline transactivator. Lentivirus was generated by transfecting HEK293FT cells with the pSLIK vectors together with packaging vectors pMDL, pRSV, and pVSV using the Polyjet DNA in vitro transfection reagent (Signagen) (24). The supernatant of culture medium containing lentivirus was collected on day 2 and day 3 post-transfection. Lentivirus was concentrated by ultracentrifugation (47,000 × g for 2 h) and resuspended in 0.2 ml of DMEM.
The construction of the pQC-Luc-IN plasmid encoding firefly luciferase (FL) has been described previously (25). Retroviral production was initiated by transiently transfecting GP-293 retroviral packing cells (Clontech), using Effectene (Qiagen) with the vectors pQC-Luc-IN and pVSVg (Clontech).
Cell Culture and Establishment of Stable Cell Lines
The human breast carcinoma cell line MDA-MB-231 (ATCC) maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS) was infected with retrovirus encoding FL and selected with G418 (400 μg/ml) to establish a stable cell line. The murine mammary carcinoma cell line 4T1 (ATCC) was transduced with lentivirus prepared from the FUGW-FL lentiviral vector (26) (kindly provided by Dr. David Piwnica-Worms from Washington University, St. Louis, MO) to simultaneously express GFP and FL. 4T1 cells expressing GFP were sorted by flow cytometry and maintained in RPMI 1640 (Invitrogen) supplemented with 10% FBS. The human mammary epithelial cell line MCF10A (ATCC) was cultured in DMEM/F-12 (Hyclone) with 10% FBS, 20 ng/ml EGF, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, and 10 μg/ml insulin. The MDA-MB-231, 4T1, and MCF10A cells were transduced with pSLIK lentiviruses encoding tetracycline-inducible EGFP or Gαt and selected with hygromycin (200–500 μg/ml) to establish stable cell lines.
Cell Proliferation Assay in Two-dimensional and Three-dimensional Cultures
For cell proliferation assays in two-dimensional culture, MDA-MB-231 (5,000 cells/well), 4T1 (2,000 cells/well), and MCF10A (2,000 cells/well) cells stably expressing EGFP or Gαt were seeded in 96-well plates in the growth medium containing 10% FBS for 24 h. Doxycycline (1 μg/ml) was then added to the medium to induce EGFP and Gαt expression. MDA-MB-231 and 4T1 cell growth was monitored by measuring the luciferase activity using a luciferase assay kit (Promega) or by counting the cell number with a hemocytometer daily over 5–6 days. MCF10A cell growth was determined by using a tetrazolium salt WST-1 cell proliferation assay kit (BioVision). To determine the effect of inhibitors on cell proliferation, MDA-MB-231, 4T1, and MCF10A cells expressing EGFP were treated with the indicated concentrations of inhibitors.
To evaluate the effect of Gαt expression or PTx on cell growth in three-dimensional cultures, cells were suspended in the complete growth medium supplemented with 2% growth factor-reduced Matrigel (BD Biosciences) and grown on top of a thin layer of Matrigel in 8-well chamber slides (27). Cells were treated with doxycycline or PTx, and the medium was changed every 3 days. On day 8 of the culture, phase-contrast images were taken, and the size of colonies was analyzed by ImageJ software. To determine the morphologies of cell colonies, cells were fixed with 4% paraformaldehyde and then stained with Alexa 568-conjugated phalloidin. Images were taken by confocal microscopy and processed by Photoshop.
Cell Migration Assay
Transwell migration of MDA-MB-231 and 4T1 cells was determined using 8-μm pore size polycarbonate membrane filters as described previously (28). The filter was coated with fibronectin (10 μg/ml) overnight at 4 °C. MDA-MB-231 cell migration was carried out at 37 °C for 4 h using a 96-well modified Boyden chamber. 4T1 cell migration was performed at 37 °C for 20 h using a 24-well transwell insert (Corning Costar). After fixation, cells that migrated to the bottom side of the filter were stained with crystal blue and quantified under a microscope. The chemotaxis index was calculated as -fold increase of the number of cells moving toward chemoattractants over medium alone.
Wound Healing Assay
The wound healing assay of tumor cells was performed as described previously (29). Wounds were created by scraping a 100% confluent cell monolayer with a 200-μl pipette tip. Cells were maintained in DMEM containing 10% FBS with 10 μg/ml mitomycin to inhibit cell proliferation. Images of wounds were taken immediately and 16 h after wounding. Wound areas were measured by ImageJ software, and wound closure was calculated as the percentage of wound area change after 16 h.
Measurement of Cytosolic Ca2+ Concentration
The cytosolic concentration of Ca2+ ([Ca2+]i) in MDA-MB-231 cells was measured as previously described (30). Briefly, cells seeded in 96-well plates were serum-starved overnight and loaded with 4 μm Fura-2/AM at room temperature for 40 min. The basal and agonist-stimulated changes in [Ca2+]i were monitored at dual excitation wavelengths 340 and 380 nm and single emission wavelength 510 nm using a Biotek Synergy 4 microplate reader. To determine the effect of EGFP and Gαt expression or PTx treatment, cells were pretreated with doxycycline (1 μg/ml) for 3 days or with PTx (200 ng/ml) overnight.
Western Blotting Analysis
Induced expression of Gαt and phosphorylation of ERKs and AKT from cell lysates and tumor tissues were determined by Western blotting using specific antibodies (28). Immunoblots were analyzed by an Odyssey infrared imaging system (LI-COR Biosciences).
Xenograft Mouse Models
All animal studies were conducted in accordance with an Institutional Animal Care and Use Committee-approved protocol at the University of Iowa. 8–10-week-old female nude BALB/c mice were used at the time of the study. In the initial experiment, MDA-MB-231 cells expressing FL and inducible EGFP or Gαt were first treated with doxycycline (1 μg/ml) for 3 days to induce EGFP or Gαt expression. 2 × 106 of these cells in 50 μl of PBS were then implanted into the right inguinal mammary fat pads of mice. After implantation, mice were given doxycycline (2 mg/ml) continuously in the drinking water, which was supplemented every 3 days. Primary tumor growth was monitored by both bioluminescence imaging (BLI) of luciferase activities and caliper measurement of tumor length (L) and width (W) once per week. The tumor volume was calculated as πLW2/6. To determine the incidence of spontaneous lung metastasis, the primary tumor was removed after the volume reached 300 mm3. Two weeks after removal of the primary tumors, lung metastasis was determined by post-mortem ex vivo BLI (25). For these analyses, mice were injected with d-luciferin intraperitoneally. 5 min postinjection, mice were euthanized, and the lung was excised and examined by BLI.
To determine the effect of EGFP or Gαt induction on the pre-established tumor growth, MDA-MB-231 cells expressing inducible EGFP and Gαt were implanted into the right and left inguinal mammary fat pads of the same mouse, respectively, and were allowed to grow for 23 days before the mice were treated with doxycycline to induce EGFP and Gαt expression. Primary tumor growth was initially monitored by BLI. After the tumors became palpable, tumor growth was determined only by caliper measurement because bioluminescence signals from both sides of the tumors interfered with each other and prevented accurate measurement by BLI.
For experimental lung metastasis formation, MDA-MB-231 cells (3 × 105 cells in 100 μl of PBS) were injected into mice via the tail vein. Mice were started with doxycycline treatment in drinking water 24 h postinjection. Lung metastasis formation was determined by BLI weekly. 9 weeks postinjection, tumor nodules formed on the surface of the isolated lung were counted after the lung was fixed with 4% paraformaldehyde.
BLI
BLI was performed as reported previously (25). Briefly, mice were anesthetized and injected with 100 μl of d-luciferin (15 mg/ml in PBS) through retro-orbital tissue. Imaging was obtained with a Xenogen IVIS200 system between 2 and 5 min after injection. The bioluminescent data were analyzed by the software Living Image (Xenogen) and expressed as photon flux (photons/s).
Immunofluorescence Staining
Resected tumors were fixed with 4% paraformaldehyde, dehydrated with 20% sucrose solution, embedded in Tissue Tek O.C.T. compound, and snap-frozen in liquid nitrogen. Tissues were sectioned at 5-μm intervals and stained with primary antibodies against CD31 (1:100), F4/80 (1:100), or CD45 (1:100), followed by the Alexa 568-conjugated donkey anti rat IgG secondary antibody (1:800) (31). At least five images per section at random fields were taken by a Leica epifluorescence microscope (DMI6000B) using a ×20 lens and analyzed by the ImageJ software.
Statistical Analysis
Data are expressed as mean ± S.E. Statistical comparisons between two groups were analyzed by two-tailed Student's t test (p < 0.05 was considered significant).
RESULTS
Gβγ Signaling Promotes Malignant Mammary Tumor Cell Growth
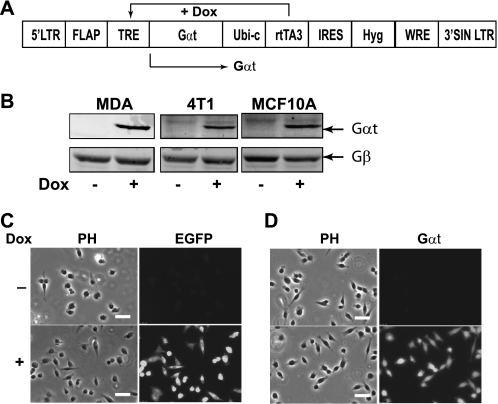
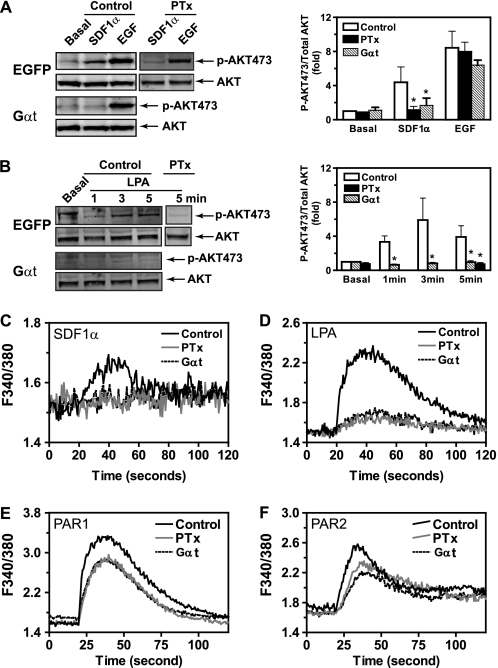
To determine if Gβγ signaling is required for breast tumor cell growth, we generated a lentiviral vector for tetracycline-inducible expression of the Gα subunit of transducin (Gαt), a scavenger of Gβγ that sequesters the Gβγ dimer to inhibit activation of downstream targets (Fig. 1A). A lentiviral vector encoding tetracycline-regulated expression of EGFP was also generated as control. These vectors were used to generate three stable cell lines using malignant cell lines MDA-MB-231 and 4T1 and the non-transformed human mammary epithelial cell line MCF10A. The ability of these cells to express EGFP and Gαt in a doxycycline-inducible manner was confirmed by Western blotting and immunofluorescence staining (Fig. 1, B–D) (data not shown). To verify that Gαt acts as a selective inhibitor of Gβγ signaling, we first evaluated the effect of induced Gαt expression in MDA-MB-231 cells on AKT phosphorylation stimulated by GPCR agonists SDF1α and LPA and the growth factor receptor agonist EGF. In this cell line, SDF1α- and LPA-mediated AKT activation is blocked by PTx (Fig. 2, A and B), implicating the involvement of Gi/o and Gβγ subunits in this activation process because Gβγ released from activated Gi/o is known to activate AKT via PI3Ks (32). In contrast, EGF stimulates AKT phosphorylation via receptor tyrosine kinases, which is insensitive to PTx treatment (Fig. 2A). Inducible expression of Gαt blocked SDF1α- and LPA-stimulated but not EGF-induced AKT phosphorylation (Fig. 2, A and B). In contrast, Gαt had no effect on the inhibition of cAMP production by SDF1α stimulation (supplemental Fig. S1), which is mediated via Gαi/o. Similarly, Gαt did not affect isoproterenol-stimulated cAMP production, which is mediated by Gαs (supplemental Fig. S1). These findings suggest that Gαt selectively inhibits Gβγ-mediated AKT activation. To provide further evidence that Gαt is a selective inhibitor of Gβγ, we examined the effect of Gαt expression on Ca2+ signaling elicited by GPCRs primarily coupled to either Gi/o or Gq/11 (22, 33). As expected, the increase in [Ca2+]i stimulated by SDF1α and LPA, which primarily activate PLCβ via Gβγ released from the activated Gi/o, was largely inhibited by PTx treatment and induced Gαt expression (Fig. 2, C and D). In contrast, Ca2+ signaling stimulated by PAR1 and PAR2, which activate PLCβ via Gαq/11 subunits (23), was less sensitive to either PTx or Gαt (Fig. 2, E and F). Similar findings were obtained in 4T1 cells expressing Gαt (data not shown). Collectively, these data indicate that Gαt is a selective inhibitor of Gβγ.
FIGURE 1.
Induced expression of EGFP and Gαt in MDA-MB-231, 4T1, and MCF10A stable cell lines. A, schematic of the lentiviral vector used for doxycycline-induced expression of Gαt. B, expression of Gαt in MDA-MB-231, 4T1, and MCF10A cells transduced with pSLIK-Gαt lentivirus. Gαt expression was induced by doxycycline (1 μg/ml) for 3 days and analyzed by Western blotting. C and D, induced expression of EGFP (C) and Gαt (D) in MDA-MB-231 cells by doxycycline (Dox) treatment. Gαt expression was revealed by staining cells with anti-Gαt antibody, followed by Alexa-488-conjugated secondary antibody. Phase contrast (PH) and fluorescent images are shown. Scale bar, 50 μm.
FIGURE 2.
Gαt selectively inhibits Gβγ-mediated signaling. A and B, effects of induced Gαt expression on SDF1α-stimulated (A) and LPA-stimulated (B) AKT phosphorylation. MDA-MB-231 cells were treated with doxycycline for 3 days to induce EGFP (Control) or Gαt expression. AKT phosphorylation stimulated by SDF1α (50 nm), LPA (10 μm), and EGF (100 ng/ml) was then determined by Western blotting analyses. To determine the effect of PTx, MDA-MB-231 cells expressing EGFP were pretreated with PTx (200 ng/ml) overnight. Representative images are shown on the left, and quantitative data are shown on the right (n = 4–5). C–F, effects of induced Gαt expression on Ca2+ signaling stimulated by SDF1α (C), LPA (D), PAR1 (E), and PAR2 (F) agonist peptides. MDA-MB-231 cells expressing inducible Gαt were treated with either buffer (Control), PTx overnight (PTx), or doxycycline for 3 days to induce Gαt expression (Gαt) and then stimulated with SDF1α (50 nm), LPA (10 μm), PAR1 (10 μm), and PAR2 (10 μm) agonist peptides. The increase in [Ca2+]i was measured using Fura-2/AM. Representative data from 3–4 independent experiments are shown. *, p < 0.05 indicates significance versus control.
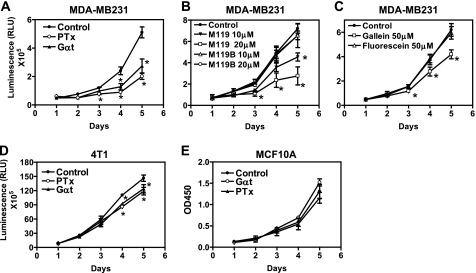
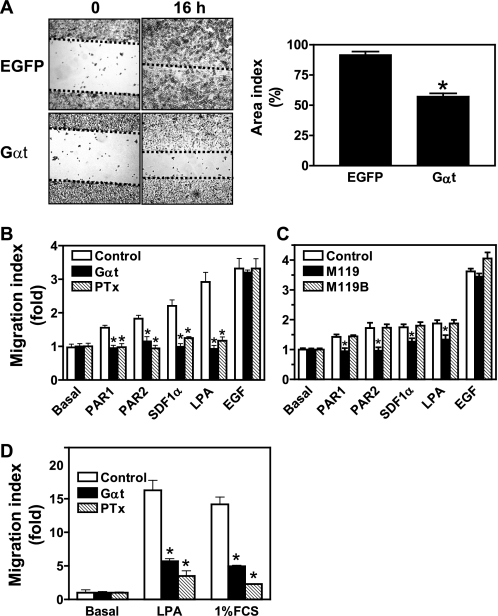
To study the effect of blocking Gβγ signaling on breast tumor cell growth, we analyzed the growth rate of cells expressing either inducible EGFP or Gαt in the growth medium containing 10% FBS. The growth rate of MDA-MB-231 and 4T1 cells was determined by measuring the increase in the luciferase activities, because these cells stably express a FL, and there is a linear correlation between luciferase activity and cell number (data not shown). Moreover, expression of EGFP or Gαt altered neither the level of luciferase expression nor its activities (data not shown). The growth rate of MCF10A cells was determined by a tetrazolium salt WST-1-based approach. As shown in Fig. 3, A–E, compared with EGFP expression, induced Gαt expression resulted in significant inhibition of MDA-MB-231 and 4T1 cell growth but had little effect on MCF10A cell proliferation. The failure of Gαt to inhibit MCF10 growth is not due to the different assays used because similar results were obtained when cell growth was evaluated by counting the cell number or when the tetrazolium salt WST-1-based assay was used to measure MDA-MB-231 cell growth (supplemental Fig. S2). Moreover, it cannot be attributed to the lower level of Gαt expression because Gαt was expressed at comparable levels between MCF10A, MDA-MB2–231, and 4T1 cells, and these cell lines expressed comparable levels of endogenous Gβγ (Fig. 1B). PTx treatment recapitulated the effect of Gαt expression on cell growth (Fig. 3, A, D, and E). Similarly, treatment with small molecular inhibitors of Gβγ, M119 and gallein, but not their inactive analogs M119B and fluorescein (34–35), decreased MDA-MB-231 but not MCF10A cell growth (Fig. 3, B and C) (data not shown). Taken together, these findings indicate that free Gβγ released from Gi/o proteins is critical for the proliferation of malignant breast tumor cells but is dispensable for the growth of non-transformed mammary epithelial cells.
FIGURE 3.
Blocking Gβγ signaling inhibits breast tumor cell growth. Effects of Gαt expression and treatment with PTx or small molecule inhibitors of Gβγ on the proliferation of MDA-MB-231 (A–C), 4T1 (D), and MCF10A (E) in the complete growth medium containing 10% FBS. The dose of PTx used was 200 ng/ml. The indicated concentrations of M119, M119B, gallein, and fluorescein were added to the growth medium and replenished every day. *, p < 0.05 indicates significance versus control. n = 4–6. Error bars, S.E.
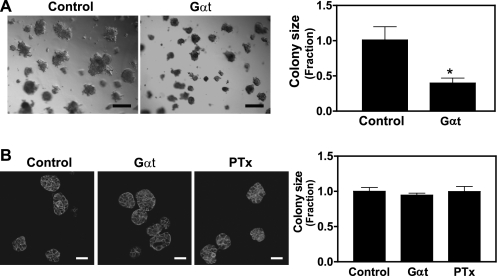
The interaction of breast tumor cells with the extracellular matrix may modulate their morphology, growth, and malignance (36). To determine if the role of Gβγ in breast tumor cell growth is influenced by extracellular matrix, we further evaluated the effect of blocking Gβγ signaling on MDA-MB-231 and MCF10A growth in Matrigel. As reported previously (37), under these culture conditions, MDA-MB-231 cells formed large, loose, and disorganized colonies, similar to invasive tumors (Fig. 4A). In contrast, MCF10A formed small and organized acini (Fig. 4B) (27). Induced Gαt expression decreased the average size of MDA-MB-231 cell colonies by 70% (Fig. 4A). In contrast, neither the size nor the morphology of MCF10A acini was affected by Gαt expression or PTx treatment (Fig. 4B). These results provide further support for the critical role of Gβγ in promoting malignant growth of breast tumor cells.
FIGURE 4.
Effects of blocking Gβγ signaling on MDA-MB-231 and MCF10A cell growth in three-dimensional culture. MDA-MB-231 (A) and MCF10A (B) cells were cultured in matrigel in the absence (Control) or presence of PTx (200 ng/ml) or doxycycline to induce Gαt expression (Gαt) for 8 days. MCF10 cells were fixed and stained with Alexa568-conjugated phalloidin. Representative phase-contrast images of MDA-MB-231 (A) and fluorescent images of MCF10A (B) cells are shown on the left. Quantitative data showing the size of MDA-MB-231 (A) and MCF10A (B) colonies are shown on the right. Data are expressed as the fraction of the size of the colonies derived from the control cells. Scale bar in A, 200 μm. Scale bar in B, 50 μm. *, p < 0.05 indicates significance versus control. n = 3.
Blockage of Gβγ Signaling Inhibited Breast Tumor Cell Migration
Breast tumor cells overexpress a plethora of GPCRs that may mediate their metastatic spread to distant organs (38). To determine if Gβγ plays a role in breast tumor cell migration, we examined the effect of blocking Gβγ signaling on MDA-MB-231 and 4T1 cell migration. As shown in Fig. 5A, induced Gαt expression significantly decreased serum-induced migration of MDA-MB-231 cells in a wound healing assay. The decreased wound closure by Gαt is unlikely to be due to the reduced cell proliferation because the assay was conducted in the presence of 10 μg/ml mitomycin, which completely abolished cell proliferation (data not shown). In line with these findings, transwell migration assays also showed that Gαt expression abolished MDA-MB-231 cell migration induced by an array of chemoattractants, including SDF1α, LPA, PAR1, and PAR2 agonist peptides and 1% FBS (Fig. 5B). The inhibitory effect of Gαt on cell migration was mirrored by PTx treatment. Neither Gαt nor PTx affected EGF-stimulated cell migration (Fig. 5B). The Gβγ inhibitor M119, but not the inactive analog M119B, exerted similar inhibitory effects on MDA-MB-231 cell migration (Fig. 5C). Similarly, Gαt and PTx attenuated LPA- and 1% FCS-stimulated 4T1 cell migration (Fig. 5D). Taken together, these findings demonstrate that Gβγ subunits, which are probably released from activated Gi/o proteins, play a central role in transmitting signals from multiple chemoattractant agents to promote breast tumor cell migration. In support of this notion, overexpression of Gβ1γ2 is sufficient to induce increased MDA-MB-231 cell migration even in the absence of GPCR stimulation (supplemental Fig. S3).
FIGURE 5.
Inhibition of Gβγ signaling blocks breast tumor cell migration. MDA-MB-231 (A–C) and 4T1 cells (D) were treated with either PTx overnight, doxycycline for 3 days to induce Gαt expression (Gαt), or small molecule inhibitors of Gβγ for 30 min to block Gβγ signaling. The effects on cell migration were determined by the ability of cells to close a wound in a wound healing assay (A) or to migrate toward a concentration gradient of chemoattractants, SDF1α (100 nm), LPA (1 μm), PAR1 (10 μm), and PAR2 (10 μm) agonist peptides, 1% FBS, and EGF (100 ng/ml), in the transwell assays. *, p < 0.05 indicates significance versus control. n = 3–5. Error bars, S.E.
Gβγ Signaling Contributes to Breast Tumor Growth in Vivo
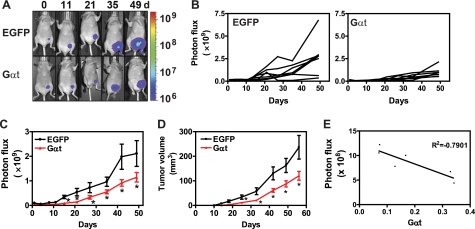
To evaluate the contribution of Gβγ signaling to breast tumor growth in vivo, we employed a xenograft mouse model of breast tumor using MDA-MB-231 cells constitutively expressing FL and inducible EGFP or Gαt. In the initial experiment, MDA-MB-231 cells were pretreated with doxycycline for 3 days to induce EGFP or Gαt expression prior to their injection into the mammary fat pads of nude mice. After cell implantation, doxycycline was continuously administered to mice in the drinking water to sustain EGFP and Gαt expression. BLI studies showed that photon flux from Gαt-expressing MDA-MB-231 cells at the primary implantation site increased more slowly than EGFP-expressing cells. Whereas EGFP-expressing cells displayed a 4-fold increase in photon flux 15 days after cells were inoculated into mice, photon flux from Gαt-expressing cells did not show a significant increase until 27 days after cell implantation (Fig. 6, B and C). Because EGFP- and Gαt-expressing cells have a comparable level of luciferase activity, and Gαt expression altered neither luciferase expression nor its activities, these findings indicate that blocking Gβγ signaling via Gαt inhibited the early growth and/or survival of breast cancer cells at the primary tumor sites. After the establishment of primary tumors, mice inoculated with Gαt-expressing cells constantly displayed a significantly lower photon flux at all time points than those inoculated with EGFP-expressing cells (Fig. 6, A–C), suggesting that Gαt also delayed tumor progression. Consistent with these data, measurement of tumor volume by calipers also showed that MDA-MB-231 tumors expressing Gαt have a significantly slower growth rate than those expressing EGFP (Fig. 6D). We observed an inverse correlation between the expression level of Gαt in the tumor and the level of photon flux (Fig. 6E), suggesting that the degree of tumor suppression is correlated to the level of inhibition of Gβγ signaling by Gαt. Collectively, these findings support the critical role of Gβγ signaling in breast tumor development.
FIGURE 6.
Blocking Gβγ signaling via induced Gαt expression inhibits tumor growth in mice. MDA-MB-231 cells expressing luciferase and inducible EGFP or Gαt were first treated with doxycycline for 3 days and then implanted into the inguinal mammary fat pads of nude mice (n = 7). Tumor growth was monitored by bioluminescence imaging and caliper measurement. A, representative bioluminescence images at the indicated times postinjection with MDA-MB-231 cells expressing EGFP or Gαt. Each set of images was taken from the same mouse. B and C, individual (B) and average tumor growth rate (C) as reflected by total photon flux. D, tumor growth rate as determined by caliper measurement. E, inverse correlation between the level of Gαt expression and the size of tumors as reflected by total photon influx. The level of Gαt expression was normalized to the total amount of protein loaded. *, p < 0.05 indicates significance versus EGFP control. Error bars, S.E.
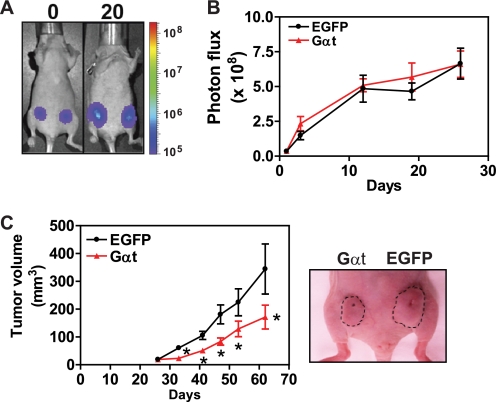
To further evaluate the role of Gβγ signaling in tumor progression, we inoculated the same number of MDA-MB-231 cells expressing EGFP or Gαt into the right and left side mammary fat pads of the same nude mouse, respectively, and then allowed them to form palpable tumors without inducing EGFP and Gαt expression for 23 days. Over this period, EGFP- and Gαt-expressing cells had the same growth rate, as determined by BLI (Fig. 7, A and B). However, 1 week after the administration of doxycycline to induce EGFP and Gαt expression, Gαt-expressing cells formed significantly smaller tumors than EGFP-expressing cells (Fig. 7C). At the end of the experiment (day 63), the resected tumors derived from Gαt-expressing cells had about half the weight of those derived from EGFP-expressing cells (data not shown). These data demonstrate that Gβγ signaling is critical not only for the initial establishment but also for the later progression of MDA-MB-231 tumors in nude mice.
FIGURE 7.
Induced Gαt expression reduces existing tumor growth. MDA-MB-231 cells expressing luciferase and inducible EGFP or Gαt were implanted to the right and left inguinal mammary fat pads of mice (n = 7), respectively, and were allowed to grow for 23 days before the mice were treated with doxycycline to induce EGFP and Gαt expression. Tumor growth was monitored by bioluminescence imaging and caliper measurement. Shown are representative bioluminescence images (A) and average tumor growth rate (B) before EGFP and Gαt induction at the indicted times postinjection. C, tumor growth rate after induced EGFP and Gαt expression, as determined by caliper measurement. Right, a representative image of one mouse bearing tumors derived from EGFP- and Gαt-expressing cells. The size of the tumors is indicated. *, p < 0.05 indicates significance versus EGFP control. Error bars, S.E.
Gβγ Signaling Is Required for Breast Tumor Metastasis in Vivo
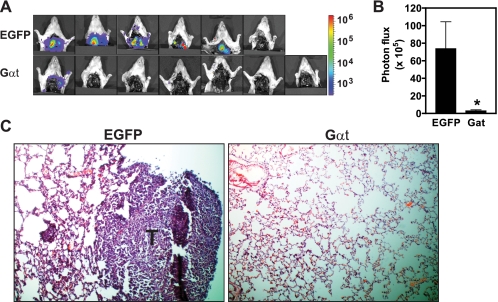
After primary tumor formation, MDA-MB-231 breast tumor cells spontaneously develop metastases, predominantly affecting the lungs. To define the role of Gβγ signaling in these processes, we monitored metastasis formation in mice inoculated with MDA-MB-231 cells and treated with doxycycline continuously to induce sustained EGFP and Gαt expression. To detect metastases, we surgically removed the primary tumors expressing EGFP or Gαt when their volume reached ∼300 mm3 and then performed BLI studies 2 weeks after tumor resection. To verify the formation of lung metastasis, we also performed ex vivo imaging of the isolated lung and histological analyses of lung sections. One of seven mice bearing EGFP-expressing tumors died in the cage before their tumors reached ∼300 mm3, thus excluding it from examination for lung metastasis. However, five of six remaining mice developed lung metastases (Fig. 8, A–C). In contrast, only one of seven mice bearing Gαt-expressing tumors developed lung metastases (Fig. 8, A–C). These findings indicate that Gβγ signaling is required for spontaneous metastasis of breast cancer cells.
FIGURE 8.
Induced Gαt expression prevents spontaneous lung metastasis. As indicted in the legend to Fig. 6, mice were inoculated with the indicated MDA-MB-231 cells. Tumors were removed after reaching a volume of 300 mm3. Lung metastasis was monitored by postmortem ex vivo BLI 2 weeks after removal of the primary tumor (A and B) and verified by histological analysis of lung sections (C). Bioluminescence (A) and representative H&E staining (C) images and mean intensity of lung metastasis signal (B), as reflected by the total photon flux, are shown. n = 7. *, p < 0.05 indicates significance versus EGFP control. Error bars, S.E.
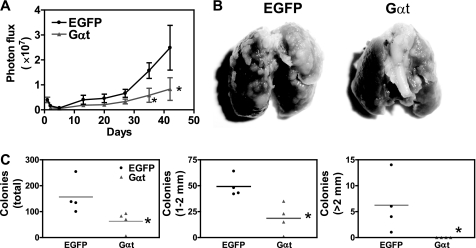
To further determine if the growth of tumor cells still relies on Gβγ signaling after they metastasize to distant organs, we injected MDA-MB-231 cells expressing EGFP or Gαt intravenously into the tail vein of nude mice and then evaluated the ability of these cells to colonize in the lung. BLI analysis of lungs indicates that a similar number of EGFP- and Gαt-expressing tumor cells were inoculated into the lung because initial bioluminescence at 3 h postinjection was comparable for both cell lines (Fig. 9A). For the first 5 days postinjection, decreases in bioluminescence were comparable for both tumor cell lines, indicating that the initial inefficiency of metastasis formation in the lung was not affected by Gαt. However, at the later time points, tumor cells expressing Gαt progressed much slower than the control cells expressing EGFP (Fig. 9A). At 9 weeks postinjection, the lungs isolated from the remaining mice injected with Gαt-expressing cells showed a significantly lower total number of surface tumor colonies than those from mice injected with EGFP-expressing cells (Fig. 9, B and C). Moreover, when tumor colonies were counted based on their size, it was found that Gαt-expressing cells developed smaller tumor colonies than EGFP-expressing cells (Fig. 9C).
FIGURE 9.
Induced Gαt expression reduced tumor formation in the experimental lung metastasis model. MDA-MB-231 cells were injected into the mice (n = 6) via the tail vein to generate lung metastases. Mice were treated with doxycycline to induce EGFP or Gαt expression 1 day postinjection. Tumor formation in the lung was monitored by BLI weekly (A). At week 9, mice were sacrificed, and the surface tumors on the isolated lung were counted and grouped based on their size (1–2 mm or bigger than 2 mm in diameter) (C). Representative images of the lungs from mice injected with the indicated tumor cells are shown in B. T, tumor cells. *, p < 0.05 indicates significance versus EGFP control. Error bars, S.E.
Blockade of Gβγ Signaling Attenuates Leukocyte Infiltration and Tumor Angiogenesis
Increased angiogenesis and chronic leukocyte infiltration have been causally associated with tumor progression (9). To determine if Gβγ signaling plays a role in these processes, we performed histological analyses of tumor sections prepared from the primary tumors.
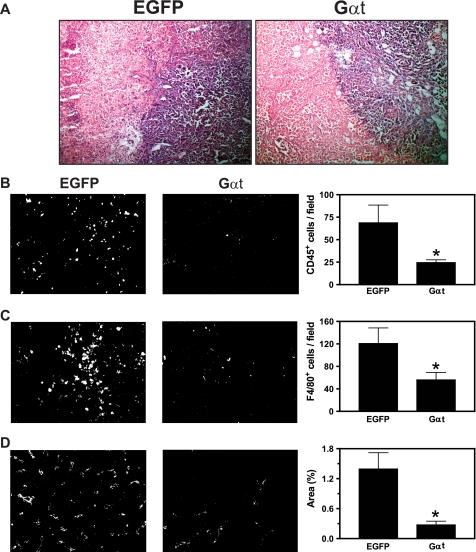
H&E staining analysis showed that in the tumor tissues, Gαt-expressing tumor cells form a cluster with a well defined boundary (Fig. 10A). Although EGFP-expressing tumor cells also form clusters, their margins are uneven and interspersed with leukocytes and other stromal cells (Fig. 10A). Consistent with this notion, immunofluorescence staining of the common leukocyte marker CD45 and macrophage-selective marker F4/80 showed a significant reduction in leukocyte infiltration in the tumors derived from Gαt-expressing cells (Fig. 10, B and C). Moreover, these tumors had reduced blood vessel density as determined by staining with the endothelial cell marker CD31 (Fig. 10D).
FIGURE 10.
Inhibition of Gβγ signaling reduced leukocyte infiltration and angiogenesis. Frozen tumor sections were prepared from mice inoculated with MDA-MB-231 cells as indicated in the legend to Fig. 7. After fixation, sections were stained with H&E (A), CD45 for common leukocytes (B), F4/80 for macrophages (C), or CD31 for endothelia (D) followed by staining with Alexa568-conjugated secondary antibody. Fluorescence images were taken from five randomly chosen fields of each section, and the positively stained cells were quantified using ImageJ software. Representative images of the indicated tumor sections and the quantitative data are shown in the left and right of each panel, respectively. *, p < 0.05 indicates significance versus EGFP control. n = 7. Error bars, S.E.
DISCUSSION
In this study, we demonstrate that Gβγ signaling promotes tumor growth and metastasis in experimental models of breast cancer. Blocking Gβγ signaling by Gαt or small molecular inhibitors M119 and gallein significantly attenuated serum-induced MDA-MB-231 and 4T1 cell growth. Treatment of tumor cells with PTx produced a similar inhibitory effect on cell growth, suggesting that serum contains mitogenic factors, such as LPA, that primarily act through the Gi/o class of G proteins to promote tumor growth. The residual cell growth in the presence of PTx and Gβγ inhibitors is probably mediated by growth factors contained in the serum. The inhibition of Gβγ signaling by Gαt was also effective in delaying the initial tumor formation and limiting the rate of tumor growth in nude mice. By using an inducible expression system, we were also able to show that sequestering Gβγ via Gαt retarded preexisting tumor growth. Of particular interest are the findings that inhibiting Gβγ signaling has little effect on the growth of the non-transformed mammary epithelial cell line MCF10A. These data suggest that the role of Gβγ in cell proliferation may be manifested in breast tumor cells. In line with these findings, it has been shown previously that the coupling of chemokine receptor CXCR4 to G proteins is impaired in MCF10A or non-invasive breast cancer cells (39). Thus, targeting Gβγ may achieve a differential inhibitory effect on the growth of aggressive tumor cells versus normal cells. Although inhibiting Gβγ signaling alone only results in ∼50% decrease in tumor growth rate, combination treatment with other antitumor agents may have synergistic inhibitory effects on tumor growth. This notion has been supported by the study of anti-Gα inhibitors (17).
Our studies also established a critical role for Gβγ signaling in mediating tumor cell migration induced by a wide array of GPCR ligands. The role of Gβγ in breast tumor cell migration is specific to GPCR stimulation and not EGF-mediated stimulation of tyrosine kinase receptors. Moreover, GPCRs appear to stimulate breast tumor cell migration primarily via a Gi/o-dependent signaling pathway. This is demonstrated by the finding that tumor cell migration stimulated by all of the tested agonists is inhibited via PTx, which uncouples Gi/o proteins from their cognate receptors by ADP-ribosylation of Gαi/o (40, 41). Because the blockade of Gβγ signaling generates the same inhibitory effect as PTx treatment on tumor cell migration, it implies that breast tumor cell migration is mediated primarily by Gβγ subunits released from the activated Gi/o proteins. These findings are interesting because several of the agonists we tested are able to activate multiple G protein subtypes via GPCRs. This is evident from the findings that increased intracellular Ca2+ stimulated by PAR1 and PAR2 agonist peptides is less sensitive to PTx and the sequestration of free Gβγ subunits via Gαt, implying the involvement of PTx-insensitive Gαq/11-mediated PLCβ activation (Fig. 2, E and F). However, despite the lack of inhibition on Ca2+ signaling, Gβγ blockade still abolished PAR-mediated cell migration, suggesting that Ca2+ signaling is not essential for cell migration. Indeed, chelation of intracellular Ca2+ with BAPTA-AM had no effect on the migration of MDA-MB-231 cells stimulated by LPA or the PAR1 agonist peptide (supplemental Fig. S4). These findings are contrary to several previous reports indicating a role of Ca2+ signaling in the migration of MDA-MB-231 cells. However, these studies used several other stimuli, including EGF, SDF1α, VEGF, and serum to induce cell migration through filter membrane or endothelial monolayer (42–45). Thus, it is possible that the role of Ca2+ signaling in tumor cell migration depends on the type of stimuli used and the assay conditions. Previous work suggests that, in addition to Gi/o, GPCRs may signal through other G protein subtypes to regulate cell migration. For example, Gα12/13 controls leukocyte migration via a RhoA-dependent signaling mechanism (46–47). Up-regulation of Gα12 proteins has been associated with enhanced metastasis of breast tumor (48). Recently, Gαq-dependent signaling pathways were also shown to regulate chemotaxis of neutrophils and dendritic cells by a subset of chemokines (49), although it remains unknown if Gq signaling is involved in tumor cell migration. Based on studies carried out in different cell types, all of the agonists that we tested have the ability to activate multiple G protein subtypes, including Gi/o, G12/13, and Gq proteins. Theoretically, in addition to Gi/o proteins, these agonists could promote breast tumor cell migration via the G12/13- and Gq-dependent signaling pathways. Nevertheless, the fact that the blockade of Gβγ signaling is sufficient to abrogate breast tumor cells by these ligands underscores the importance of Gβγ signaling in breast tumor cell migration.
Our in vivo studies have provided compelling evidence that Gβγ signaling promotes breast tumor metastasis. This is evident from the findings that Gαt expression in MDA-MB-231 cells dramatically alleviated the formation of spontaneous lung metastases from primary tumors and decreased the formation of tumor colonies in the experimental lung metastasis model. Tumor metastasis is a complicated multistep process involving tumor cell local invasion, intravasation into and extravasation out of blood and lymphatic vessels, colonization at distant organs, and angiogenesis (50). The exact step whereby Gβγ signaling impinges on tumor metastasis remains unclear. However, the fact that the blockade of Gβγ signaling attenuated tumor growth in the experimental lung metastasis model indicates that Gβγ signaling is also required in the later steps of metastatic cascade, such as extravasation, migration, growth, and replication of tumor cells at the distant organs (51). These findings are consistent with the diverse functions of GPCRs, such as chemokine receptors CXCR4 and CXCR2, LPA receptors, and PARs, in regulating many aspects of tumor metastasis, including tumor cell proliferation, migration, and invasion and induction of angiogenesis at the tumor site (3, 52–54).
Gβγ regulates the activity of diverse signaling molecules, including PI3Ks, the small GTPases Rac and Cdc42, and PLCβ (55). The importance of these signaling molecules in cell motility and survival has been well documented (55). Recently, it has been shown that Gβγ mediates breast cancer cell migration and invasion via a Rac-dependent signaling pathway (20). By using inhibitors of PI3Ks and PLC, we were able to show that serum-induced breast cancer cell growth and GPCR-stimulated tumor cell migration depend on the enzymatic activities of PI3Ks and PLC (data not shown). Thus, it is likely that Gβγ regulates breast tumor progression via diverse signaling pathways. Further studies to define the downstream signaling pathways of Gβγ that promote tumor progression and metastasis will provide new insights into the molecular basis of breast cancer and identify new targets for anticancer therapy.
Apart from mediating the proliferative, migratory, and invasive properties of breast tumor cells via their overexpressed GPCRs, Gβγ signaling may also regulate the tumor microenvironment because Gβγ blockade attenuated leukocyte infiltration and angiogenesis in primary tumors. This effect may be mediated by the inhibition of cytokines and chemokines production from tumor cells, infiltrating leukocytes and other stromal cells. Thus, these findings suggest that targeting Gβγ could have the dual beneficial effect of acting on both the tumor cells and the cancer-supporting microenvironment to prevent tumor progression.
In summary, the data presented here establish a critical function for Gβγ in breast tumor growth and metastasis and indicate that targeting Gβγ could represent a novel strategy to block tumor progression linked to multiple GPCRs. Given that enhanced GPCR signaling is associated with initiation and progression of many types of cancer (3), these findings have implications for the treatment of other types of tumors.
This work was supported, in whole or in part, by National Institutes of Health, NCI, Grant R01CA115438. The work was also supported by American Cancer Society Grant IRG-77-004-31 (to S. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- GPCR
- G protein-coupled receptor
- BLI
- bioluminescence imaging
- FL
- firefly luciferase
- LPA
- lysophosphatidic acid
- PAR
- protease-activated receptor
- PTx
- pertussis toxin
- EGFP
- enhanced green fluorescent protein.
REFERENCES
- 1. Coughlin S. S., Ekwueme D. U. (2009) Cancer Epidemiol. 33, 315–318 [DOI] [PubMed] [Google Scholar]
- 2. Jemal A., Siegel R., Xu J., Ward E. (2010) CA Cancer J. Clin. 60, 277–300 [DOI] [PubMed] [Google Scholar]
- 3. Dorsam R. T., Gutkind J. S. (2007) Nat. Rev. Cancer 7, 79–94 [DOI] [PubMed] [Google Scholar]
- 4. Schmid B. C., Rudas M., Rezniczek G. A., Leodolter S., Zeillinger R. (2004) Breast Cancer Res. Treat. 84, 247–250 [DOI] [PubMed] [Google Scholar]
- 5. Cabioglu N., Sahin A., Doucet M., Yavuz E., Igci A. O., Yildirim E., Aktas E., Bilgic S., Kiran B., Deniz G., Price J. E. (2005) Clin. Exp. Metastasis 22, 39–46 [DOI] [PubMed] [Google Scholar]
- 6. Cabioglu N., Yazici M. S., Arun B., Broglio K. R., Hortobagyi G. N., Price J. E., Sahin A. (2005) Clin. Cancer Res. 11, 5686–5693 [DOI] [PubMed] [Google Scholar]
- 7. Even-Ram S., Uziely B., Cohen P., Grisaru-Granovsky S., Maoz M., Ginzburg Y., Reich R., Vlodavsky I., Bar-Shavit R. (1998) Nat. Med. 4, 909–914 [DOI] [PubMed] [Google Scholar]
- 8. Panupinthu N., Lee H. Y., Mills G. B. (2010) Br. J. Cancer 102, 941–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ali S., Lazennec G. (2007) Cancer Metastasis Rev. 26, 401–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verástegui E., Zlotnik A. (2001) Nature 410, 50–56 [DOI] [PubMed] [Google Scholar]
- 11. Mantovani A., Allavena P., Sica A., Balkwill F. (2008) Nature 454, 436–444 [DOI] [PubMed] [Google Scholar]
- 12. Ruffell B., DeNardo D. G., Affara N. I., Coussens L. M. (2010) Cytokine Growth Factor Rev. 21, 3–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Balkwill F. (2004) Nat. Rev. Cancer 4, 540–550 [DOI] [PubMed] [Google Scholar]
- 14. Wu X., Lee V. C., Chevalier E., Hwang S. T. (2009) Curr. Pharm. Des. 15, 742–757 [DOI] [PubMed] [Google Scholar]
- 15. Liebmann C. (2004) Curr. Pharm. Des. 10, 1937–1958 [DOI] [PubMed] [Google Scholar]
- 16. Oldham W. M., Hamm H. E. (2008) Nat. Rev. Mol. Cell Biol. 9, 60–71 [DOI] [PubMed] [Google Scholar]
- 17. Prévost G. P., Lonchampt M. O., Holbeck S., Attoub S., Zaharevitz D., Alley M., Wright J., Brezak M. C., Coulomb H., Savola A., Huchet M., Chaumeron S., Nguyen Q. D., Forgez P., Bruyneel E., Bracke M., Ferrandis E., Roubert P., Demarquay D., Gespach C., Kasprzyk P. G. (2006) Cancer Res. 66, 9227–9234 [DOI] [PubMed] [Google Scholar]
- 18. Ayoub M. A., Damian M., Gespach C., Ferrandis E., Lavergne O., De Wever O., Banères J. L., Pin J. P., Prévost G. P. (2009) J. Biol. Chem. 284, 29136–29145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bookout A. L., Finney A. E., Guo R., Peppel K., Koch W. J., Daaka Y. (2003) J. Biol. Chem. 278, 37569–37573 [DOI] [PubMed] [Google Scholar]
- 20. Kirui J. K., Xie Y., Wolff D. W., Jiang H., Abel P. W., Tu Y. (2010) J. Pharmacol. Exp. Ther. 333, 393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu S., Murph M., Panupinthu N., Mills G. B. (2009) Cell Cycle 8, 3695–3701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi J. W., Herr D. R., Noguchi K., Yung Y. C., Lee C. W., Mutoh T., Lin M. E., Teo S. T., Park K. E., Mosley A. N., Chun J. (2010) Annu. Rev. Pharmacol. Toxicol. 50, 157–186 [DOI] [PubMed] [Google Scholar]
- 23. Soh U. J., Dores M. R., Chen B., Trejo J. (2010) Br. J. Pharmacol. 160, 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shin K. J., Wall E. A., Zavzavadjian J. R., Santat L. A., Liu J., Hwang J. I., Rebres R., Roach T., Seaman W., Simon M. I., Fraser I. D. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13759–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Drake J. M., Gabriel C. L., Henry M. D. (2005) Clin. Exp. Metastasis 22, 674–684 [DOI] [PubMed] [Google Scholar]
- 26. Smith M. C., Luker K. E., Garbow J. R., Prior J. L., Jackson E., Piwnica-Worms D., Luker G. D. (2004) Cancer Res. 64, 8604–8612 [DOI] [PubMed] [Google Scholar]
- 27. Debnath J., Muthuswamy S. K., Brugge J. S. (2003) Methods 30, 256–268 [DOI] [PubMed] [Google Scholar]
- 28. Chen S., Lin F., Shin M. E., Wang F., Shen L., Hamm H. E. (2008) Mol. Biol. Cell 19, 3909–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang C. C., Park A. Y., Guan J. L. (2007) Nat. Protoc. 2, 329–333 [DOI] [PubMed] [Google Scholar]
- 30. McLaughlin J. N., Shen L., Holinstat M., Brooks J. D., Dibenedetto E., Hamm H. E. (2005) J. Biol. Chem. 280, 25048–25059 [DOI] [PubMed] [Google Scholar]
- 31. Weydert C. J., Esser A. K., Mejia R. A., Drake J. M., Barnes J. M., Henry M. D. (2009) Cancer Biol. Ther. 8, 720–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stephens L., Smrcka A., Cooke F. T., Jackson T. R., Sternweis P. C., Hawkins P. T. (1994) Cell 77, 83–93 [DOI] [PubMed] [Google Scholar]
- 33. Roland J., Murphy B. J., Ahr B., Robert-Hebmann V., Delauzun V., Nye K. E., Devaux C., Biard-Piechaczyk M. (2003) Blood 101, 399–406 [DOI] [PubMed] [Google Scholar]
- 34. Bonacci T. M., Mathews J. L., Yuan C., Lehmann D. M., Malik S., Wu D., Font J. L., Bidlack J. M., Smrcka A. V. (2006) Science 312, 443–446 [DOI] [PubMed] [Google Scholar]
- 35. Lehmann D. M., Seneviratne A. M., Smrcka A. V. (2008) Mol. Pharmacol. 73, 410–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weigelt B., Bissell M. J. (2008) Semin. Cancer Biol. 18, 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Park C. C., Zhang H., Pallavicini M., Gray J. W., Baehner F., Park C. J., Bissell M. J. (2006) Cancer Res. 66, 1526–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taborga M., Corcoran K. E., Fernandes N., Ramkissoon S. H., Rameshwar P. (2007) Mini. Rev. Med. Chem. 7, 245–251 [DOI] [PubMed] [Google Scholar]
- 39. Holland J. D., Kochetkova M., Akekawatchai C., Dottore M., Lopez A., McColl S. R. (2006) Cancer Res. 66, 4117–4124 [DOI] [PubMed] [Google Scholar]
- 40. West R. E., Jr., Moss J., Vaughan M., Liu T., Liu T. Y. (1985) J. Biol. Chem. 260, 14428–14430 [PubMed] [Google Scholar]
- 41. Ui M., Katada T. (1990) Adv. Second Messenger Phosphoprotein Res. 24, 63–69 [PubMed] [Google Scholar]
- 42. Price J. T., Tiganis T., Agarwal A., Djakiew D., Thompson E. W. (1999) Cancer Res. 59, 5475–5478 [PubMed] [Google Scholar]
- 43. Lee T. H., Avraham H. K., Jiang S., Avraham S. (2003) J. Biol. Chem. 278, 5277–5284 [DOI] [PubMed] [Google Scholar]
- 44. Lee B. C., Lee T. H., Avraham S., Avraham H. K. (2004) Mol. Cancer Res. 2, 327–338 [PubMed] [Google Scholar]
- 45. Yang S., Zhang J. J., Huang X. Y. (2009) Cancer Cell 15, 124–134 [DOI] [PubMed] [Google Scholar]
- 46. Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H. R. (2003) Cell 114, 201–214 [DOI] [PubMed] [Google Scholar]
- 47. Tan W., Martin D., Gutkind J. S. (2006) J. Biol. Chem. 281, 39542–39549 [DOI] [PubMed] [Google Scholar]
- 48. Kelly P., Moeller B. J., Juneja J., Booden M. A., Der C. J., Daaka Y., Dewhirst M. W., Fields T. A., Casey P. J. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shi G., Partida-Sánchez S., Misra R. S., Tighe M., Borchers M. T., Lee J. J., Simon M. I., Lund F. E. (2007) J. Exp. Med. 204, 2705–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nguyen D. X., Bos P. D., Massagué J. (2009) Nat. Rev. Cancer 9, 274–284 [DOI] [PubMed] [Google Scholar]
- 51. Chambers A. F., Groom A. C., MacDonald I. C. (2002) Nat. Rev. Cancer 2, 563–572 [DOI] [PubMed] [Google Scholar]
- 52. Mills G. B., Moolenaar W. H. (2003) Nat. Rev. Cancer 3, 582–591 [DOI] [PubMed] [Google Scholar]
- 53. Nierodzik M. L., Karpatkin S. (2006) Cancer Cell 10, 355–362 [DOI] [PubMed] [Google Scholar]
- 54. Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L., Weinberg R. A. (2005) Cell 121, 335–348 [DOI] [PubMed] [Google Scholar]
- 55. Smrcka A. V. (2008) Cell Mol. Life Sci. 65, 2191–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]