Abstract
Acid sphingomyelinase (aSMase) generates the bioactive lipid ceramide (Cer) from hydrolysis of sphingomyelin (SM). However, its precise roles in regulating specific sphingolipid-mediated biological processes remain ill defined. Interestingly, the aSMase gene gives rise to two distinct enzymes, lysosomal sphingomyelinase (L-SMase) and secretory sphingomyelinase (S-SMase) via alternative trafficking of a shared protein precursor. Previously, our laboratory identified Ser508 as a crucial residue for the constitutive and regulated secretion of S-SMase in response to inflammatory cytokines, and demonstrated a role for S-SMase in formation of select cellular Cer species (Jenkins, R. W., Canals, D., Idkowiak-Baldys, J., Simbari, F., Roddy, P., Perry, D. M., Kitatani, K., Luberto, C., and Hannun, Y. A. (2010) J. Biol. Chem. 285, 35706–35718). In the present study using a chemokine/cytokine screen, we identified the chemokine CCL5 (formerly known as RANTES) as a candidate-specific downstream target for aSMase. Regulation of CCL5 by aSMase was subsequently validated using both loss-of-function and gain-of-function models indicating that aSMase is both necessary and sufficient for CCL5 production. Interestingly, cells deficient in acid ceramidase (aCDase) also exhibited defects in CCL5 induction, whereas cells deficient in sphingosine kinase-1 and -2 exhibited higher levels of CCL5, suggesting that sphingosine and not sphingosine 1-phosphate (S1P) is responsible for the positive signal to CCL5. Consistent with this, co-expression of aSMase and aCDase was sufficient to strongly induce CCL5. Taken together, these data identify a novel role for aSMase (particularly S-SMase) in chemokine elaboration by pro-inflammatory cytokines and highlight a novel and shared function for aSMase and aCDase.
Keywords: Chemokines, Glycoprotein Secretion, Inflammation, Lysosomes, Sphingolipid, Acid Ceramidase, Acid Sphingomyelinase
Introduction
Once considered inert structural components of biological membranes, sphingolipids (SPLs)2 are now recognized as biologically active molecules that have been implicated in the regulation of numerous cellular processes (1). Ceramide (Cer, N-acylsphingosine) occupies a central position in SPL metabolism, serving as a metabolic precursor to more complex SPLs, such as sphingomyelin (SM) and the glycosphingolipids (1, 2). Cer can also be deacylated by ceramidases (CDase) to form sphingosine (Sph) (3), which in turn can be phosphorylated by sphingosine kinases (SphK) to generate sphingosine 1-phosphate (S1P) (4). Although Cer has been shown to play an important role in cellular stress signaling and cell death (5, 6), S1P acts primarily as a pro-survival signal promoting growth, migration, and angiogenesis (7). As such, enzymes of SPL metabolism determine the relative levels of Cer, Sph, and S1P; however, the precise roles and regulation of these enzymes are incompletely understood.
Acid sphingomyelinase (aSMase, SMPD1) is a soluble acid hydrolase that cleaves the phosphodiester bond of SM to form Cer. Mutations in the gene encoding aSMase give rise to Niemann-Pick disease (NPD) types A and B, a lysosomal storage disease with multiorgan involvement (8). In addition to its importance in preserving lysosomal function, aSMase has also been reported to be regulated by various cellular stressors to promote Cer formation (6). Consequently, the aSMase/Cer pathway is considered a major component of the acute cellular stress response (9). Although primarily a lysosomal protein, aSMase can also be secreted extracellularly through alternative trafficking of a shared protein precursor, giving rise to secretory aSMase (S-SMase) (10, 11). Notably, S-SMase secretion is enhanced by inflammatory cytokines (10) and serum levels of S-SMase are increased in mouse models of inflammation (12), as well as a number of human disease states characterized by a pro-inflammatory state (13–15).
Both SPLs and SPL metabolic enzymes have been linked to multiple inflammatory pathways (16, 17), including cytokine-induced eicosanoid signaling (18, 19), up-regulation of adhesion molecules (20), and regulation of cytokine elaboration (21). However, the role for aSMase in inflammatory processes is less well understood; indeed, aSMase has been reported to exhibit both pro- and anti-inflammatory properties. For example, peritoneal macrophages from aSMase−/− mice produce higher levels of TNF-α in response to LPS challenge (22), whereas T-lymphocytes from aSMase−/− mice exhibit defective elaboration of IFN-γ (23), and microglia from aSMase−/− mice are unable to produce IL-1β following purinergic stimulation (24). Moreover, cells and tissues from NPD patients and aSMase−/− mice exhibit basal elevations in inflammatory markers (25–27). Taken together, this suggests that chronic perturbation of the sphingolipid metabolism may underlie dysregulation of the inflammatory response. However, another contributing factor is that aSMase-null cells lack both S-SMase and L-SMase. In particular, the lack of tools available to address the contribution of S-SMase and L-SMase has limited study on the specific roles of these enzymes.
Recently, we described a role for S-SMase in cellular Cer formation using a mutant form of aSMase (S508A) that retained L-SMase function in the absence of S-SMase secretion (28). However, the biological significance of this novel S-SMase/Cer pathway was not explored. Here, we now identify the chemokine CCL5 (formerly known as RANTES) as a novel effector of the S-SMase/Cer pathway using both gain-of-function and loss-of-function analyses. Further analysis demonstrated a requirement for acid ceramidase (aCDase), but not SphK in the up-regulation of CCL5. Based on these findings, we demonstrate a novel role for the S-SMase/Cer pathway in the regulation of the chemokine CCL5 and highlight a novel and shared function of aSMase and aCDase in the inflammatory response.
EXPERIMENTAL PROCEDURES
Materials
MCF7 breast carcinoma cells were obtained from ATCC (Manassas, VA). RPMI culture medium, fetal bovine serum, and Blasticidin S HCl were obtained from Invitrogen. Anti-V5 mouse monoclonal antibody was from Invitrogen. HRP-labeled secondary antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Enhanced chemiluminescence kit was from ThermoScientific (Rockford, IL). Interleukin-1β was purchased from R&D Systems (Minneapolis, MN). Tumor necrosis factor-α was obtained from Peprotech (Rocky Hill, NJ). Myriocin was obtained from Alexis Biochemicals/Enzo Life Sciences (Plymouth Meeting, PA). Fumonisin B1 was obtained from Acros Organics (Geel, Belgium). The sphingosine kinase inhibitor SKI-II was from Calbiochem. Porcine brain sphingomyelin, d-erythro-sphingosine, and d-erythro-sphingosine-1-phosphate were from Avanti Polar Lipids (Alabaster, AL). Desipramine, brefeldin A, and all other chemicals were obtained from Sigma.
Cell Culture
MCF7 cells were grown in RPMI 1640, supplemented with l-glutamine and 10% (v/v) fetal bovine serum. Monoclonal transfectants were isolated as described previously (29). Stable overexpression was verified by Western blotting and confocal microscopy. Cells were maintained in 10% FBS/RPMI plus 7 μg/ml of Blasticidin S-HCl. Cells were maintained at less than 80% confluence under standard incubator conditions (humidified atmosphere, 95% air, 5% CO2, 37 °C). Cells plated for experiments were cultured in the absence of Blasticidin. Control dermal fibroblasts (from the unaffected mother of a child with Lesch-Nyhan, Ctl, GM02226), Niemann-Pick type A dermal fibroblasts (NPD-1, GM13205, and NPD-2, GM16195), and Farber disease dermal fibroblasts (FD-1, GM02315, and FD-2, GM20017) were obtained from the Coriell Cell Repository. Cells were maintained in high-glucose DMEM supplemented with 10% (v/v) fetal bovine serum without antibiotics. Control (L-N), NPD-1, FD-1, and FD-2 fibroblasts were used from passage 10 to 16 and NPD-2 cells from passage 15 to 20. MCF7 cells were treated with IL-1β and TNF-α in serum-free conditions (unless otherwise indicated), whereas dermal fibroblasts and MEFs were cultured and treated in serum-replete media.
Generation and Immortalization of Mouse Embryonic Fibroblasts
MEFs were generated from SphK1+/−, and SphK2+/− littermate matings in a C57BL/6J/129Sv background. Primary MEFs were harvested from E13.5 embryos as described earlier (30, 31). Primary wild-type, SphK1−/−, and SphK2−/− MEFs (passages 3–4) were immortalized by retroviral infection with pBabe-hygro DD p53 expressing dominant-negative p53 (32) and selected with 150 μg/ml of hygromycin for 14 days. MEFs were maintained in high-glucose DMEM supplemented with 10% (v/v) fetal bovine serum without antibiotics. For genotyping, genomic DNA was extracted from MEFs using a DNAEasy kit (Qiagen). For each cell line, 2 μl of DNA was combined with 1 μl each of three primers for either SphK1 or SphK2 and 20 μl of PCR Platinum SuperMix (Invitrogen) for a total reaction volume of 25 μl for PCR. The following primers (10 μm) from Integrated DNA Technologies (Coralville, IA) were used for SphK1: 5′-TGTCACCCATGAACCTGCTGTCCCTG-3′, 5′-AGAAGGCACTGGCTCCTCCAGAGGA-3′, and 5′-TCGTGCTTTACGGTATCGCCGCTCCC-3′, and the following primers were used for SphK2, 5′-GCACCCAGTGTGAATCGAGC-3′, 5′-TCTGGAGACGGGCTGCTTTA-3′, and 5′-CGCTATCAGGACATAGCGTT-3′. PCR was performed on a Biometra Thermocycler T3000 with the following reaction conditions: for SphK1, 94 °C, 0.5 min; 55 °C, 0.5 min; 72 °C, 6.5 min for 30 cycles; and for SphK2, 95 °C, 0.5 min; 55 °C, 0.5 min; 72 °C, 2 min for 40 cycles. PCR products were run on a 2% agarose gel and visualized via UV transillumination. For SphK1, a 310-bp band indicated the WT SphK1 allele, and a band at 390 bp represented a knockout or neo SphK1 allele. For SphK2, a 680-bp band indicated the WT SphK2 allele, and a band at 310 bp represented a knockout or neo SphK2 allele.
Plasmids and Transient Transfection
pEF6.V5/His.aSMaseWT and pEF6.V5/His.aSMaseS508A were previously described (33). pEF6.V5/His.LacZ is the control plasmid for the pEF6/V5-His-TOPO cloning kit (Invitrogen). pEF6.V5/His.aCDaseWT was generated using pEGFP-aCDase as a template (kindly provided by Dr. James Norris, MUSC). Using a 5′-GCTAGAGCGATGCCGGGCCGGAGTTGCGTCG-3′ primer containing nine nucleotides of the native 5′-untranslated region upstream of the initiation codon (AUG) that contains a purine (G) at −3 position, which is crucial for efficient initiation of translation and a 3′-CCAACCTATACAAGGGTCAGGGCA-5′ primer containing the last eight codons of the mRNA minus the stop codon. The amplified human aCDase fragment was then subcloned into the pEF6/V5-His TOPO-TA cloning vector. The orientation of insertion was verified by restriction mapping and sequencing. For transient transfections, cells were plated in 6-well trays (∼200,000 cells/well) and transfected with 0.25–1.0 μg of DNA using Lipofectamine 2000 (Invitrogen) (3 μl/transfection) according to the manufacturer's instructions.
RNA Interference
Small interfering RNA (siRNA) duplexes were obtained from Qiagen (Valencia, CA), and were designed against the following target sequences: aSMase, 5′-AACTCCTTTGGATGGGCCTGG-3′; SphK1, 5′-AAGGGCAAGGCCTTGCAGCTC-3′; SphK2, 5′-AACGCTTTGCCCTCACCCTTA-3′; and aCDase, 5′-AATCAACCTATCCTCCTTCAG-3′. All-Star Negative Control siRNA was from Qiagen. For siRNA transfections, cells were plated in 6-well trays (∼50,000 cells/well) and transfected with 20 nm siRNA using Oligofectamine (Invitrogen) (4 μl/transfection) according to the manufacturer's instructions. After 60 h, siRNA-Oligofectamine was removed, and fresh medium was added before treatments.
Cytokine Array and ELISA
Human Protein Profiler Cytokine Panel A, human RANTES ELISA, and murine RANTES ELISA were obtained from R&D Systems (Minneapolis, MN) and used according to the manufacturer's instructions. Densitometry for the cytokine panel was performed with NIH Image J software using the integrated density analysis function. The integrated density for all analytes was then normalized to positive controls.
Immunoprecipitation, SDS-PAGE, and Immunoblotting
V5-S-SMase and V5-S-aCDase were immunoprecipitated directly from conditioned medium, as a previously described secretion (28). Briefly, conditioned medium was first cleared of floating cells by brief centrifugation at 800 × g for 5 min (4 °C). The supernatant containing S-SMase was then incubated overnight at 4 °C with 1 μg/ml of primary antibody (V5) and 1:20 protein A/G-agarose using a circular rotator. Bound protein was isolated by centrifugation (1 min, 12,000 × g), washed three times with lysis buffer, followed by addition of 2× Laemmli buffer after which samples were boiled for 5 min. Equal volumes were subjected to SDS-PAGE (10% Tris-HCl) using the Bio-Rad Criterion system. Proteins were electrophoretically transferred to nitrocellulose membranes, blocked with PBS, 0.1% Tween 20 (PBS-T) containing 5% nonfat dried milk, and incubated overnight at 4 °C with primary antibody (V5–1:5,000) in PBS-T containing 5% nonfat dried milk. After overnight incubation with primary antibody, the blots were washed with PBS-T and incubated with the appropriate HRP-conjugated secondary antibody (1:5000) in 5% milk/PBS-T. After washing (3 times for 10 min in PBS-T), enhanced chemiluminescence was used to visualize bands.
Real Time Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
RNA was homogenized and extracted from cultured cells or mouse tissue using QIAShredder and RNeasy kits (Qiagen), respectively. cDNA was synthesized from 1.0 μg of RNA using Oligo(dT) primers (Invitrogen) using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. The standard RT-PCR included 12.5 μl of SYBR Green (Bio-Rad), 0.5 μl of 10 μm forward primer, 0.5 μl of 10 μm reverse primer, and 5.0 μl of diluted (1:10) cDNA adjusted to a final volume of 25.0 μl. The RT-PCR was performed using an iCycler (Bio-Rad) as follows: 3 min at 95 °C, followed by cycles (n = 40) consisting of a 10-s melt at 95 °C, a 45-s annealing at 54 °C, and an extension step of 45 s at 68 °C. All reactions were performed in triplicate. Primers were designed using Beacon Primer Design Software and purchased from Integrated DNA Technologies (Coralville, IA). Primers used for real time PCR are as follows: forward human CCL5, 5′-GCTGTCATCCTCATTGCTACTG-3′, reverse human CCL5, 5′-TGGTGTAGAAATACTCCTTGATGTG-3′; forward human acid ceramidase (ASAH1), 5′-TCTTCCATGATCGCAGAACGCC-3′; reverse human acid ceramidase (ASAH1), 5′-ACGGTCAGCTTGTTGAGGAC-3′; forward human β-actin, 5′-ATTGGCAATGAGCGGTTCC-3′, human reverse β-actin, 5′-GGTAGTTTCGTGGATGCCACA-3′; forward mouse CCL5, 5′-ACTCCCTGCTGCTTTGCCTAC-3′; reverse mouse CCL5, 5′-ACTTGCTGGTGTAGAAATACT-3′; forward mouse β-actin, 5′-TAAGGCCAACCGTGAAAAGATG-3′; reverse mouse β-actin, 5′-CTGGATGGCTACGTACATGGCT-3′. Threshold cycle (Ct) values for target genes were normalized to the reference gene using Q-Gene software (28) to determine the mean normalized expression.
In Vitro Acid SMase Activity Assay
The aSMase activity assay was performed as previously described (28). Briefly, porcine brain sphingomyelin (0.2 mm) mixed with [14C]sphingomyelin (radiolabeled in the choline moiety, specific radioactivity = 1.5 × 105 cpm/μl) were presented to enzyme (conditioned media or cellular extracts) in Triton X-100 micelles in a buffer containing 250 mm sodium acetate (pH 5.00) and either 2.0 mm EDTA (L-SMase) or 0.2 mm ZnCl2 (S-SMase) to a final concentration of 0.1 mm SM with either 0.1 mm ZnCl2 (S-SMase) or 1.0 mm EDTA (L-SMase). The enzyme reaction was run for 30 min at 37 °C and terminated with the addition of 1.5 ml of CHCl3:MeOH (2:1, v/v) followed by 0.4 ml of water. Samples were vortexed, centrifuged (5 min at 3,000 × g), and 0.8 ml of the aqueous/methanolic phase was removed for scintillation counting.
Sphingolipidomic Analysis
Adherent cells were washed twice with cold PBS and lysed in buffer containing 1% Triton X-100, 50 mm Tris (pH 7.4), 0.15 m NaCl, 1.0 mm EDTA, supplemented with protease and phosphatase inhibitors. Cellular homogenates (0.1–1.0 mg) were frozen at −80 °C and then submitted for sphingolipidomic analysis by reverse phase high pressure liquid chromatography coupled to electrospray ionization followed by separation by MS. Analysis of sphingoid bases, ceramides, and sphingomyelins was performed on a Thermo Finnigan TSQ 7000 triple quadrupole mass spectrometer, operating in a multiple reaction-monitoring positive ionization mode, as described (34).
Statistical Analysis
Data are represented as mean ± S.E., unless otherwise indicated. Unpaired Student's t test, one-way ANOVA with Dunnett's post test, and two-way ANOVA with Bonferroni post test statistical analyses were performed using Prism/GraphPad software with additional post test analysis using the Prism/GraphPad website.
RESULTS
CCL5 (RANTES) Is Up-regulated by Wild-type aSMase, Lack of Effect of S508A aSMase
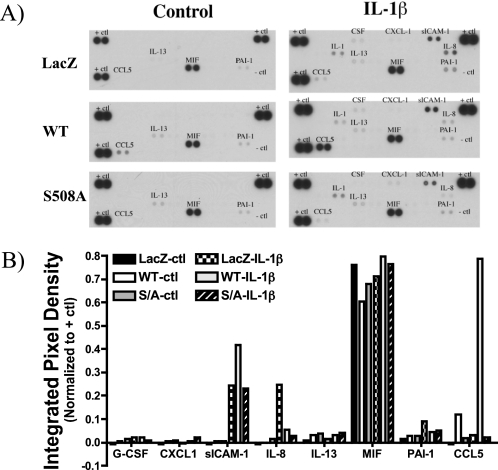
Previously, we demonstrated up-regulation of S-SMase in response to inflammatory cytokines (28). To assess the functional role of S-SMase in the inflammatory response, we utilized our previously characterized MCF7 breast carcinoma cells lines stably expressing vector control (V5-LacZ), wild-type human aSMase (V5-aSMaseWT), or the secretion-incompetent aSMase mutant (V5-aSMaseS508A). Following treatment with vehicle or IL-1β, conditioned medium was collected, and the levels of multiple cytokines and chemokines were analyzed utilizing a proteome profiler array (see “Experimental Procedures”). In resting cells, levels of inflammatory cytokines/chemokines in the medium were minimal. As expected, IL-1β (2.5 ng/ml, 18 h) increased the levels of several analytes including CCL5/RANTES, sICAM-1, IL-8/CXCL8, and plasminogen activator inhibitor-1 (PAI-1). Of these, only CCL5 was markedly elevated in aSMaseWT cells relative to LacZ cells following IL-1β treatment (Fig. 1, A and B). Moreover, CCL5 levels were significantly increased in untreated aSMaseWT cells suggesting that aSMase overexpression was sufficient to promote CCL5 up-regulation. These results suggest a novel effect for aSMase in specifically regulating the elaboration of CCL5.
FIGURE 1.
Identification of candidate effectors of the S-SMase/ceramide pathway. MCF7 breast carcinoma cells lines stably expressing vector control (V5-LacZ), wild-type aSMase (V5-aSMaseWT), or the secretion-incompetent aSMase mutant (V5-aSMaseS508A) were treated with vehicle (PBS) or IL-1β (2.5 ng/ml) for 18 h. A, cell-free conditioned medium was collected and subjected to cytokine/chemokine analysis using a Cytokine Panel Array (see “Experimental Procedures”) to identify any inflammatory mediators specifically up- or down-regulated in WT, but not S508A MCF7. B, densitometric analysis of analyte data were performed using NIH Image J as described under “Experimental Procedures.” Integrated pixel densities of the various analytes were normalized to internal positive controls (+ ctl) (n = 1).
Interestingly, the S508A aSMase-expressing cells failed to demonstrate the up-regulation of CCL5 in unstimulated cells, and did not enhance the IL-1β response (Fig. 1). Because the S508A mutant is not secreted but retains lysosomal targeting and activity, these results suggest that the effects on CCL5 are specific to S-SMase.
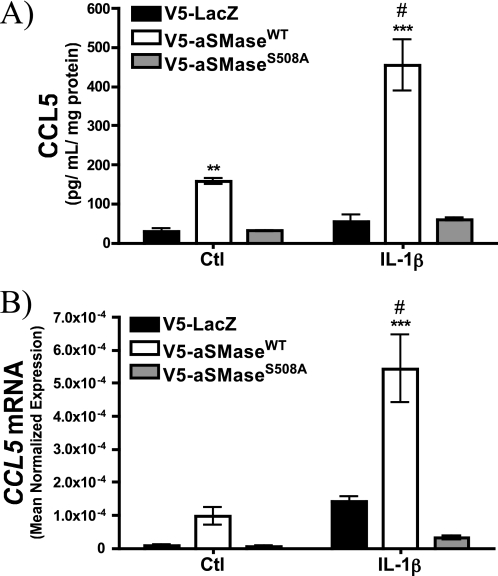
To validate these results, an ELISA specific for CCL5 was performed. Importantly, this confirmed the up-regulation of CCL5 by wild-type, but not S508A acid SMase in MCF7 cells (Fig. 2A). As increased secretion of CCL5 in MCF7 has been reported to proceed in the absence of increased transcription (35), the effect of aSMase overexpression on CCL5 mRNA was analyzed by qRT-PCR. Interestingly, CCL5 mRNA was elevated in untreated aSMaseWT cells compared with both LacZ and aSMaseS508A cells. Moreover, levels were further enhanced above control cells upon treatment with IL-1β (Fig. 2B). Surprisingly, CCL5 mRNA levels in S508A cells were diminished compared with control cells, suggesting that the S508A aSMase mutant has a dominant-negative function. Taken together, these results indicate that overexpression of WT but not S508A aSMase is sufficient to increase CCL5 mRNA and protein levels. This identifies a novel target not only of aSMase, but also specifically of S-SMase. To our knowledge, this is the first putative downstream effector of the S-SMase pathway.
FIGURE 2.
Validation of CCL5 as an effector of the S-SMase/ceramide pathway. MCF7 breast carcinoma cells lines stably expressing V5-LacZ, V5-aSMaseWT, and V5-aSMaseS508A were treated with vehicle (PBS) or IL-1β (2.5 ng/ml) for 18 h. A, cell-free conditioned medium was collected, and CCL5 protein levels were determined by ELISA. B, CCL5 mRNA levels were determined by RT-PCR in response to 18 h PBS (Ctl) or IL-1β (2.5 ng/ml) and were normalized to β-actin (see “Experimental Procedures”) (n ≥ 3; two-way ANOVA with Bonferroni post-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus LacZ; #, p < 0.05 versus untreated).
CCL5 Elaboration Is a Late Event in Response to TNF-α and IL-1β
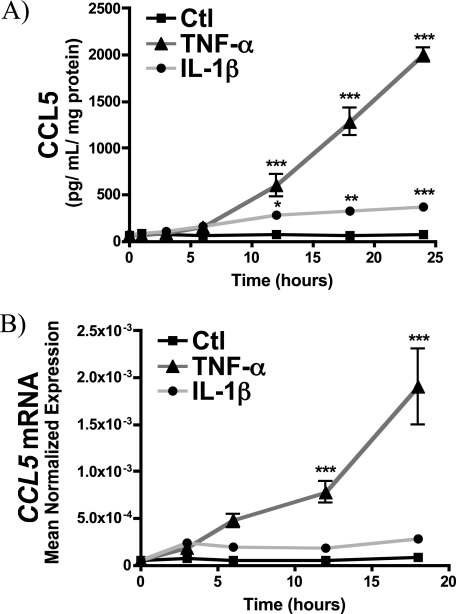
As up-regulation of S-SMase is a protracted event following inflammatory cytokine challenge (28, 36), effectors of S-SMase would be expected to exhibit a similar delay. Accordingly, to establish a temporal relationship between S-SMase secretion and CCL5 elaboration, a time course of CCL5 release in MCF7 V5-aSMaseWT MCF7 was determined following inflammatory cytokine challenge (Fig. 3). Both TNF-α and IL-1β induced a time-dependent increase in CCL5, which became statistically significant after 12 h. However, the effect of TNF-α on CCL5 was much stronger than IL-1β, which became more apparent at 18 and 24 h. Importantly, the time course and extent of CCL5 elaboration following TNF-α and IL-1β treatment closely tracked that of S-SMase secretion and Cer generation (28). These results demonstrate that the kinetics and extent of CCL5 induction in response to TNF-α and IL-1β closely track the up-regulation of S-SMase.
FIGURE 3.
Time course of CCL5 elaboration in response to TNF-α and IL-1β. A, MCF7 breast carcinoma cells lines stably expressing V5-aSMaseWT were treated with vehicle (PBS), IL-1β (2.5 ng/ml), or TNF-α (50 ng/ml) for the indicated time points. Cell-free conditioned medium were collected, and CCL5 protein levels were determined by ELISA (n ≥ 3; two-way ANOVA with Bonferroni post-test; *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus LacZ; #, p < .05 versus untreated). B, CCL5 mRNA levels were determined by RT-PCR in response to PBS (Ctl), IL-1β (2.5 ng/ml), and TNF-α (50 ng/ml) for the indicated times and normalized to β-actin (see “Experimental Procedures”) (n ≥ 3; two-way ANOVA with Bonferroni post-test; ***, p < 0.001 versus Ctl).
Acid Sphingomyelinase Is Required for CCL5 Up-regulation
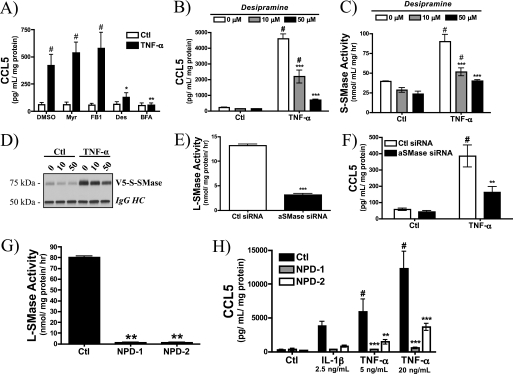
To complement the findings with aSMase overexpression, the requirement for endogenous aSMase in CCL5 regulation was evaluated. Initially, multiple inhibitors of SPL enzymes/pathways were employed to determine whether perturbation of the SPL metabolism in general could alter CCL5 elaboration (Fig. 4A). As can be seen, inhibition of de novo (myriocin (37)) and salvage pathways of ceramide formation (fumonosin B1 (38)) had no effect on TNF-α-induced CCL5 release. Importantly, the indirect inhibitor of aSMase desipramine effectively blocked CCL5 up-regulation, an effect comparable with brefeldin A, which inhibits the Golgi secretory pathway (39). Desipramine is an established in vivo inhibitor of L-SMase (40), but its impact on constitutive or regulated S-SMase secretion has not been studied. To further explore this, stable aSMaseWT MCF7 cells were utilized. As in parental MCF7 cells, desipramine strongly inhibited the elaboration of CCL5 in a dose-dependent manner in response to TNF-α in V5-aSMaseWT MCF7 (Fig. 4B). Interestingly, desipramine also inhibited TNF-α-mediated up-regulation of S-SMase (Fig. 4, C and D). This is the first report suggesting that desipramine, considered an inhibitor of L-SMase, can also influence the secretion and action of S-SMase. Nevertheless, these data support the role of the S-SMase pathway in regulation of CCL5/RANTES.
FIGURE 4.
Acid sphingomyelinase is required for CCL5 elaboration. A, parental MCF7 cells were pre-treated with vehicle (DMSO), myriocin (Myr, 100 nm), fumonisin B1 (FB1, 50 μm), desipramine (Des, 50 μm), or brefeldin A (BFA, 5 μg/ml) for 30 min before treatment with PBS (Ctl) or TNF-α (20 ng/ml) for 18 h. CCL5 levels were determined by ELISA (n = 3; two-way ANOVA with Bonferroni post-test, #, p < 0.05 versus untreated cells; *, p < 0.05 versus DMSO; **, p < 0.01 versus DMSO). B, CCL5 levels were determined in V5-aSMaseWT MCF7 pre-treated with 0, 10, and 50 μm desipramine for 30 min before treatment with PBS (Ctl) or TNF-α (20 ng/ml) for 18 h. The effect of desipramine on S-SMase activity (C) and V5-S-SMase (D) protein levels in response to 18 h TNF-α (20 ng/ml) were determined. E, L-SMase activity in MCF7 cells treated with 20 nm control siRNA or aSMase siRNA for 72 h (n = 4; unpaired t test; ***, p < 0.001). F, CCL5 levels were determined in MCF7 treated with 20 nm control siRNA or aSMase siRNA for 60 h prior to 12 h treatment with PBS (Ctl) or TNF-α (20 ng/ml) in serum-replete medium (n = 4; two-way ANOVA with Bonferroni post-test, #, p < 0.05 versus untreated cells; **, p < 0.01 versus Ctl siRNA). G, L-SMase activity in dermal fibroblasts from patients with Niemann-Pick disease type-A (NPD-1 and NPD-2) compared with control (Ctl) fibroblasts (n = 4; one-way ANOVA, Dunnett's post-test; **, p < 0.01). H, CCL5 levels were determined in Ctl, NPD-1, and NPD-2 dermal fibroblasts in response to 18 h IL-1β (2.5 ng/ml) and TNF-α (5 and 20 ng/ml) (n = 4; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; **, p < 0.01; ***, p < 0.001 versus Ctl fibroblasts).
To consolidate the inhibitor data, a siRNA approach was utilized. As expected, aSMase siRNA significantly decreased endogenous L-SMase activity (Fig. 4E). Importantly, aSMase siRNA also decreased TNF-stimulated CCL5 release by 60–70% (Fig. 4, E and F).
Finally, to prove conclusively that aSMase was required for CCL5 up-regulation, dermal fibroblasts were obtained from patients with Niemann-Pick disease type-A (NPD-1 and NPD-2). Consequently, these fibroblasts exhibit complete loss of endogenous aSMase activity (Fig. 4G). As can be seen, both NPD-1 and NPD-2 fibroblasts exhibited severe defects in CCL5 induction compared with control, aSMase-replete dermal fibroblasts (Fig. 4H). Taken together, these results confirm that functional endogenous aSMase is required for CCL5 induction by TNF-α and IL-1β.
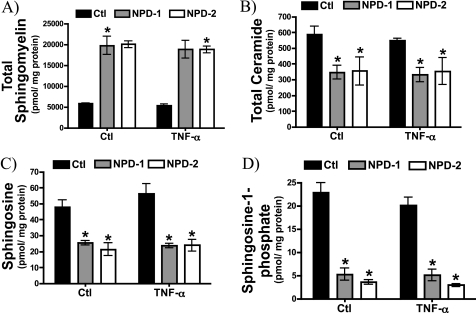
Acid SMase-deficient Niemann-Pick Dermal Fibroblasts Exhibit Defective Ceramide and Sphingoid Base Formation
The immediate metabolic product of aSMase-driven hydrolysis of SM is Cer (41); however, Cer can be metabolized to a number of other lipids. CDases deacylate Cer to form Sph (3), itself a potent signaling molecule, and Sph is further phosphorylated by SphK to form S1P (3, 42). Accordingly, to determine whether impaired CCL5 up-regulation in aSMase-deficient cells was associated with impaired formation of Cer or downstream Cer metabolites, NPD-1, and NPD-2 fibroblasts were treated with vehicle or TNF-α and submitted for sphingolipidomic analysis. As expected, both NPD-1 and NPD-2 cells displayed a marked elevation in total SM mass (Fig. 5A) and a significant decrease in total Cer levels consistent with impaired aSMase activity (Fig. 5B). Interestingly, levels of Sph (Fig. 5C) and S1P (Fig. 5D) were also dramatically reduced in both NPD cell lines. Of note, TNF-α did not produce statistically significant elevations in the mass of any of the sphingolipid analytes at the time point evaluated. This may indicate that localized elevations in specific bioactive lipids are more biologically significant than changes in mass measured in cell homogenates. Nonetheless, these results indicate that in addition to influencing SM and Cer levels, aSMase deficiency affects Cer metabolites (Sph and S1P). This raises the possibility that defective formation of any (or all) of these lipid mediators may underlie the defective CCL5 response in NPD cells.
FIGURE 5.
Defective ceramide and sphingoid base formation in Niemann-Pick dermal fibroblasts. Sphingolipids were measured in control, NPD-1, and NPD-2 fibroblasts following treatment with vehicle (PBS) or TNF-α (20 ng/ml) for 18 h. A, total sphingomyelin; B, total ceramide; C, sphingosine; and D, sphingosine 1-phosphate (n = 4; two-way ANOVA with Bonferroni post-test, *, p < 0.05 versus Ctl cells).
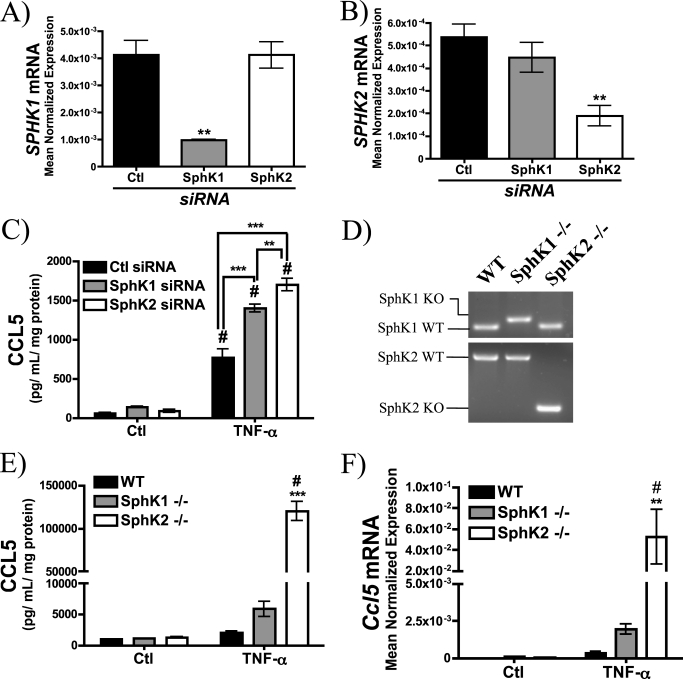
Absence of Sphingosine Kinase Promotes CCL5 Up-regulation
Previous studies have implicated the SphK/S1P pathway in the regulation of inflammatory cytokine and chemokine elaboration (43–46) including CCL5 (47–50). Thus, it became important to determine whether Sph or S1P could be responsible for aSMase-mediated CCL5 induction. Consequently, the effect of RNAi-mediated knockdown of SphK1 and SphK2 on TNF-α-mediated CCL5 release in MCF7 was determined (Fig. 6, A–C). As can be seen, SphK1 and SphK2 silencing resulted in a 1.5–2-fold elevation of CCL5 compared with control siRNA although this was to a greater extent with SphK2 siRNA. To confirm these results, MEFs from WT, SphK1−/−, and SphK2−/− mice were obtained (Fig. 6D) and CCL5 elaboration was evaluated following treatment with TNF-α. Although SphK1−/− MEFs exhibited a ∼3-fold elevation over WT MEFs, CCL5 levels in SphK2−/− MEFs were 50–60-fold higher relative to WT MEFs (Fig. 6E). Furthermore, qRT-PCR analysis demonstrated commensurate elevations in Ccl5 mRNA in SphK1−/− and SphK2−/− MEFs (Fig. 6F). This exaggerated induction of CCL5 in SphK1- and SphK2-null cells suggests that Cer or Sph, but not S1P, is required for up-regulation of CCL5 in response to TNF-α.
FIGURE 6.
Enhanced up-regulation of CCL5 in cells lacking sphingosine kinase. A, SPHK1, and B, SPHK2 mRNA levels were determined by qRT-PCR after 72 h (20 nm) siRNA treatment (n = 3; one-way ANOVA with Dunnett's post-test; **, p < 0.01). C, CCL5 levels were determined in MCF7 treated with 20 nm control siRNA, SphK1 siRNA, or SphK2 siRNA for 60 h prior to 12 h treatment with PBS (Ctl) or TNF-α (20 ng/ml) (n = 4; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; **, p < 0.01 versus Ctl siRNA). D, genotyping of WT, SphK1−/−, and SphK2−/− MEFs. E, CCL5 levels were determined in WT, SphK1−/−, and SphK2−/− MEFs in response to 18 h TNF-α (20 ng/ml) (n ≥ 3; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; ***, p < 0.001 versus WT MEFs). F, Ccl5 mRNA levels in WT, SphK1−/−, and SphK2−/− MEFs in response to 18 h TNF-α (20 ng/ml) were determined by RT-PCR and normalized to β-actin (n ≥ 3; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; **, p < 0.01 versus WT MEFs).
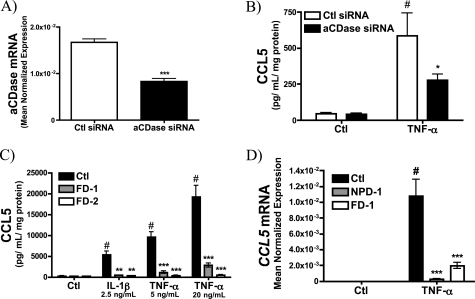
Acid Ceramidase Is Required for CCL5 Up-regulation
aCDase functions to degrade Cer within endolysosomes to generate Sph (51, 52). However, aCDase is also secreted, can interact with S-SMase (53), and thus may contribute to metabolism of the S-SMase-derived Cer. Accordingly, to determine whether aSMase-derived Cer or Sph was responsible for CCL5 up-regulation, the effect of aCDase siRNA on CCL5 production was evaluated. As can be seen, aCDase siRNA effectively decreased aCDase mRNA levels (Fig. 7A). More importantly, aCDase siRNA significantly inhibited TNF-α induction of CCL5 suggesting that aCDase plays a role in CCL5 regulation (Fig. 7B). To confirm this, dermal fibroblasts from patients with Farber disease (FD), which exhibit complete loss of aCDase activity (54), were obtained. As with the siRNA data, elaboration of CCL5 was significantly impaired in two different Farber disease cell lines (FD-1 and FD-2) compared with control cells. Moreover, this was true in response to both IL-1β and TNF-α (Fig. 7C). Finally, to determine whether aCDase was required for transcriptional up-regulation of CCL5, CCL5 mRNA was evaluated in both NPD-1 and FD-1 cells in response to TNF-α (Fig. 7D). Importantly, both NPD-1 and FD-1 cells failed to induce CCL5 mRNA in response to TNF-α, suggesting that both aSMase and aCDase are required for transcriptional induction of CCL5 presumably by promoting formation of Sph.
FIGURE 7.
Acid ceramidase is required for CCL5 elaboration. A, acid CDase mRNA levels in MCF7 cells treated with 20 nm control siRNA or aCDase siRNA for 72 h (n = 4; unpaired t test; ***, p < 0.001). B, CCL5 levels were determined in MCF7 treated with 20 nm control siRNA or aCDase siRNA for 60 h prior to the 12-h treatment with PBS (Ctl) or TNF-α (20 ng/ml) in serum-replete medium (n = 4; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; *, p < 0.05 versus Ctl siRNA). C, CCL5 levels were determined in control (Ctl) and Farber disease (FD-1 and FD-2) dermal fibroblasts in response to 18 h IL-1β (2.5 ng/ml) and TNF-α (5, 20 ng/ml) (n = 4; two-way ANOVA with Bonferroni post-test, #, p < 0.05 versus untreated cells; **, p < 0.01; ***, p < 0.001 versus Ctl fibroblasts). D, CCL5 mRNA levels in Ctl, FD-1, and FD-2 dermal fibroblasts were determined by RT-PCR in response to 18-h PBS (Ctl) or TNF-α (20 ng/ml) treatments and normalized to β-actin (n ≥ 3; two-way ANOVA with Bonferroni post-test; #, p < 0.05 versus untreated cells; ***, p < 0.001 versus Ctl fibroblasts).
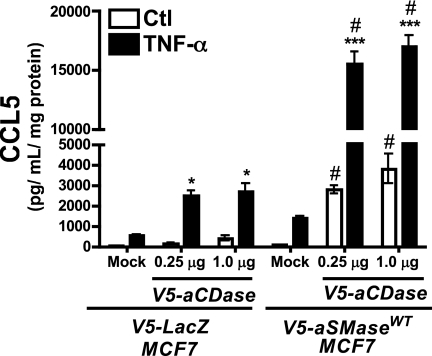
Co-expression of aSMase and aCDase Is Sufficient to Induce CCL5
To determine whether the concerted action of aSMase and aCDase was sufficient to promote CCL5 up-regulation, V5-aCDase was transiently transfected into vector control (LacZ) and aSMaseWT MCF7 stable cell lines. Notably, aCDase transfection into control cells resulted in enhanced elaboration of CCL5 in response to TNF-α (Fig. 8) further supporting a role for this enzyme in CCL5 regulation. More strikingly, transfection of aCDase into aSMaseWT cells resulted in a dramatic 13-fold up-regulation of CCL5 over mock-transfected TNF-α-treated control cells (LacZ-mock + TNF-α, 525.5 ± 67.5 pg/ml/mg of protein; aSMase + aCDase + TNF-α, 6988.1 ± 951.4 pg/ml/mg of protein). Moreover, co-expression of aSMase and aCDase was sufficient to increase levels of CCL5 nearly 40-fold (mock-transfected aSMase-Ctl, 93.7 ± 10.4 pg/ml/mg of protein; aSMase + aCDase-Ctl, 3804.1 ± 728.7 pg/ml/mg of protein) even in the absence of an inflammatory stimulus. Taken together, these results indicate that both aSMase and aCDase contribute to CCL5 production and that co-expression of aSMase and aCDase is sufficient to promote CCL5 up-regulation.
FIGURE 8.
Co-expression of acid sphingomyelinase and acid ceramidase is sufficient to increase CCL5. CCL5 levels were determined in V5-LacZ and V5-aSMaseWT MCF7 cells transfected with 0 (mock), 0.25, or 1.0 μg of V5-aCDase plasmid DNA before treatment with vehicle (PBS) or TNF-α (20 ng/ml) for 18 h (n ≥ 3; two-way ANOVA with Bonferroni post-test, *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus Ctl; #, p < 0.05 versus mock-transfected cells).
DISCUSSION
In this study we set out to identify a downstream effector of the S-SMase/Cer pathway using our S508A aSMase mutant as a novel tool to distinguish the roles of S-SMase and L-SMase. Using these cells, we identified the β-chemokine CCL5 (previously known as RANTES) as a novel effector of the S-SMase/Cer pathway. However, upon further investigation we identified the Cer metabolite Sph as the candidate lipid for the regulation of CCL5, and not Cer itself. Generation of Sph from S-SMase-derived Cer required the action of aCDase. Importantly, the conversion of Sph to S1P was not required for CCL5 up-regulation, as CCL5 levels were significantly elevated in cells lacking SphK, in particular SphK2. Taken together, these results demonstrate a novel role for both aSMase and aCDase in chemokine elaboration, and identify the first effector of the S-SMase/Cer pathway.
CCL5: A Novel Target of aSMase/S-SMase Signaling
Although aSMase has been implicated in various physiological and pathophysiological processes (41, 55), the precise contribution of S-SMase and L-SMase has remained unclear because of the lack of molecular tools to dissect out their individual roles (56). Previously, we have characterized an S508A aSMase mutant as secretion incompetent although still maintaining L-SMase activity (28). Utilizing the S508A aSMase mutant as a novel tool to evaluate the role of S-SMase, the chemokine CCL5 was identified as a candidate effector of the S-SMase/Cer pathway. Subsequently, pharmacological inhibitors of aSMase (i.e. desipramine), RNAi, and cells lacking aSMase activity (i.e. NPD fibroblasts) were used to demonstrate that endogenous aSMase is required for induction of CCL5 in response to pro-inflammatory cytokines.
Notably, up-regulation of S-SMase and CCL5 are both protracted events in the inflammatory cascade (36, 57). Importantly, the time course of CCL5 up-regulation tracked that of S-SMase up-regulation (28), indicating a close temporal association between activation of the S-SMase/Cer pathway and CCL5 induction. Taken together, these results demonstrate a novel role for aSMase in chemokine elaboration and identify CCL5 as a unique target of the S-SMase/Cer pathway. To our knowledge, this is the first proposed target of the S-SMase/Cer pathway.
Regulation of CCL5 by Sphingolipids
Several studies have investigated the role of SPLs in the regulation of inflammatory cytokines and chemokines (7), including CCL5 (47–49), and the SphK/S1P pathway has been most closely associated with inflammatory responses (16). However, the role of other SPL pathways in the elaboration of selective cytokines is becoming more apparent (21, 58). The results of the present study demonstrate that both aSMase and aCDase are required for induction of CCL5 in the pro-inflammatory response (Fig. 9). Furthermore, overexpression of aSMase and aCDase is sufficient to drive CCL5 up-regulation, presumably through the generation of Sph. Impaired conversion of Sph to S1P, primarily in cells lacking SphK2−/− resulted in overproduction of CCL5, providing additional support that Sph provides the primary signal to induce CCL5.
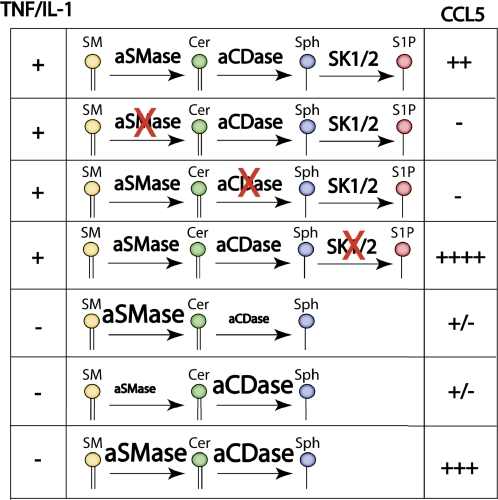
FIGURE 9.
Schematic representation of the regulation of CCL5 by sphingolipids. In cells with aSMase, aCDase, and SphK1/2, up-regulation of CCL5 in response to TNF-α or IL-1β proceeds normally. In cells, lacking either aSMase or aCDase, up-regulation of CCL5 is impaired and Sph levels are diminished. On the other hand, cells lacking SphK1/2 exhibited significantly higher levels of CCL5 following inflammatory challenge and exhibit elevated Sph levels. In the absence of an inflammatory stimulus, overexpression of either aSMase or aCDase can modestly increase CCL5 production, but co-expression of aSMase and aCDase is sufficient for a dramatic up-regulation of CCL5 albeit without significant changes in Sph mass.
Efforts to demonstrate a relationship between cellular Sph levels and CCL5 induction were met with limited success (supplemental Figs. S1–S4). This is most likely due to the presence of the majority of cellular Sph in the lysosomes (greater than 90%) (18), which then acts as a “reservoir” for Sph, but also serves to mask the detection of significant changes in other compartments. Further complicating this analysis, exogenous Sph (and S1P) were unable to recapitulate the induction of CCL5 seen with other manipulations (supplemental Figs. S5 and S6). Although SPL signaling processes at the plasma membrane may be more sensitive to exogenous SPLs (59), at this time tools to evaluate specific lipid alterations in other regions of the cell are lacking, thus limiting further analysis. Because of the limited utility of these simple pharmacologic methods, we relied heavily on molecular approaches to begin to define the specific lipid(s) downstream of the S-SMase action that drove CCL5 up-regulation. Based on this molecular triangulation, Sph represents the most likely lipid mediator, although given our limited capacity to interrogate the compartment-specific SPL metabolism we cannot confirm a role for Sph at this time.
The dramatic hyper-induction of CCL5 in SphK2−/− MEFs (>50-fold) suggests that aSMase-derived Sph, and not S1P, drives CCL5 up-regulation, but also provides a novel distinction between the functional roles of SphK1 and SphK2. Although the precise mechanism whereby SPLs modulate CCL5 mRNA levels was not investigated, SphK2 was recently shown to modulate histone acetylation thereby influencing gene transcription (60). Interestingly, chromatin remodeling is an essential step in CCL5 induction in T lymphocytes (61), and inhibition of HDACs has been shown to prevent up-regulation of CCL5 (62). Future studies will be required to determine the precise mechanism whereby specific SPLs modulate CCL5 mRNA levels.
Importantly, regulation of CCL5 varies among different cell types (63). Although first described in T-lymphocytes (64), it is now well established that multiple cells produce CCL5 including platelets (65), vascular smooth muscle cells (66), epithelial cells (67), and fibroblasts (68). Although some cell types store CCL5 in granules ready for release (e.g. CD8+ T lymphocytes (69) and platelets (65)), CCL5 induction in most cells requires transcriptional up-regulation of CCL5 mRNA (70). Several constitutive and regulated transcription factors participate in CCL5 transcriptional activation (71), including NF-κB (72) and STAT-3 (66). For example, up-regulation of CCL5 in primary T-lymphocytes occurs over 3 to 5 days requiring a unique complement of transcription factors that are subject to multiple levels of regulation (61, 63, 73, 74). CCL5 is then stored in granules and can be released upon T-cell receptor activation. Notably, release of CCL5 from activated T-lymphocyte granules does not appear to require aSMase (23), suggesting that up-regulation of CCL5 mRNA, rather than secretion itself, is regulated by SPLs. Future studies will be required to elucidate the mechanisms by which SPLs regulate CCL5 mRNA levels.
Compartmentalization of Sphingolipid Signaling
Several lines of evidence indicate an essential role for SPLs, in particular Sph, in the up-regulation of CCL5 following TNF-α or IL-1β treatments. However, these results also suggest that regulation of CCL5 requires compartment-specific generation of bioactive lipids. First, impaired up-regulation of CCL5 in MCF7 expressing the secretion-incompetent S508A mutant suggests the involvement of S-SMase (and not L-SMase). Second, modulation of the SPL metabolism in general was insufficient to influence CCL5 elaboration, as inhibition of de novo SPL biosynthesis or the ceramide salvage pathway with myrocin and fumonisin B1, respectively, did not alter TNF-α-induced CCL5 elaboration. Third, up-regulation of CCL5 was sensitive to desipramine, a cationic amphiphilic drug that accumulates in acidic organelles and inhibits both aSMase and aCDase function (40, 75), and prevented S-SMase secretion in response to TNF-α. Fourth, overproduction of CCL5 was evident in SphK2−/− MEFs, to a far greater degree than SphK1−/− MEFs. SphK2 localizes to the nucleus and ER, whereas SphK1 is primarily cytosolic (76–78), suggesting that the site of Sph (and S1P) formation determines its biological function. Fifth, addition of exogenous Sph was insufficient to induce CCL5, and exogenous S1P was unable to restore elevated CCL5 to normal levels in SphK2−/− MEFs (supplemental Figs. S5 and S6). Taken together, these results suggest the involvement of a specific subcellular compartment, with a defined complement of SPL enzymes (i.e. aSMase, aCDase, and SphK2) functioning to regulate the levels of CCL5. Identifying the specific cellular/subcellular site of Sph generation will be crucial for future mechanistic studies, and is an ongoing project in our laboratory.
Biological Role of the aSMase/aCDase/CCL5 Axis
Although the results of this study indicate that both aSMase and aCDase are necessary and sufficient to induce CCL5, the precise role of aSMase and aCDase in CCL5-mediated biological processes is not known. CCL5 (RANTES) is a C-C type (β-) chemokine that promotes the chemotaxis of a variety of cells, including monocytes, T-lymphocytes, dendritic cells, basophils, eosinophils, and mast cells to sites of inflammation (79, 80). CCL5 is important for physiologic inflammatory responses, as failure to up-regulate CCL5 leads to impaired viral clearance (81) and defective type IV hypersensitivity responses because of impaired T-cell function (82).
In patients lacking aSMase or aCDase (i.e. patients with NPD or FD, respectively), dysregulation of inflammatory responses is often observed, but alterations in CCL5 levels or CCL5-mediated responses have not been determined. However, altered levels of various chemokines have been detected both in patients with NPD and in aSMase−/− mice, including MIP-1α, CCL2, and CCL18 (25–27). In FD (also known as Farber lipogranulomatosis), chronic inflammation resulting from leukocyte dysregulation leads to the formation of pathologic granulomas (83). Finally, excessive elaboration of CCL5 has also been implicated in several disease states, including allograft transplant rejection (84), asthma (85), rheumatoid arthritis (86), atherosclerosis (87, 88), endometriosis (89), and several cancers (90–93). Thus, whereas the roles of aSMase and aCDase in CCL5-mediated biological processes have yet to be defined, given the multiple roles attributed to the action of CCL5, interventions aimed at inhibiting aSMase and/or aCDase may prove useful in the modulation of CCL5 and CCL5-mediated disease processes.
Conclusion
In this study we identify aSMase (S-SMase) and aCDase as novel regulators of the chemokine CCL5, previously known as RANTES. Furthermore, SphK2 appears to be a negative regulator of CCL5, as loss of SphK2 results in overproduction of CCL5. As CCL5 has been associated with several diseases, including atherosclerosis, asthma, and cancer, modulation of SPLs may represent a novel approach to modify CCL5 elaboration to better manage and treat a variety of different human diseases.
Acknowledgments
Special thanks to the Medical University of South Carolina Analytical Lipidomics Core (Dr. Jacek Bielawski, Barbara Rembiesa, Justin Snider, and Jason Pierce) and the Medical University of South Carolina Synthetic Lipidomics Core (Dr. Alicja Bielawska and Nalini Mayroo). We also express thanks to Dr. Leah Siskind (Medical University of South Carolina) for careful reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P01 CA097132 (to Y. A. H.) from the NCI, MSTP Training Grant GM08716 (to R. W. J.), Grant GM062887 from the NIGMS (to L. M. O.), Grant CA097132 from the NCI (to L. M. O.), American Heart Association Pre-doctoral Fellowship AHA 081509E (to R. W. J.), a Medical University of South Carolina Hollings Cancer Center Abney Foundation Scholarship (to R. W. J.), Ministerio de Educacion y Ciencia (Spain) Predoctoral Fellowship AP2006-02190 (to F. S.), and the Japan Society for the Promotion of Science Grant-in-aid for Young Scientists (Start-up) 21890144 (to K. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6.
- SPL
- sphingolipid
- aSMase
- acid sphingomyelinase
- S-SMase
- secretory sphingomyelinase
- L-SMase
- lysosomal sphingomyelinase
- aCDase
- acid ceramidase
- SphK1
- sphingosine kinase 1
- SphK2
- sphingosine kinase 2
- SM
- sphingomyelin
- Cer
- ceramide
- Sph
- sphingosine
- S1P
- sphingosine 1-phosphate
- NPD
- Niemann-Pick disease
- FD
- Farber disease
- MEF
- mouse embryonic fibroblast
- ANOVA
- analysis of variance
- qRT
- quantitative reverse transcription.
REFERENCES
- 1. Hannun Y. A., Obeid L. M. (2008) Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 2. Bartke N., Hannun Y. A. (2009) J. Lipid Res. 50, (suppl.) S91–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mao C., Obeid L. M. (2008) Biochim. Biophys. Acta 1781, 424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hait N. C., Oskeritzian C. A., Paugh S. W., Milstien S., Spiegel S. (2006) Biochim. Biophys. Acta 1758, 2016–2026 [DOI] [PubMed] [Google Scholar]
- 5. Grassmé H., Riethmüller J., Gulbins E. (2007) Prog. Lipid Res. 46, 161–170 [DOI] [PubMed] [Google Scholar]
- 6. Gulbins E., Li P. L. (2006) Am. J. Physiol. Regul. Integr. Comp. Physiol. 290, R11–26 [DOI] [PubMed] [Google Scholar]
- 7. Snider A. J., Alexa Orr Gandy K., Obeid L. M. (2010) Biochimie 92, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schuchman E. H. (2007) J. Inherit. Metab. Dis. 30, 654–663 [DOI] [PubMed] [Google Scholar]
- 9. Zeidan Y. H., Hannun Y. A. (2010) Curr. Mol. Med. 10, 454–466 [DOI] [PubMed] [Google Scholar]
- 10. Schissel S. L., Keesler G. A., Schuchman E. H., Williams K. J., Tabas I. (1998) J. Biol. Chem. 273, 18250–18259 [DOI] [PubMed] [Google Scholar]
- 11. Schissel S. L., Schuchman E. H., Williams K. J., Tabas I. (1996) J. Biol. Chem. 271, 18431–18436 [DOI] [PubMed] [Google Scholar]
- 12. Wong M. L., Xie B., Beatini N., Phu P., Marathe S., Johns A., Gold P. W., Hirsch E., Williams K. J., Licinio J., Tabas I. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8681–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Doehner W., Bunck A. C., Rauchhaus M., von Haehling S., Brunkhorst F. M., Cicoira M., Tschope C., Ponikowski P., Claus R. A., Anker S. D. (2007) Eur. Heart J. 28, 821–828 [DOI] [PubMed] [Google Scholar]
- 14. Claus R. A., Bunck A. C., Bockmeyer C. L., Brunkhorst F. M., Lösche W., Kinscherf R., Deigner H. P. (2005) FASEB J. 19, 1719–1721 [DOI] [PubMed] [Google Scholar]
- 15. Takahashi T., Abe T., Sato T., Miura K., Takahashi I., Yano M., Watanabe A., Imashuku S., Takada G. (2002) J. Pediatr. Hematol. Oncol. 24, 401–404 [DOI] [PubMed] [Google Scholar]
- 16. Nixon G. F. (2009) Br. J. Pharmacol. 158, 982–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pettus B. J., Chalfant C. E., Hannun Y. A. (2004) Curr. Mol. Med. 4, 405–418 [DOI] [PubMed] [Google Scholar]
- 18. Zeidan Y. H., Pettus B. J., Elojeimy S., Taha T., Obeid L. M., Kawamori T., Norris J. S., Hannun Y. A. (2006) J. Biol. Chem. 281, 24695–24703 [DOI] [PubMed] [Google Scholar]
- 19. Pettus B. J., Kitatani K., Chalfant C. E., Taha T. A., Kawamori T., Bielawski J., Obeid L. M., Hannun Y. A. (2005) Mol. Pharmacol. 68, 330–335 [DOI] [PubMed] [Google Scholar]
- 20. Clarke C. J., Truong T. G., Hannun Y. A. (2007) J. Biol. Chem. 282, 1384–1396 [DOI] [PubMed] [Google Scholar]
- 21. Kitatani K., Sheldon K., Anelli V., Jenkins R. W., Sun Y., Grabowski G. A., Obeid L. M., Hannun Y. A. (2009) J. Biol. Chem. 284, 12979–12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rozenova K. A., Deevska G. M., Karakashian A. A., Nikolova-Karakashian M. N. (2010) J. Biol. Chem. 285, 21103–21113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herz J., Pardo J., Kashkar H., Schramm M., Kuzmenkina E., Bos E., Wiegmann K., Wallich R., Peters P. J., Herzig S., Schmelzer E., Krönke M., Simon M. M., Utermöhlen O. (2009) Nat. Immunol. 10, 761–768 [DOI] [PubMed] [Google Scholar]
- 24. Bianco F., Perrotta C., Novellino L., Francolini M., Riganti L., Menna E., Saglietti L., Schuchman E. H., Furlan R., Clementi E., Matteoli M., Verderio C. (2009) EMBO J. 28, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dhami R., Passini M. A., Schuchman E. H. (2006) Mol. Ther. 13, 556–564 [DOI] [PubMed] [Google Scholar]
- 26. Dhami R., He X., Gordon R. E., Schuchman E. H. (2001) Lab. Invest. 81, 987–999 [DOI] [PubMed] [Google Scholar]
- 27. Brinkman J., Wijburg F. A., Hollak C. E., Groener J. E., Verhoek M., Scheij S., Aten J., Boot R. G., Aerts J. M. (2005) J. Inherit. Metab. Dis. 28, 13–20 [DOI] [PubMed] [Google Scholar]
- 28. Jenkins R. W., Canals D., Idkowiak-Baldys J., Simbari F., Roddy P., Perry D. M., Kitatani K., Luberto C., Hannun Y. A. (2010) J. Biol. Chem. 285, 35706–35718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zeidan Y. H., Wu B. X., Jenkins R. W., Obeid L. M., Hannun Y. A. (2008) FASEB J. 22, 183–193 [DOI] [PubMed] [Google Scholar]
- 30. Neumann C. A., Krause D. S., Carman C. V., Das S., Dubey D. P., Abraham J. L., Bronson R. T., Fujiwara Y., Orkin S. H., Van Etten R. A. (2003) Nature 424, 561–565 [DOI] [PubMed] [Google Scholar]
- 31. Cao J., Schulte J., Knight A., Leslie N. R., Zagozdzon A., Bronson R., Manevich Y., Beeson C., Neumann C. A. (2009) EMBO J. 28, 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hahn W. C., Dessain S. K., Brooks M. W., King J. E., Elenbaas B., Sabatini D. M., DeCaprio J. A., Weinberg R. A. (2002) Mol. Cell. Biol. 22, 2111–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zeidan Y. H., Hannun Y. A. (2007) J. Biol. Chem. 282, 11549–11561 [DOI] [PubMed] [Google Scholar]
- 34. Bielawski J., Szulc Z. M., Hannun Y. A., Bielawska A. (2006) Methods 39, 82–91 [DOI] [PubMed] [Google Scholar]
- 35. Soria G., Yaal-Hahoshen N., Azenshtein E., Shina S., Leider-Trejo L., Ryvo L., Cohen-Hillel E., Shtabsky A., Ehrlich M., Meshel T., Keydar I., Ben-Baruch A. (2008) Cytokine 44, 191–200 [DOI] [PubMed] [Google Scholar]
- 36. Marathe S., Schissel S. L., Yellin M. J., Beatini N., Mintzer R., Williams K. J., Tabas I. (1998) J. Biol. Chem. 273, 4081–4088 [DOI] [PubMed] [Google Scholar]
- 37. Miyake Y., Kozutsumi Y., Nakamura S., Fujita T., Kawasaki T. (1995) Biochem. Biophys. Res. Commun. 211, 396–403 [DOI] [PubMed] [Google Scholar]
- 38. Wang E., Norred W. P., Bacon C. W., Riley R. T., Merrill A. H., Jr. (1991) J. Biol. Chem. 266, 14486–14490 [PubMed] [Google Scholar]
- 39. Orci L., Tagaya M., Amherdt M., Perrelet A., Donaldson J. G., Lippincott-Schwartz J., Klausner R. D., Rothman J. E. (1991) Cell 64, 1183–1195 [DOI] [PubMed] [Google Scholar]
- 40. Hurwitz R., Ferlinz K., Sandhoff K. (1994) Biol. Chem. Hoppe Seyler 375, 447–450 [DOI] [PubMed] [Google Scholar]
- 41. Jenkins R. W., Canals D., Hannun Y. A. (2009) Cell. Signal. 21, 836–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tani M., Ito M., Igarashi Y. (2007) Cell. Signal. 19, 229–237 [DOI] [PubMed] [Google Scholar]
- 43. Oskeritzian C. A., Alvarez S. E., Hait N. C., Price M. M., Milstien S., Spiegel S. (2008) Blood 111, 4193–4200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhi L., Leung B. P., Melendez A. J. (2006) J. Cell. Physiol. 208, 109–115 [DOI] [PubMed] [Google Scholar]
- 45. Billich A., Urtz N., Reuschel R., Baumruker T. (2009) Int. J. Biochem. Cell Biol. 41, 1547–1555 [DOI] [PubMed] [Google Scholar]
- 46. Chandru H., Boggaram V. (2007) Gene 391, 150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roviezzo F., Del Galdo F., Abbate G., Bucci M., D'Agostino B., Antunes E., De Dominicis G., Parente L., Rossi F., Cirino G., De Palma R. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11170–11175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kawata T., Ishizuka T., Tomura H., Hisada T., Dobashi K., Tsukagoshi H., Ishiwara M., Kurose H., Mori M., Okajima F. (2005) Biochem. Biophys. Res. Commun. 331, 640–647 [DOI] [PubMed] [Google Scholar]
- 49. Billich A., Bornancin F., Mechtcheriakova D., Natt F., Huesken D., Baumruker T. (2005) Cell. Signal. 17, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 50. Méndez-Samperio P., Pérez A., Trejo A. (2007) Cell. Immunol. 249, 94–100 [DOI] [PubMed] [Google Scholar]
- 51. Zeidan Y. H., Jenkins R. W., Korman J. B., Liu X., Obeid L. M., Norris J. S., Hannun Y. A. (2008) Curr. Drug Targets 9, 653–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Park J. H., Schuchman E. H. (2006) Biochim. Biophys. Acta 1758, 2133–2138 [DOI] [PubMed] [Google Scholar]
- 53. He X., Okino N., Dhami R., Dagan A., Gatt S., Schulze H., Sandhoff K., Schuchman E. H. (2003) J. Biol. Chem. 278, 32978–32986 [DOI] [PubMed] [Google Scholar]
- 54. Koch J., Gärtner S., Li C. M., Quintern L. E., Bernardo K., Levran O., Schnabel D., Desnick R. J., Schuchman E. H., Sandhoff K. (1996) J. Biol. Chem. 271, 33110–33115 [DOI] [PubMed] [Google Scholar]
- 55. Smith E. L., Schuchman E. H. (2008) FASEB J. 22, 3419–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tabas I. (1999) Chem. Phys. Lipids 102, 123–130 [DOI] [PubMed] [Google Scholar]
- 57. Hao S., Baltimore D. (2009) Nat. Immunol. 10, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kitatani K., Sheldon K., Rajagopalan V., Anelli V., Jenkins R. W., Sun Y., Grabowski G. A., Obeid L. M., Hannun Y. A. (2009) J. Biol. Chem. 284, 12972–12978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Canals D., Jenkins R. W., Roddy P., Hernández-Corbacho M. J., Obeid L. M., Hannun Y. A. (2010) J. Biol. Chem. 285, 32476–32485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., Spiegel S. (2009) Science 325, 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ahn Y. T., Huang B., McPherson L., Clayberger C., Krensky A. M. (2007) Mol. Cell. Biol. 27, 253–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lindström T. M., Mohan A. R., Johnson M. R., Bennett P. R. (2008) Mol. Pharmacol. 74, 109–121 [DOI] [PubMed] [Google Scholar]
- 63. Krensky A. M., Ahn Y. T. (2007) Nat. Clin. Pract. Nephrol. 3, 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schall T. J., Jongstra J., Dyer B. J., Jorgensen J., Clayberger C., Davis M. M., Krensky A. M. (1988) J. Immunol. 141, 1018–1025 [PubMed] [Google Scholar]
- 65. Kameyoshi Y., Dörschner A., Mallet A. I., Christophers E., Schröder J. M. (1992) J. Exp. Med. 176, 587–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kovacic J. C., Gupta R., Lee A. C., Ma M., Fang F., Tolbert C. N., Walts A. D., Beltran L. E., San H., Chen G., St Hilaire C., Boehm M. (2010) J. Clin. Invest. 120, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Casola A., Henderson A., Liu T., Garofalo R. P., Brasier A. R. (2002) Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1280–1290 [DOI] [PubMed] [Google Scholar]
- 68. Maune S., Berner I., Sticherling M., Kulke R., Bartels J., Schröder J. M. (1996) Rhinology 34, 210–214 [PubMed] [Google Scholar]
- 69. Catalfamo M., Karpova T., McNally J., Costes S. V., Lockett S. J., Bos E., Peters P. J., Henkart P. A. (2004) Immunity 20, 219–230 [DOI] [PubMed] [Google Scholar]
- 70. Nelson P. J., Kim H. T., Manning W. C., Goralski T. J., Krensky A. M. (1993) J. Immunol. 151, 2601–2612 [PubMed] [Google Scholar]
- 71. Miyamoto N. G., Medberry P. S., Hesselgesser J., Boehlk S., Nelson P. J., Krensky A. M., Perez H. D. (2000) J. Neuroimmunol. 105, 78–90 [DOI] [PubMed] [Google Scholar]
- 72. Schall T. J. (1991) Cytokine 3, 165–183 [DOI] [PubMed] [Google Scholar]
- 73. Ortiz B. D., Nelson P. J., Krensky A. M. (1997) Immunol. Today 18, 468–471 [DOI] [PubMed] [Google Scholar]
- 74. Ortiz B. D., Krensky A. M., Nelson P. J. (1996) Mol. Cell. Biol. 16, 202–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Elojeimy S., Holman D. H., Liu X., El-Zawahry A., Villani M., Cheng J. C., Mahdy A., Zeidan Y., Bielwaska A., Hannun Y. A., Norris J. S. (2006) FEBS Lett. 580, 4751–4756 [DOI] [PubMed] [Google Scholar]
- 76. Maceyka M., Sankala H., Hait N. C., Le Stunff H., Liu H., Toman R., Collier C., Zhang M., Satin L. S., Merrill A. H., Jr., Milstien S., Spiegel S. (2005) J. Biol. Chem. 280, 37118–37129 [DOI] [PubMed] [Google Scholar]
- 77. Okada T., Ding G., Sonoda H., Kajimoto T., Haga Y., Khosrowbeygi A., Gao S., Miwa N., Jahangeer S., Nakamura S. (2005) J. Biol. Chem. 280, 36318–36325 [DOI] [PubMed] [Google Scholar]
- 78. Spiegel S., Milstien S. (2007) J. Biol. Chem. 282, 2125–2129 [DOI] [PubMed] [Google Scholar]
- 79. Appay V., Rowland-Jones S. L. (2001) Trends Immunol. 22, 83–87 [DOI] [PubMed] [Google Scholar]
- 80. Levy J. A. (2009) J. Immunol. 182, 3945–3946 [DOI] [PubMed] [Google Scholar]
- 81. Tyner J. W., Uchida O., Kajiwara N., Kim E. Y., Patel A. C., O'Sullivan M. P., Walter M. J., Schwendener R. A., Cook D. N., Danoff T. M., Holtzman M. J. (2005) Nat. Med. 11, 1180–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Makino Y., Cook D. N., Smithies O., Hwang O. Y., Neilson E. G., Turka L. A., Sato H., Wells A. D., Danoff T. M. (2002) Clin. Immunol. 102, 302–309 [DOI] [PubMed] [Google Scholar]
- 83. Ehlert K., Frosch M., Fehse N., Zander A., Roth J., Vormoor J. (2007) Pediatr. Rheumatol. Online J. 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nelson P. J., Krensky A. M. (2001) Immunity 14, 377–386 [DOI] [PubMed] [Google Scholar]
- 85. Alam R., York J., Boyars M., Stafford S., Grant J. A., Lee J., Forsythe P., Sim T., Ida N. (1996) Am. J. Respir. Crit. Care Med. 153, 1398–1404 [DOI] [PubMed] [Google Scholar]
- 86. Boiardi L., Macchioni P., Meliconi R., Pulsatelli L., Facchini A., Salvarani C. (1999) Clin. Exp. Rheumatol. 17, 419–425 [PubMed] [Google Scholar]
- 87. von Hundelshausen P., Weber K. S., Huo Y., Proudfoot A. E., Nelson P. J., Ley K., Weber C. (2001) Circulation 103, 1772–1777 [DOI] [PubMed] [Google Scholar]
- 88. Koenen R. R., von Hundelshausen P., Nesmelova I. V., Zernecke A., Liehn E. A., Sarabi A., Kramp B. K., Piccinini A. M., Paludan S. R., Kowalska M. A., Kungl A. J., Hackeng T. M., Mayo K. H., Weber C. (2009) Nat. Med. 15, 97–103 [DOI] [PubMed] [Google Scholar]
- 89. Lebovic D. I., Chao V. A., Taylor R. N. (2004) J. Clin. Endocrinol. Metab. 89, 1397–1401 [DOI] [PubMed] [Google Scholar]
- 90. Mori N., Krensky A. M., Ohshima K., Tomita M., Matsuda T., Ohta T., Yamada Y., Tomonaga M., Ikeda S., Yamamoto N. (2004) Int. J. Cancer 111, 548–557 [DOI] [PubMed] [Google Scholar]
- 91. Mrowietz U., Schwenk U., Maune S., Bartels J., Küpper M., Fichtner I., Schröder J. M., Schadendorf D. (1999) Br. J. Cancer 79, 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Moran C. J., Arenberg D. A., Huang C. C., Giordano T. J., Thomas D. G., Misek D. E., Chen G., Iannettoni M. D., Orringer M. B., Hanash S., Beer D. G. (2002) Clin. Cancer Res. 8, 3803–3812 [PubMed] [Google Scholar]
- 93. Kapoor S. (2008) J. Hum. Genet. 53, 377–378 [DOI] [PubMed] [Google Scholar]