Abstract
In contrast to the NADPH oxidases Nox1 and Nox2, which generate superoxide (O2˙̄), Nox4 produces hydrogen peroxide (H2O2). We constructed chimeric proteins and mutants to address the protein region that specifies which reactive oxygen species is produced. Reactive oxygen species were measured with luminol/horseradish peroxidase and Amplex Red for H2O2 versus L-012 and cytochrome c for O2˙̄. The third extracytosolic loop (E-loop) of Nox4 is 28 amino acids longer than that of Nox1 or Nox2. Deletion of E-loop amino acids only present in Nox4 or exchange of the two cysteines in these stretches switched Nox4 from H2O2 to O2˙̄ generation while preserving expression and intracellular localization. In the presence of an NO donor, the O2˙̄-producing Nox4 mutants, but not wild-type Nox4, generated peroxynitrite, excluding artifacts of the detection system as the apparent origin of O2˙̄. In Cos7 cells, in which Nox4 partially localizes to the plasma membrane, an antibody directed against the E-loop decreased H2O2 but increased O2˙̄ formation by Nox4 without affecting Nox1-dependent O2˙̄ formation. The E-loop of Nox4 but not Nox1 and Nox2 contains a highly conserved histidine that could serve as a source for protons to accelerate spontaneous dismutation of superoxide to form H2O2. Mutation of this but not of four other conserved histidines also switched Nox4 from H2O2 to O2˙̄ formation. Thus, H2O2 formation is an intrinsic property of Nox4 that involves its E-loop. The structure of the E-loop may hinder O2˙̄ egress and/or provide a source for protons, allowing dismutation to form H2O2.
Keywords: Heme, Oxygen Radicals, Reactive Oxygen Species (ROS), Superoxide Dismutase (SOD), Superoxide Ion
Introduction
The class of Nox protein NADPH oxidases is a group of enzymes whose sole known function is the production of reactive oxygen species (ROS).2 The enzyme family is named for the enzymatically active transmembrane protein Nox. All seven Nox proteins share highly conserved structural features; the C-terminal dehydrogenase domain contains binding sites for FAD and NADPH. The N-terminal transmembrane region consists of six α-helical transmembrane domains that contain four conserved histidine residues, located in the third and fifth transmembrane helices that coordinate two hemes. Electron transfer occurs from NADPH to oxygen via FAD and the two heme groups, with the second heme group reducing molecular oxygen (1, 2). Because heme is an obligate 1-electron donor, it is generally accepted that the superoxide anion (O2˙̄) is the initial reduction product of oxygen, although the latter can also react with a second O2˙̄ to form hydrogen peroxide (H2O2) plus oxygen.
Despite these similarities, Nox proteins differ in their mode of activation, their interaction with the small transmembrane protein p22phox, and the requirement for additional maturation and activation factors. The most extensively studied NADPH oxidase isoform is the phagocyte Nox2 (previously termed gp91phox), which depends on p22phox and whose activation requires assembly with the cytosolic regulatory subunits p47phox and p67phox (3), along with GTP-loaded Rac1 or Rac2. Similar to Nox2, the homologue Nox1 requires regulatory subunits. Unlike all the other Nox proteins, Nox4 is constitutively active and is independent of cytosolic activator proteins or regulatory domains (4, 5). Another interesting difference between Nox1/2 and Nox4 is that Nox1 and Nox2 produce primarily O2˙̄, whereas most studies report that Nox4 generates H2O2 (4, 6, 7).
Given that the prosthetic groups are identical and the core protein structures are very similar among the Nox proteins, it has been suggested that the failure to detect O2˙̄ formation by Nox4 is a consequence of its intracellular location, resulting in problems detecting O2˙̄ within the cell and the inability of this ion to pass freely through the membrane. Indeed, although a significant fraction of Nox1 and Nox2 is located at the plasma membrane and thus would reduce extracellular oxygen, Nox4 is localized predominantly to intracellular membranes where any generated O2˙̄ might be cryptic. In fact, Nox4 protein has been reported in mitochondria (8), the nucleus (9), the cytoskeleton (10), and the endoplasmic reticulum (11). Thus, it was plausible to propose that O2˙̄ generated in these compartments must undergo dismutation to leave the cell as the freely diffusible H2O2. However, Nox4 in some cells resides in part in the plasma membrane but unexpectedly still produces H2O2 without any detectable O2˙̄ (12). Moreover, a careful analysis of intracellular ROS formation using overexpressed Nox4 failed to detect Nox4-mediated O2˙̄ production using ESR spin traps and the dihydroethidium method (13). In contrast, Nox4 was able to reduce nitro blue tetrazolium. However, the site of the electron efflux from Nox4 to nitro blue tetrazolium has not yet been determined but is potentially via the FAD-containing dehydrogenase domain, which is known to catalyze the direct reduction of various dyes (14). Based on the above, it has to be concluded that although mechanistically, heme reduction of oxygen must initially generate O2˙̄, Nox4 releases H2O2 without releasing free O2˙̄. The molecular basis for this potentially physiologically important difference is unclear.
Based on the presence of six transmembrane α-helical domains, the current model for NADPH oxidases predicts that the N- as well as C-terminal parts of the protein reside in the cytosol, giving rise to two intracellular loops (B- and D-loop) and three loops oriented away from the cytosol and toward the extracellular space or intracellular compartments (A-, C-, and E-loop). So far, little work has been devoted to the extracellular loops. Although asparagines within these regions are glycosylated in Nox2, to our knowledge, no mutations leading to chronic granulomatous disease have been reported for these loops. Also, the functional significance of glycosylation is somewhat uncertain as unlike human Nox2, the murine enzyme does not undergo this modification and glycosylation has not been reported for Nox1. Based upon their proximity to the site of oxygen reduction by the B heme, we hypothesized that differences in the extracellular loops are responsible for the unique ability of Nox4 to release H2O2 rather than O2˙̄.
EXPERIMENTAL PROCEDURES
Sequence Alignment
Nox sequences were aligned using the online program ClustalW2 from the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI) (15).
Generation and Transfection of Mutant Nox Constructs
Plasmids encoding human full-length Nox1, Nox4, and p22phox were kindly provided by T. Leto (National Institutes of Health, Bethesda, MD). The plasmids coding for mouse Noxa1 and Noxo1 were generous gifts of B. Banfi (Iowa University, Iowa City, IA). The plasmids coding for the Nox4 deletion mutants were generated by overlap extension PCR. The plasmids coding for the cysteine and histidine mutants of Nox1 and Nox4 were generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All cloned plasmids were confirmed with DNA sequencing. Transient transfection of HEK293 or Cos7 cells (ATCC, Manassas, VA) was performed with Lipofectamine 2000 (Invitrogen, Paisley, UK) according to the manufacturer's instructions. HEK293 cells stably expressing Nox4 Δ218–235 + Δ264–273 were generated by transfection with linearized plasmid with Lipofectamine 2000 and subsequent antibiotic selection.
Determination of ROS by Chemiluminescence Measurement
HEK293 or Cos7 cells were seeded on 3.5-cm dishes and transiently transfected. After 24 h, EDTA-detached cells were suspended in 500 μl of HEPES-modified Tyrode's solution containing the appropriate chemiluminescence enhancer. As enhancer for H2O2, luminol (Sigma; 100 μm) in combination with horseradish peroxidase (HRP; Sigma; 1 units/ml) were used. As enhancer for O2˙̄, L-012 (WAKO Chemicals, Richmond, VA, 200 μm) was used. As enhancer for peroxynitrite (ONOO−), luminol without HRP was used, and DetaNONOate (Enzo Life Sciences, Plymouth Meeting, PA; 100 μm) was added as NO donor to facilitate the formation of ONOO− from O2˙̄.
Determination of ROS by Amplex Red Measurement
HEK293 cells were grown on 12-well plates and transiently transfected. 24 h later, cells were washed once with phosphate-buffered saline (PBS) and subsequently incubated in PBS containing 100 μm Amplex Red and 0.25 units/ml HRP. After 30 min, the supernatant was transferred to a 96-well plate, and H2O2-dependent oxidation of Amplex Red was measured in a microplate fluorometer (excitation 540 nm, emission 580 nm). H2O2 formation was quantified by a standard curve of known H2O2 concentrations.
Determination of Reactive Oxygen Species by Cytochrome c Reduction
HEK293 cells were seeded on 12-well plates and transiently transfected. 24 h later, cells were washed once in HEPES-modified Tyrode's solution containing cytochrome c (from horse heart; Sigma; 1 mg/ml) and then incubated in this solution in the presence or absence of superoxide dismutase (Sigma, 100 units/ml) at 37 °C. After 30–60 min, supernatants were transferred to fresh tubes on ice, and the superoxide dismutase-inhibitable reduction of cytochrome c was quantified in a spectrophotometer (Uvikon, Kontron Instruments). Data were normalized to the isosbestic points at 542 and 558 nm, and O2˙̄ formation was calculated with the aid of the molar extinction coefficient of 21 mm−1 cm−1.
Western Blot Analysis
24 h after transfection, cells were incubated for 8 h with the proteasome inhibitor MG132 (10 μm, Calbiochem) to stabilize and increase the Nox expression. Cell lysis, SDS-PAGE, and Western blot were carried out (16). The following antibodies were used: anti-Nox1 (Santa Cruz Biotechnology: Mox-1 H-15); anti-Nox4 (generated by us (17)); anti-β-actin (Sigma); and anti-Erk1/2 and anti-phosphorylated Erk1/2 (Cell Signaling).
Confocal Fluorescence Microscopy
HEK293 cells stably expressing Nox4 Δ218–235 + Δ264–273 were seeded on μ-dishes (ibidi, Martinsried, Germany). When the cells reached ∼80% confluence, they were incubated for 8 h with the translation inhibitor anisomycin (Calbiochem; 20 μm) to reduce potential localization of the protein to the endoplasmic reticulum due to de novo synthesis. Imaging was carried out as described (16) with the following antibodies: anti-Nox4 (generated by us) and anti-GRP78 (Santa Cruz Biotechnology) for endoplasmic reticulum staining.
Statistical Analysis
Unless otherwise stated, all data shown are mean ± S.E. Statistical significance was determined by one-way analysis of variance followed by Newman-Keuls post hoc test or by paired or unpaired t test, if appropriate.
RESULTS
Role of the Length of the E-loop of Nox4
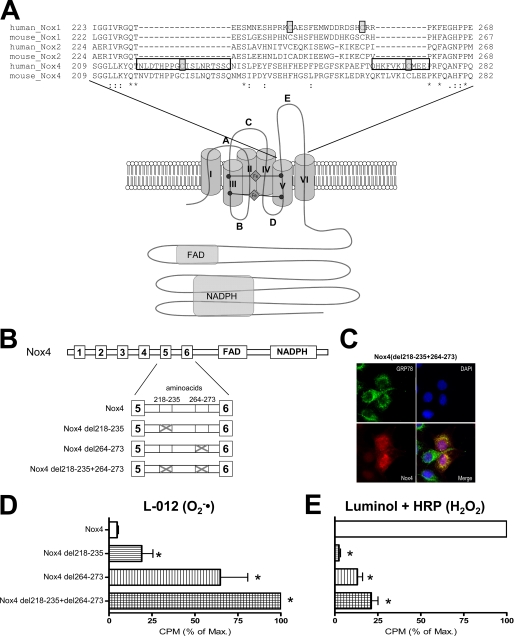
To identify a structural basis for the H2O2 formation of Nox4, we compared this protein with Nox1 and Nox2. An alignment of the amino acid sequences of human and mouse Nox1, Nox2, and Nox4 revealed that the E-loop of Nox4 is 28 amino acids longer than the E-loop of Nox1 and Nox2 (Fig. 1A) as a consequence of two insertions of 18 and 10 amino acids, respectively. The sequence within the two insertions is similar among Nox4s from different species, suggesting a conserved Nox4-specific function. We deleted these Nox4-specific E-loop regions so as to approximate the length of the E-loop in Nox1 and Nox2 (Fig. 1B). These mutations, which did not affect the endoplasmic reticulum localization of Nox4 (Fig. 1C), had a strong effect on ROS production by Nox4; the mutants produced very little H2O2 when compared with native Nox4 and instead released O2˙̄ (Fig. 1, D and E). The effect was more pronounced in the double deletion mutant, which lacked both sequence insertions, but deletion of amino acids 264–273 was almost equally effective. Although the Nox4 deletion mutants produced O2˙̄, their O2˙̄ formation rate was still lower than that of native Nox1 when equimolar amounts of the expression plasmid were transfected (data not shown). Given that Nox4 del218–235+264–273 colocalized with the heat-shock protein GRP78 (Fig. 1C) and thus is expressed in the endoplasmic reticulum-like native Nox4, we conclude that the type of ROS formed by Nox proteins is an intrinsic feature of the protein and not a consequence of the intracellular localization.
FIGURE 1.
Function of the E-loop of Nox4. A, schematic illustration of the Nox protein and alignment of the amino acid sequence of the E-loop of human and murine Nox1, Nox2, and Nox4. B, schematic illustration of Nox4 and the deletion constructs generated. Boxes represent transmembrane domains (1–6), FAD binding site (FAD), and NADPH binding site (NADPH). Illustrations are not to scale. C, confocal microscopic fluorescent image of Nox4 del218–235+264–273 (red) stably expressed in HEK293 cells counterstained with GRP78 (green) and DAPI. D and E, determination of ROS production of HEK293 cells transiently transfected with the plasmids indicated. O2˙̄ generation was determined by L-012 (D), and H2O2 formation was determined by luminol + HRP chemiluminescence (E). ROS production was normalized against Nox protein expression determined by Western blot analysis. To facilitate better comparison of the constructs, the ROS formation of the most active construct was set to 100% (corresponding to mean 271,217 cpm (D) and 426,172 cpm (E)) n ≥ 3, mean ± S.E., *, p < 0.05 versus wild-type construct. % of Max., percentage of maximum.
Role of the Cysteines in the E-loop of Nox Proteins
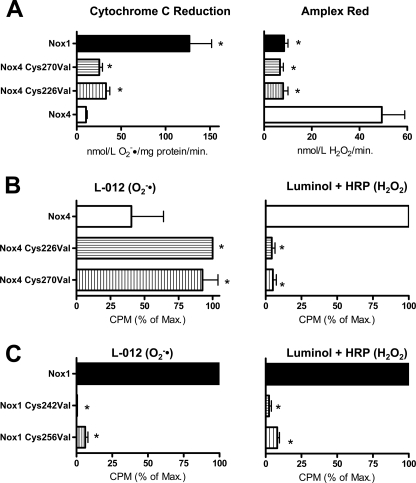
The E-loops of Nox4 as well as Nox1 and Nox2 both contain two conserved cysteines, although they are located in different positions in the two isoforms. In Nox4, both are lost upon the construction of the deletion mutants of Nox4. As the cysteines might form a disulfide bridge to maintain the integrity of the E-loop, we studied their role for the function of Nox4. Mutation of the cysteines to valines had a similar, although less pronounced, effect on ROS production by Nox4 as the deletion constructs (Fig. 2, A and B), as demonstrated by chemiluminescence as well as cytochrome c reduction and Amplex Red oxidation. A plasmid harboring the mutation of both Nox4 cysteines together showed a more pronounced effect on the switching of the ROS type generated than the single mutations (data not shown). Mutation of the two cysteines in Nox1 decreased the overall activity of the enzyme but did not change the type of ROS released (Fig. 2C). These data show that even minor alterations of the E-loop of Nox4 switch the protein from H2O2 to O2˙̄ production and that the integrity of the E-loop is also essential for O2˙̄ formation of Nox1.
FIGURE 2.
Role of the cysteines in the E-loop of Nox4 and Nox1. A–C, determination of ROS production of HEK293 cells transiently transfected with the plasmids indicated. O2˙̄ generation was determined by superoxide dismutase-sensitive cytochrome c reduction (A, left) or by L-012 chemiluminescence (B and C, left). H2O2 formation was determined by Amplex Red oxidation (A, right) or luminol + HRP chemiluminescence (B and C, right). The Nox1 plasmid was cotransfected with Noxo1 and Noxa1. Normalization was as in Fig. 1, n ≥ 3, mean ± S.E. *, p < 0.05 versus wild-type construct. 100% corresponds to mean 35,800 and 711,785 cpm (B, left and right) and to mean 734,017 and 59,462 cpm (C, left and right). % of Max., percentage of maximum.
Nox4 E-loop Mutants but Not Native Nox4 Form Peroxynitrite in the Presence of an NO Donor
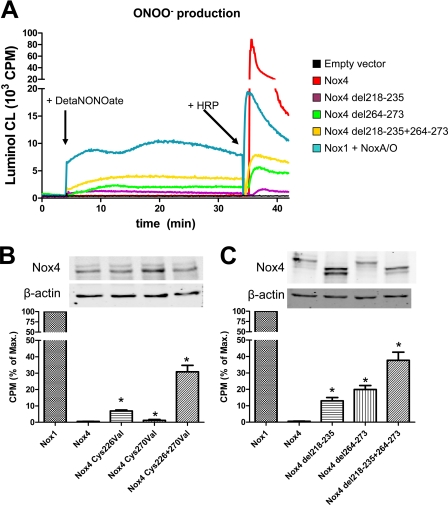
To address whether the H2O2 formation of Nox4 occurs via free intermediate formation of O2˙̄ and to further exclude that H2O2 formation is a consequence of some artifact of the detection system, we studied ROS formation in the presence of an NO donor using luminol chemiluminescence. We hypothesized that the failure to detect O2˙̄ by various methods may have resulted from a combination of poor access to the site of formation of O2˙̄ and/or relatively slow rates of reaction of the probe with O2˙̄. NO, on the other hand, is known to react at a near diffusion controlled rate with O2˙̄ to yield ONOO−, and its small size should allow better access to the site of ROS generation when compared with larger probes. Therefore, if any free O2˙̄ is formed by Nox4, it should react rapidly with NO to yield ONOO−, which is able to oxidize luminol in the absence of HRP. Indeed, for overexpressed Nox1, the DetaNONOate-induced luminol signal was easily detected, whereas overexpressed Nox4 failed to produce any luminol signal. These data exclude that significant amounts of O2˙̄ are released by Nox4. Importantly, similar to the other assays that detected O2˙̄ (above), the E-loop mutants of Nox4 produced ONOO− (Fig. 3).
FIGURE 3.
Effect of mutations in the E-loop on the ONOO− production of Nox4. A–C, representative recording (A) and statistical analysis (B and C) of ONOO− production of HEK293 cells transiently transfected with the plasmids indicated as well as protein expression of the constructs. ROS formation was determined by luminol chemiluminescence. After reading the cells in the absence of NO, the NO donor DetaNONOate (100 μmol/liter) was administered (first arrow). The subsequent signal was considered to arise from ONOO− formation. At the end of the experiment, horseradish peroxidase (+HRP) was added (second arrow) to determine H2O2 formation. The Nox1 plasmid was cotransfected with Noxo1 and Noxa1. Normalization was as in Fig. 1. Exemplary Western blots of the construct are shown in B and C. n ≥ 3, mean ± S.E., *, p < 0.05 versus wild-type construct. 100% corresponds to mean 7625 and 7818 cpm (B and C, respectively). % of Max., percentage of maximum.
Binding of an Antibody to the E-loop Affects H2O2 Production of Nox4
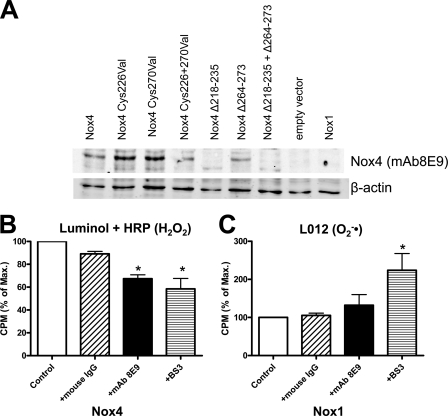
A general problem with mutational analyses is that local changes in structure may affect the function of an enzyme at a remote site by some allosteric modulations. We therefore sought a second approach to interfere with the E-loop. Recently, a monoclonal antibody directed against this part of the protein was generated (mAb8E9) (18). When compared with an antibody that binds in the NADPH binding site (Fig. 3C), Western blot analysis confirmed that the antibody mAb8E9 binds the E-loop as labeling was lost in the Nox4 mutants (Fig. 4). As the antibody can only directly interact with extracellular portions of Nox4, this part of the study was carried out in Cos7 cells, which exhibit some plasma membrane expression of this NADPH oxidase (12).
FIGURE 4.
Effect of an antibody directed against the E-loop on the H2O2 formation of Nox1 and Nox4. A, representative Western blot of HEK293 cells transfected with the plasmids indicated probed with the antibody mAB8E9, which is directed against the E-loop of Nox4. B and C, determination of H2O2 production by luminol + HRP chemiluminescence and O2˙̄ production in Cos7 cells transiently transfected with wild-type Nox4 or Nox1 (cotransfected with Noxo1 and Noxa1). The following substances indicated were added to the cells prior to measurements: monoclonal antibody directed against the E-loop (mAb8E9: 1 μg/ml), normal control mouse IgG (1 μg/ml), and the surface cross-linker bis-sulfosuccinimidyl suberate (BS3, 1 mmol/liter). Normalization was as in Fig. 1. n ≥ 3, mean ± S.E., *, p < 0.05 versus control. 100% corresponds to mean 1390 and 1355 cpm (B and C, respectively). % of Max., percentage of maximum.
The addition of the mAb8E9 decreased H2O2 production by Nox4-transfected Cos7 cells by ∼20% (Fig. 4), and in a pilot study, mAb8E9 also increased the O2˙̄ formation of Nox4 (+28 ± 11%, n = 2). Importantly, a nonspecific mouse IgG control at the same concentration had no effect on ROS formation by Nox4, and also, mAb8E9 did not alter O2˙̄ production in Nox1-overexpressing cells. The interpretation of the signaling is, however, difficult as the fraction of plasma membrane-localized Nox4 to total Nox4 is unknown. To estimate the amount of Nox4 accessible by this approach, we treated the cells with the non-cell-permeable cross-linker bis-sulfosuccinimidyl suberate (1 mm), assuming that this should destroy the Nox4 activity at the plasma membrane, and indeed, Nox4 protein became undetectable by Western blot after cross-linking (data not shown). With respect to ROS production, bis-sulfosuccinimidyl suberate was equally effective as mAb8E9 in reducing H2O2 formation by Nox4. Surprisingly, the compound increased O2˙̄ produced by Nox1, suggesting that indeed alterations of the extracytosolic loop directly affect the efficacy of O2˙̄ formation (Fig. 4).
Western blot analysis of the Nox4 deletion constructs (Fig. 3C) indicated that the protein Nox4 del218–235 might be unstable as it was detected with a molecular mass of 6 kDa less than the calculated mass. As, however, ROS measurements of the other deletion constructs and the cysteine mutants in the same region yielded similar results, the reduced stability of this single construct might be of lesser importance for the interpretation of the data.
Role of Extracytoplasmic Histidines for H2O2 Production by Nox4
Our data so far suggest that Nox4 directly generates H2O2 without the formation of significant amounts of a free O2˙̄ intermediate. To generate H2O2, Nox4 either has to act as a dioxygenase (which could not occur from the heme itself) or has to facilitate the transfer of an electron from one O2˙̄ to a second, in effect acting as a superoxide dismutase. We speculate that histidine residues might play a role in generating H2O2 by serving as a source for protons or by binding a metal, as is needed for catalysis by all known superoxide dismutase enzymes.
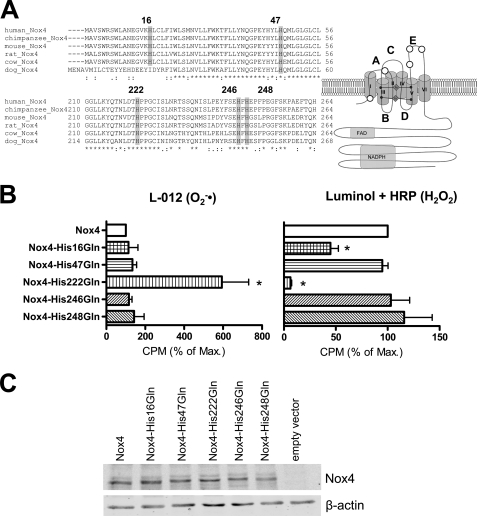
Structure alignment revealed that in addition to those that are directly involved in heme ligation, Nox4 contains five additional highly conserved histidines in its membrane integrated part (Fig. 5A), and interestingly, three of these are located in the E-loop. We mutated these five histidines to glutamine and studied protein expression and ROS formation by the mutants. The mutations of His-47, His-246, and His-248 had no effect on ROS production, whereas H16Q reduced H2O2 formation by Nox4 by ∼50% without affecting O2˙̄ formation. In contrast, Nox4-H222Q, which carries the mutation at the beginning of the E-loop, produced almost no H2O2 and instead released O2˙̄ (Fig. 5B). The expression of the histidine mutants was similar to that of native Nox4 as determined by Western blot analysis (Fig. 5C). These observations not only confirm that the E-loop is central for H2O2 formation but also indicate that specific features of the E-loop are required for the process.
FIGURE 5.
Role of conserved histidines on the ROS formation of Nox4. A, sequence alignment of Nox4 proteins from different species for conserved histidines on the extracytoplasmic side of the protein and schematic representation of the localization of the conserved histidines within the proteins (white circles). B, ROS production of HEK293 cells transiently transfected with the plasmids indicated. O2˙̄ generation was determined by L-012 chemiluminescence (left panel), and H2O2 formation was assessed by luminol + HRP chemiluminescence (right panel). Normalization was as in Fig. 1, n ≥ 3, mean ± S.E. *, p < 0.05 versus wild-type construct. 100% corresponds to mean 7647 (left) and 82,425 cpm (right). C, Western blot analysis of the expression of the different histidine mutants in HEK293 cells.
Mutation of the Cysteines in the E-loop Prevents Nox4-induced Erk1/2 Phosphorylation
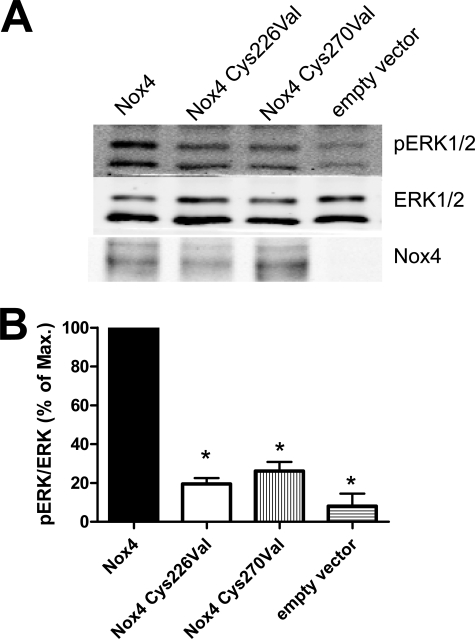
In the final step, we sought to demonstrate that the type of ROS released by Nox4 is physiologically relevant. Unlike Nox2 and Nox1, Nox4 activity has previously been linked to Erk1/2 phosphorylation (17). We therefore investigated the activation of this MAP kinase in response to the overexpression of Nox4 versus Nox4 cysteine mutants. Although overexpression of native Nox4 induced a robust Erk1/2 phosphorylation in HEK293 cells, this effect was not observed with the cysteine mutants, although they were expressed to a similar level and total Erk1/2 expression was not affected by this approach (Fig. 6). These data indicate that the direct formation of H2O2 by Nox4 has important biological consequences for Nox-dependent signal transduction.
FIGURE 6.
Effect of mutations in the E-loop of Nox4 on Erk1/2 phosphorylation. HEK293 cells were transiently transfected with the plasmids indicated and subsequently subjected to SDS-PAGE and Western blot analysis with the antibodies indicated. A and B, representative blots (A) and statistical analysis (B) of the ratio of phosphorylated to non-phosphorylated Erk1/2 (pERK1/2). To allow better comparison, the signal of wild-type Nox4 was set to 100%. n ≥ 3, mean ± S.E. *, p < 0.05 versus Nox4 construct. % of Max., percentage of maximum.
DISCUSSION
In this work, we studied the structural basis for H2O2 formation by Nox4. We identified the extended E-loop of the protein as an essential structural feature for this process and showed that alterations of the E-loop switch Nox4 from an H2O2 into an O2˙̄-producing enzyme. The ROS product generated appears to be sensitive to minor structural perturbations in the E-loop because binding by a monoclonal antibody or cross-linking at the cell surface impaired H2O2 formation by Nox4 but not O2˙̄ formation by Nox1. H2O2 formation appeared to be an intrinsic function of Nox4 as we could not detect O2˙̄ formation, based on the formation of ONOO− in the presence of an NO donor. In agreement with such a scenario, we identified a highly conserved histidine in the E-loop, which might serve as source of protons or as a binding site for metals to provide superoxide dismutase activity to Nox4.
Based on their similarity to Nox2 and the obligate 1-electron transfer from heme iron, all Nox NADPH oxidases should primarily produce O2˙̄ (1). The H2O2 formation by Nox4 was therefore initially interpreted as artifact but occurring as a result of the use of reagents that detected exclusively extracellular ROS generated by an intracellular enzyme. Mutational analysis (16), intracellular probes for ROS detection (13), and comparative expression studies in cells exhibiting different localizations of Nox4 (12), however, demonstrated that the type of ROS produced by the enzyme is independent of its localization. This suggests that Nox NADPH oxidases can directly release either O2˙̄ or H2O2 and that this feature is dependent on structural properties of the individual Nox enzyme.
Nox4, indeed, is not the only Nox homologue that primarily produces H2O2. Under physiological conditions, the Duox enzymes also generate H2O2 exclusively (19, 20). The molecular mechanisms leading to Duox-dependent H2O2 formation are not clear but appear to be distinct from that of Nox4. Based on homologue searches, it was suggested that Duox enzymes might bear an intrinsic superoxide dismutase activity localized to the peroxidase homology domain of these proteins (21), but recently, it was shown that although structurally similar to peroxidases, mammalian Duox enzymes are not able to directly dismutate O2˙̄ (22). Rather, the type of ROS generated appears to depend on the maturation state of the Duox enzymes. In the absence of the maturation factors Duoxa1 or Duoxa2, the Duox enzymes generate O2˙̄, and only after interaction with the Duoxa proteins does H2O2 formation becomes apparent (23). This process also involves relocalization of the Duox-Duoxa complex from the endoplasmic reticulum to the plasma membrane, and it is not clear whether localization impacts the observed reduced oxygen species (24). The enzymatic activity of the Duox enzymes is independent of p22phox, whereas the maturation of Nox2 (25) as well as the activity of most of the Nox proteins (with the exception of Nox5) is dependent on p22phox (7). Thus, it is not an understatement that, by analogy with the Duox-Duoxa proteins, p22phox was referred to as a maturation factor for the Nox proteins (24). Divergent from this concept, it was observed that alteration of the maturation factors for the Duox proteins switch them from H2O2 to O2˙̄ formation (24), whereas so far no mutation in p22phox was reported to induce such a transition for Nox4 (7). Interestingly, based on truncation experiments, it can be concluded that at least some different regions of p22phox interact with Nox1/2 when compared with Nox4 (7), but an in-depth molecular analysis has not yet been carried out.
As for Nox4, a molecular explanation for H2O2 formation by Duox is unclear. The lack of superoxide dismutase activity in the peroxidase domain may suggest that the enzymes can produce H2O2 directly like dioxygenases or that whatever O2˙̄ is produced remains cryptic and inaccessible to assay reagents prior to its dismutation to form H2O2, for example, due to either spontaneous dismutation or transfer to a second, so far unidentified, interacting protein like a superoxide dismutase. Obviously, these mechanisms could also account for the H2O2 formation of Nox4. The localization of Nox4 differs among cell types, which is suggestive of the presence of interacting proteins directing the protein to different destinations, but so far, little has been published to support this view. The p22phox-interacting protein Poldip2 has been shown to affect Nox4 expression and activity, but it has not been reported that it alters the type of ROS formed by Nox4 (26). A direct interaction of Nox4 with one of the superoxide dismutase isoforms seems unlikely due to the orientation of the O2˙̄ exit site to the extracytosolic compartment. Indeed, by co-immunoprecipitation experiments, we found no evidence for such an interaction.3
On the basis of the present data, we suggest that H2O2 formation is an intrinsic function of Nox4. Thus, either the enzyme has endogenous superoxide dismutase activity or it acts as a dioxygenase and directly forms H2O2. For both processes, histidines and cysteines could be important. Superoxide dismutase activity requires the presence of metal cofactors such as zinc, manganese, or iron, which are coordinated by these amino acids. Indeed, zinc in CuZn-superoxide dismutase is coordinated by three histidines (27), and the zinc in zinc finger proteins is coordinated by two histidines and two cysteines. However, at least two coordinating histidines are likely to be required to form a metal binding site, and our mutational analysis of Nox4 provided evidence for only a single histidine (His-222) of importance. Such a scenario might, however, be viable if Nox4 forms a homodimer in the membrane, as has been suggested for Nox5 (35). According to this scenario, the metal binding site would be formed at the interface between the subunits. Interestingly, also p22phox contains a single histidine, and this is not required for the function of Nox2 (28). This histidine is located in a region important for Nox4-p22phox interaction (7) but appears to be located in the transmembrane region of the protein directed more toward the cytosolic side. Thus, it seems unlikely that this amino acid of p22phox has a specific function for ROS formation by Nox4. Interestingly, His-222 of Nox4 is embedded in the sequence THPPGC, and deletion of this stretch or mutation of the Cys-226 also switched Nox4 from H2O2 to O2˙̄ formation. Given that the two prolines in this sequence should force a 90° deviation in the secondary structure of the sequence, the Cys-226 is likely to be in close proximity to His-222 and therefore might contribute to metal coordination. Unfortunately, we are currently unable to purify sufficient amounts of Nox4 to test whether it indeed contains such metals. To our knowledge, no specific chelators are available that are compatible with living cells and that might deplete the metal from the E-loop without extracting the heme iron. With EDTA, at least, we were unable to switch Nox4 from H2O2 to O2˙̄ generation.4
Despite this speculation about metal coordination, histidine might also directly accelerate the formation of H2O2 in the absence of a metal. This amino acid is frequently involved in enzyme catalysis, where it acts as a proton donor (29). The spontaneous rate of dismutation of superoxide is high, provided that the reaction is between O2˙̄ and its protonated form HO2· (pKa 4.9). The reported rate constant for the process is ∼1 × 108 m−1 s−1, 7 orders of magnitude higher than the spontaneous dismutation rate for two superoxide anions (30). Thus, we suggest that His-222 could serve as a proton donor for a superoxide anion to form the perhydroxyl radical, which would accelerate spontaneous O2˙̄ dismutation by many orders of magnitude. Although this rate is still at least 200-fold less than that catalyzed by superoxide dismutases, it is likely to be more than sufficient to keep up with the turnover rate of Nox4, which has been reported to be around 200 min−1 (14).
Another possible role for the E-loop (not mutually exclusive with the above) is to form a physical structure that slows the egress of O2˙̄ and/or HO2· from its site of formation at the heme, allowing it to accumulate and accelerating the spontaneous dismutation by virtue of increasing the local concentration of O2˙̄. Because the rate of spontaneous dismutation of superoxide is a function of the square of its concentration, creation of a “cage” at the site of generation would be expected to accelerate the spontaneous dismutation rate, particularly in the presence of a proton donor. Such a mechanism would extend the retention time of O2˙̄ and potentially allow for the collision of two O2˙̄ molecules before O2˙̄ can be released into solution. Relative to Nox1 and Nox2, Nox4 shows significantly increased length of the extended E-loop, consistent with this concept. The cysteines Cys-226 and Cys-270 might form a disulfide bridge to stabilize such a structure. According to such a model, any manipulation at the E-loop (e.g. the truncation experiments, the cysteine, and the His-222 mutations) would affect the conformation of the E-loop, thereby accelerating the exit of O2˙̄. The data obtained with the antibody mAb8E9 appear to support this concept. Although the exact effect of the antibody binding to the E-loop is unknown, it was able to decrease H2O2 to the same extent as an extracytoplasmic cross-linking agent, which should also interfere with the structure of the E-loop. Our observation of an inability of Nox4 to form peroxynitrite in the presence of an NO donor would argue that if O2˙̄ is formed as an intermediate, either its half-life has to be unusually short or its access to the NO donor must be impeded by the native protein structure. Thus, according to the “caged superoxide” scenario, O2˙̄ is not released free into solution, where it can react with NO.
Structurally, H2O2- and O2˙̄-forming enzymes are usually quite different, and xanthine oxidase is one of the few examples of an enzyme capable of producing both types of ROS (31). The enzymatic mechanisms underlying this function are, however, completely different from the Nox proteins. Recently, a mutant of xanthine oxidase was constructed with increased O2˙̄ and decreased H2O2 formation (32), which was achieved by increasing the redox potential of FAD so that the rate constant of electron transfer from FADH* onto dioxygen was increased (32). Usually, dioxygen is reduced by FADH2 in xanthine oxidase, and as this reaction in the native enzyme is much faster than the single electron transfer from FADH*, native xanthine oxidase produces much less O2˙̄ than H2O2 (33). In this context, it is important to emphasize that H2O2 formation indeed usually occurs at FAD sites and does not require the heme present in Nox proteins. Heme proteins such as cytochrome P450 monooxygenases, the NO synthase enzymes, and also the Nox enzymes in contrast produce O2˙̄. Thus, one could speculate that H2O2 formation by Nox4 occurs at the FAD site. Up to now, no evidence has been presented to suggest that the FAD in Nox enzymes is able to directly reduce oxygen. Expression of the isolated dehydrogenase domain of Nox4 demonstrated that this domain of Nox4 is able to reduce cytochrome c and several dye electron acceptors (14) but does not form H2O2 directly.5
From the physiological point of view, the functional role of Nox4 is still incompletely understood, and thus, it is also unclear whether the type of ROS generated by Nox4 impacts function. In overexpression experiments, Nox4 activated a different set of MAP kinases than Nox2, with a particularly strong activation of the Erk1/2 pathway (17). As Erk1/2 phosphorylation was attenuated in the present study by switching Nox4 from H2O2 to O2˙̄, it could be inferred that this action is indeed predominately mediated by H2O2. In line with this concept, it was recently shown that Nox4-derived H2O2 oxidizes Ras (34), which would subsequently activate the Erk1/2 pathway.
In conclusion, we have demonstrated that Nox4 directly produces H2O2 by a mechanism involving the relatively large E-loop of the enzyme and that the mechanism requires Cys-226 and Cys-270 as well as His-222 of this loop. Functionally, Erk1/2 activation by Nox4 required H2O2 formation and was not observed with O2˙̄-generating Nox4 mutants. Structural information on the transmembrane region of Nox4, however, will be needed to ultimately clarify the exact mechanism of H2O2 formation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA084138, R01 CA105116, and P01 ES011163 (to J. D. L.) and the Leducq Foundation and the British Heart Foundation (to A. M. S.). This work was also supported by grants from the Deutsche Forschungsgemeinschaft (SFB815/TP1 and SFB834/TPA2) and the Excellence Cluster Cardio-pulmonary System (ECCPS).
I. Takac, unpublished observation.
R. Brandes, unpublished observation.
Y. Nishimoto and J. D. Lambeth, unpublished observation.
- ROS
- reactive oxygen species.
REFERENCES
- 1. Bedard K., Krause K. H. (2007) Physiol. Rev. 87, 245–313 [DOI] [PubMed] [Google Scholar]
- 2. Vignais P. V. (2002) Cell Mol. Life Sci. 59, 1428–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nauseef W. M. (2004) Histochem. Cell Biol. 122, 277–291 [DOI] [PubMed] [Google Scholar]
- 4. Ambasta R. K., Kumar P., Griendling K. K., Schmidt H. H., Busse R., Brandes R. P. (2004) J. Biol. Chem. 279, 45935–45941 [DOI] [PubMed] [Google Scholar]
- 5. Geiszt M., Kopp J. B., Várnai P., Leto T. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8010–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martyn K. D., Frederick L. M., von Loehneysen K., Dinauer M. C., Knaus U. G. (2006) Cell. Signal. 18, 69–82 [DOI] [PubMed] [Google Scholar]
- 7. von Löhneysen K., Noack D., Jesaitis A. J., Dinauer M. C., Knaus U. G. (2008) J. Biol. Chem. 283, 35273–35282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Block K., Gorin Y., Abboud H. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 14385–14390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kuroda J., Nakagawa K., Yamasaki T., Nakamura K., Takeya R., Kuribayashi F., Imajoh-Ohmi S., Igarashi K., Shibata Y., Sueishi K., Sumimoto H. (2005) Genes Cells 10, 1139–1151 [DOI] [PubMed] [Google Scholar]
- 10. Hilenski L. L., Clempus R. E., Quinn M. T., Lambeth J. D., Griendling K. K. (2004) Arterioscler. Thromb. Vasc. Biol. 24, 677–683 [DOI] [PubMed] [Google Scholar]
- 11. Chen K., Kirber M. T., Xiao H., Yang Y., Keaney J. F., Jr. (2008) J. Cell Biol. 181, 1129–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. von Löhneysen K., Noack D., Wood M. R., Friedman J. S., Knaus U. G. (2010) Mol. Cell. Biol. 30, 961–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Serrander L., Cartier L., Bedard K., Banfi B., Lardy B., Plastre O., Sienkiewicz A., Fórró L., Schlegel W., Krause K. H. (2007) Biochem. J. 406, 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nisimoto Y., Jackson H. M., Ogawa H., Kawahara T., Lambeth J. D. (2010) Biochemistry 49, 2433–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 16. Helmcke I., Heumüller S., Tikkanen R., Schröder K., Brandes R. P. (2009) Antioxid. Redox Signal. 11, 1279–1287 [DOI] [PubMed] [Google Scholar]
- 17. Anilkumar N., Weber R., Zhang M., Brewer A., Shah A. M. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 18. Zhang L., Nguyen M. V., Lardy B., Jesaitis A. J., Grichine A., Rousset F., Talbot M., Paclet M. H., Qian G., Morel F. (2011) Biochimie 93, 457–468 [DOI] [PubMed] [Google Scholar]
- 19. Geiszt M., Witta J., Baffi J., Lekstrom K., Leto T. L. (2003) FASEB J. 17, 1502–1504 [DOI] [PubMed] [Google Scholar]
- 20. Forteza R., Salathe M., Miot F., Forteza R., Conner G. E. (2005) Am. J. Respir. Cell Mol. Biol. 32, 462–469 [DOI] [PubMed] [Google Scholar]
- 21. Leto T. L., Geiszt M. (2006) Antioxid. Redox. Signal. 8, 1549–1561 [DOI] [PubMed] [Google Scholar]
- 22. Meitzler J. L., Ortiz de Montellano P. R. (2009) J. Biol. Chem. 284, 18634–18643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ameziane-El-Hassani R., Morand S., Boucher J. L., Frapart Y. M., Apostolou D., Agnandji D., Gnidehou S., Ohayon R., Noël-Hudson M. S., Francon J., Lalaoui K., Virion A., Dupuy C. (2005) J. Biol. Chem. 280, 30046–30054 [DOI] [PubMed] [Google Scholar]
- 24. Morand S., Ueyama T., Tsujibe S., Saito N., Korzeniowska A., Leto T. L. (2009) FASEB J. 23, 1205–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. DeLeo F. R., Burritt J. B., Yu L., Jesaitis A. J., Dinauer M. C., Nauseef W. M. (2000) J. Biol. Chem. 275, 13986–13993 [DOI] [PubMed] [Google Scholar]
- 26. Lyle A. N., Deshpande N. N., Taniyama Y., Seidel-Rogol B., Pounkova L., Du P., Papaharalambus C., Lassègue B., Griendling K. K. (2009) Circ. Res. 105, 249–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strange R. W., Antonyuk S., Hough M. A., Doucette P. A., Rodriguez J. A., Hart P. J., Hayward L. J., Valentine J. S., Hasnain S. S. (2003) J. Mol. Biol. 328, 877–891 [DOI] [PubMed] [Google Scholar]
- 28. Yu L., Quinn M. T., Cross A. R., Dinauer M. C. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 7993–7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Holliday G. L., Mitchell J. B., Thornton J. M. (2009) J. Mol. Biol. 390, 560–577 [DOI] [PubMed] [Google Scholar]
- 30. Marklund S. (1976) J. Biol. Chem. 251, 7504–7507 [PubMed] [Google Scholar]
- 31. Nishino T., Okamoto K., Eger B. T., Pai E. F., Nishino T. (2008) FEBS J. 275, 3278–3289 [DOI] [PubMed] [Google Scholar]
- 32. Asai R., Nishino T., Matsumura T., Okamoto K., Igarashi K., Pai E. F., Nishino T. (2007) J. Biochem. 141, 525–534 [DOI] [PubMed] [Google Scholar]
- 33. Harris C. M., Massey V. (1997) J. Biol. Chem. 272, 8370–8379 [DOI] [PubMed] [Google Scholar]
- 34. Wu R. F., Ma Z., Liu Z., Terada L. S. (2010) Mol. Cell. Biol. 30, 3553–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kawahara T., Jackson H. M., Smith S. M., Simpson S. M., Lambeth J. D. (2011) Biochemistry, in press [DOI] [PMC free article] [PubMed] [Google Scholar]