Abstract
Diabetes affects many organs including the stomach. Altered number and function of interstitial cells of Cajal (ICC), the gastrointestinal pacemaker cells, underlie a number of gastrointestinal motility disorders, including diabetic gastroparesis. In the muscle layers, ICC selectively express Ano1, thought to underlie classical Ca2+-activated Cl− currents. Mice homozygous for Ano1 knock-out exhibit abnormal ICC function and motility. Several transcripts for Ano1 are generated by alternative splicing of four exons. Here, we report expression levels of transcripts encoded by alternative splicing of Ano1 gene in gastric muscles of patients with diabetic gastroparesis and nondiabetic control tissues. Expression of mRNA from two alternatively transcribed exons are significantly different between patients and controls. Furthermore, patients with diabetic gastroparesis express mRNA for a previously unknown variant of Ano1. The 5′ end of this novel variant lacks exons 1 and 2 and part of exon 3. Expression of this variant in HEK cells produces a decreased density of Ca2+-activated Cl− currents that exhibit slower kinetics compared with the full-length Ano1. These results identify important changes in expression and splicing of Ano1 in patients with diabetic gastroparesis that alter the electrophysiological properties of the channel. Changes in Ano1 expression in ICC may directly contribute to diabetic gastroparesis.
Keywords: Calcium, Chloride Channels, Diabetes, Gene Transcription, RNA Splicing, Interstitial Cell of Cajal, Calcium-activated Chloride Channel, Gastric Motility, Patch Clamp
Introduction
Ca2+-activated Cl− channels are essential for a variety of physiological functions including sensation (1), smooth muscle contraction (2), and secretion (3). Recently Ano1 (also known as TMEM16A) was identified as a Ca2+-activated Cl− channel expressed in epithelial cells of many organs including lung (4), salivary glands, and kidney (5, 6). The voltage- and Ca2+-dependent properties of the heterologously expressed channel suggest that it is responsible for a significant subset of “classical” endogenous Ca2+-activated Cl− currents. It may also have a role in the pathology of several tumors, including gastrointestinal stromal tumors, where Ano1 is highly expressed (7).
In the muscle layers of the gastrointestinal tract, Ano1 is selectively expressed in the interstitial cells of Cajal (ICC)2 (8) and appears to play an important role in the electrical activity of ICC (9, 10). ICC are required for normal gastrointestinal motility. Abnormalities in both the number and function of ICC are associated with motility disorders such as slow transit constipation and gastroparesis, in particular diabetic gastroparesis (see Ref. 11 for a review). Gastroparesis is a disorder of the stomach characterized by a delay in gastric emptying of solids and/or liquids in the absence of mechanical obstruction. Nausea and vomiting are frequent symptoms associated with delayed gastric emptying. Gastroparesis is a well recognized complication of diabetes, but the pathophysiology of this disease is still not understood. Multiple cellular defects have been reported in diabetic gastroparesis (12) with the most common defect being the loss or altered function of ICC both in animal models (13, 14) and in human studies (15–17).
Pacemaker activity of ICC (9) and generation of slow waves (10) have been postulated to depend on Ano1 function. The Ano1 gene contains 26 exons, and several transcripts of Ano1 have been identified that are generated by alternative inclusion/skipping of coding regions (segments named a, b, c, and d in Ref. 4), resulting in proteins containing eight transmembrane spanning domains, which range in size from 840 to 1008 amino acids (4). It has been suggested that segment a, corresponding to exon 1 and part of exon 2, is skipped when an alternate promoter is used (18); the identity of segment a as a distinct splice variant is not clear. The resulting protein lacks the first 116 amino acids. The other segments, whose inclusion/skipping is due to alternative splicing of single exons, correspond to exon 6b (segment b), localized also in the N terminus of the protein, and exons 13 (segment c) and 15 (segment d), localized in the first intracellular loop. Furthermore, splicing of exons 6b, 13, and 15 is differentially processed across various organs (18). Exons 6b and 13 were shown to influence the Ca2+ sensitivity and voltage dependence of Ano1 currents, respectively (18). The other alternatively spliced exons have not been functionally characterized. All isoforms of Ano1 examined to date have produced chloride currents when expressed in HEK cells including the isoform without all four alternative segments (4). Recently, Ferrera et al. (18) failed to identify transcripts lacking segment a. They concluded that its inclusion/exclusion could be under the control of an alternate promoter and restricted to particular physiological or pathological conditions.
Alteration of the equilibrium between different Ano1 proteins could potentially have physiological and pathological effects. Splicing is the process by which exons are joined to form mRNA through the removal of introns contained in the precursor heterogeneous nuclear RNA. Alongside its established role in expansion of transcriptome and proteome complexity, alternative splicing is also emerging as a major mechanism for modulation of gene expression (19). Likewise, the use of alternative promoters can vary the expression and function of proteins (20). We hypothesized that the relative expression levels of Ano1 variants are associated with diabetic gastroparesis, a disease involving well established changes in ICC, an Ano1-expressing cell type. In this study, we show significant differences in expression of mRNA from alternatively spliced exons in patients with diabetic gastroparesis when compared with age- and sex-matched nondiabetic controls. We also identify mRNA for a novel variant of Ano1 with an alternative 5′ end that is expressed in patients with diabetic gastroparesis. Expression of this isoform of Ano1 results in altered current densities and activation kinetics compared with expression of the full-length isoform. We conclude that regulation of Ano1 expression is complex and results in different electrophysiological properties of Ca2+-activated Cl− currents in diseased tissue.
EXPERIMENTAL PROCEDURES
Tissue Collection
Full thickness gastric biopsies were collected from the anterior aspect of the stomach. Tissue was obtained from 12 patients with diabetic and 8 with idiopathic gastroparesis undergoing surgery for placement of a gastric stimulator following institutional review board-approved protocols (Table 1 and supplemental Table S1, respectively). Control tissue was from 12 age- and sex-matched nondiabetic and 7 diabetic patients undergoing duodenal switch gastric bypass surgery following institutional review board-approved protocols (Table 1 and supplemental Table S1, respectively). The location of the tissue for the controls was matched to the patients. All of the patients with gastroparesis had delayed gastric emptying on a standardized radionuclide gastric emptying test, and the diabetic patients presented either type 1 or type 2 diabetes.
TABLE 1.
Demographics of patients who provided tissue
| Nondiabetic control |
Diabetic gastroparesis |
||||
|---|---|---|---|---|---|
| Sex | Age | Sex | Age | Type | |
| 1 | Female | 49 | Female | 49 | 1 |
| 2 | Male | 64 | Male | 68 | 2 |
| 3 | Male | 36 | Male | 34 | 1 |
| 4 | Female | 40 | Female | 35 | 1 |
| 5 | Female | 48 | Female | 50 | 1 |
| 6 | Male | 39 | Male | 39 | 1 |
| 7 | Female | 37 | Female | 33 | 1 |
| 8 | Female | 30 | Female | 30 | 2 |
| 9 | Female | 26 | Female | 25 | 1 |
| 10 | Female | 58 | Female | 55 | 2 |
| 11 | Female | 52 | Female | 50 | 2 |
| 12 | Male | 58 | Male | 56 | 2 |
Immunofluorescence
Tissue was fixed in 4% paraformaldehyde (Sigma-Aldrich) in 1× phosphate buffer for 24 h and then washed five times over 1 h in 0.1 m PBS. After overnight incubation at 4 °C in a solution containing 30% sucrose (Sigma-Aldrich) in 1× PBS, the specimens were cut in cross-section (parallel to the circular muscle) and frozen in Tissue-Tek OCT compound (Electron Microscopy Sciences). 12-μm cryostat sections were cut, and slides were stored at −80 °C until use. For immunofluorescence staining, the sections were warmed up to room temperature and rinsed twice in 1× PBS followed by a blocking step for nonspecific antibody binding with a solution of 1× PBS containing 10% normal donkey serum (Jackson ImmunoResearch Lab) and 0.3% Triton X-100 (Pierce) for an hour at room temperature. Antibodies against Kit (Lab Vision Corporation) were diluted to 500 ng/ml in 1× PBS, 5% normal donkey serum, 0.3% Triton X-100 and incubated overnight at 4 °C. Next, the slides were rinsed in 1× PBS three times, followed by a 1-h incubation with Cy-3-conjugated secondary antibodies diluted in 1× PBS and 0.5% Tween 20. For Ano1 staining, the sections were subjected to antigen retrieval, carried out by the steamer method with the use of preheated antigen retrieval solution (Dako) for 15 min. After washing in PBS, tissues were subjected to a blocking step for 2 h at room temperature in 0.3% Triton X-100 plus 1% BSA in PBS, followed by incubation overnight at 4 °C with an in-house primary antibody for Ano1 diluted to 160 ng/ml in blocking solution. The next day, the slides were rinsed in 1× PBS three times, followed by 1 h of incubation with Cy-5-conjugated secondary antibodies diluted in blocking solution. Finally, the slides were rinsed three times in 1× PBS and mounted in SlowFade Gold with DAPI (Invitrogen).
To quantify ICC bodies, two or three nonadjacent slides were analyzed per patient. The fields were selected using an automated stage. ICC cell bodies were counted from 38–40 fields per patient with 40× or 60× objectives on an Olympus FV300 confocal microscope. Before counting, the images were renamed using a random number generator. An ICC body was defined as a Kit-positive structure with a DAPI positive nucleus within the structure. Mast cells were excluded by their larger, more circular appearance and brighter fluorescence.
Quantitative RT-PCR
For RNA extraction, the tissue collected as described above was immediately frozen in liquid nitrogen, and RNA was extracted using the RNA-Bee solution (Tel-Test Inc.) according to the manufacturer's instructions, followed by an in-column clean-up with RNeasy kit (Invitrogen). RT was performed using SuperScript VILO enzyme (Invitrogen), and the reaction protocol consisted of annealing at 25 °C for 10 min, followed by one cycle at 42 °C for 60 min and terminated by incubation at 85 °C for 5 min. The cDNA was then used for real time PCR using the Prism 7000 Sequence Detector (Applied Biosystems) with RT2 SYBR Green/ROX Master Mix (SABioscience) and primers specific for the exon of interest. The standard protocol used was the following: 10 min at 95 °C to activate the Taq polymerase followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Melting curves generated with a dissociation protocol were used to ensure the specificity of primers for a single product for each reaction. Plasmid DNA containing the inserted PCR products were constructed to verify by sequencing the identity of the products and used to generate standard curves of concentration of plasmid DNA versus threshold cycle number for each product of interest with the aim to convert cycle number to transcript copy number.
The expression of the exons of interest was normalized to the expression of Ano1, measured by using primers annealing in a region of the gene not subjected to alternative splicing. The sequences of all the primers used for the quantitative PCR experiments are listed in supplemental Table S2.
Construction of Expression Vectors for Ano1 Isoforms and Transfection in HEK293 Cells
The expression vector containing the full-length Ano1 (Ano1) was constructed as previously described (4). The other Ano1 isoforms were constructed by site-directed mutagenesis introducing the desired mutations on the full-length expression vector using the QuikChange XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All of the constructs were verified by sequencing.
The Ano1Δ1,2(5′) isoform was constructed by introducing a point mutation in the starting ATG causing a G to C base change in position 3 in exon 1. This resulted in a substitution of the starting methionine with an isoleucine. The next ATG, in-frame with the expression vector promoter, is in position 349 in exon 2, right after the end of segment a.
A similar mutation from ATG to ATC in position 349 was introduced to construct Ano1Δ1,2,3(5′) isoform identified in patients with diabetic gastroparesis. The next ATG in-frame is in position 526 in exon 3, corresponding to the one predicted as possible translational start site in position 866 by the NetStart server (see supplemental Fig. S2).
LipofectamineTM 2000 reagent (Invitrogen) was used to transiently co-transfect vectors containing green fluorescent protein pEGFP-C1 (Clontech) and the Ano1 isoform of interest into HEK293 cells (ATCC). Transfected cells were identified by fluorescence microscopy prior to patch clamping.
Electrophysiology
Electrodes were prepared by pulling KG-12 glass on a Sutter P-97 puller (Sutter Instrument Co.), fire polishing tips to 2–5 MΩ, and then coating glass with R6101 compound (Dow Corning). HEK293 cells expressing the following constructs: full-length Ano1 (Ano1), Ano1Δ1,2(5′), or Ano1Δ1,2,3(5′) and co-transfected fluorescent marker GFP, were patch-clamped (Axopatch 200B, Digidata 1322A, and pCLAMP 9 software; Molecular Devices) in standard whole cell configuration in a glucose-free N-methyl d-glucamine extracellular solution (149.2 mm NMDG+, 4.74 mm K+, 2.54 mm Ca2+, 0.01 mm Gd3+, 159 mm Cl−, 5 mm HEPES, pH 7.35; osmolality, 290 mmol/kg) with CsCl and 500 or 50 nm free Ca2+ in the pipette (145 mm Cs+, 5 mm Na+, 5 mm Mg2+, 1.27 or 0.3 mm Ca2+, 162.5 or 160.6 mm Cl−, 2 mm EGTA, 5 mm HEPES, pH 7.2; osmolality, 290 mmol/kg). Free Ca2+ was calculated using an online software tool. Under these conditions, Cs+, NMDG+, and Gd3+ block K+, Na+, and nonselective cation currents, and equimolar Cl− results in a reversal potential at 0 mV.
The cells were held at −60 mV between 1-s 10-mV voltage steps from −100 to +120 mV. Start-to-start time between sweeps was 5 s. The data were analyzed using Clampfit and Excel (Microsoft). In current-voltage (I-V) relationships (see Figs. 5B and 7B and supplemental Fig. S3B), Cl− currents at 1 s are shown as a fraction of total cell capacitance (pA/pF). Nontransfected cells displayed currents <2 pA/pF in high or low free Ca2+ (data not shown). Activation or deactivation kinetics (Figs. 6 and 8 and supplemental Fig. S3, C and D) were fitted from 0.050 to 1.000 s or from 1.050 to 1.250 s, respectively, by a one-term weighted exponential function: f(t) = K0(f1e−t/τ(1)) + C, where τ1 was the time constant of activation or deactivation. Significance was determined by a two-tailed unpaired Student's t test. A p value less than 0.05 was considered significant.
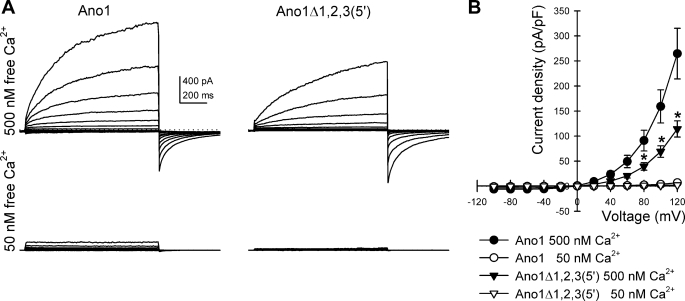
FIGURE 5.
Ano1Δ1,2(5′) does not show altered current density. A, outwardly rectifying chloride currents recorded from HEK293 cells transfected with full-length Ano1 or with Ano1 isoform without exon 1 and the 5′ end of exon 2 (Ano1Δ1,2(5′)) at a free intracellular Ca2+ concentration of 50 and 500 nm showing the Ca2+ dependence of the chloride currents. Dotted line, 0 pA. B, current-voltage graph for chloride currents at 1 s showing a difference in the current density of the channel at 500 (filled symbols) and 50 nm (empty symbols) calcium concentrations, but no significant difference between the full-length (circle) and mutated (triangle) isoforms of Ano1 tested (n = 10, p < 0.01 low to high free Ca2+ for each isoform, p > 0.05 between isoforms at high free Ca2+).
FIGURE 7.
Ano1Δ1,2,3(5′) shows altered current density at high voltages. A, outwardly rectifying chloride currents recorded from HEK293 cells transfected with full-length Ano1 or with Ano1 isoform with alternative 5′ end (Ano1Δ1,2,3(5′)) at a free intracellular Ca2+ concentration of 50 and 500 nm showing that the Ca2+ dependence of the chloride currents was preserved. Dotted line, 0 pA. B, current-voltage graph for the chloride currents at 1 s showing a difference in current density at 500 (filled symbols) and 50 nm (empty symbols) Ca2+ concentrations. At 500 nm, significant differences were observed between the full-length (circle) and Ano1Δ1,2,3(5′) isoform of Ano1 (triangle) tested at voltages higher than 80 mV (n = 8–10). *, p < 0.05.
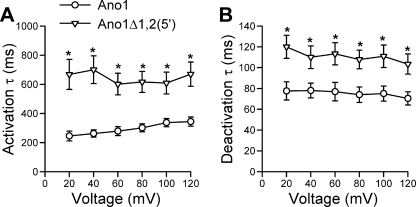
FIGURE 6.
Ano1Δ1,2(5′) shows altered kinetics. Time constant (τ) of activation (A) and of deactivation (B) are significantly slower in the Ano1Δ1,2(5′) isoform (triangle) than in the full-length Ano1 construct (circle) at voltage steps from −60 mV holding potential to 20–120 mV (n = 10). *, p < 0.01.
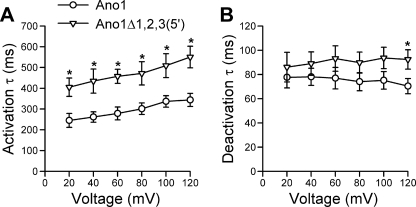
FIGURE 8.
Ano1Δ1,2,3(5′) shows altered kinetics. A, time constant (τ) of activation was significantly slower in Ano1Δ1,2,3(5′) (triangle) than in the full-length control (circle) at voltage steps 20 through 120 mV (n = 8–10 each). *, p < 0.05. B, significant differences were observed in the time constant of deactivation only at 120 mV (n = 8–10). *, p < 0.05.
Single channel recordings were made with an Axon Instruments 200B patch clamp amplifier with an accompanying Cyberamp. The current records were sampled at 10 kHz and filtered through a 2000-Hz analog 8-pole Bessel filter. The electrodes were pulled from Schott 8250 glass using a two stage pull on the Sutter puller. For the measurements, both the electrode and the bath were filled with a solution containing 150 mm NMDG Cl, 2.5 mm CaCl2, and 5 mm HEPES. The fine-tipped electrodes when filled had resistances in the 23–30-megaOhm range. The recordings were made from inside-out patches with the electrode positioned very near the surface of the bathing medium so as to reduce capacitance and noise. When required, the records were additionally filtered at 500 Hz using a digital eight-pole Bessel filter.
Rapid Amplification of cDNA Ends
The rapid amplification of cDNA ends (RACE) technique was used to isolate full-length 5′ ends of Ano1 mRNA, and it was performed using the SMARTerTM RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. Briefly, total RNA was extracted from control and patient tissues as described above and cleaned up using the in-column treatment of the RNeasy kit (Invitrogen) according to directions. 1 μg of RNA was reverse-transcribed into first strand cDNA using SMARTer II A oligonucleotide by incubation at 42 °C for 90 min, followed by 70 °C for 10 min. The resulting cDNA was diluted to 100 μl with Tricine-EDTA buffer and used for a PCR performed using a universal primer mix from the kit, which binds to the 3′ ends of first-strand cDNA, in combination with a primer specific for exon 3 of Ano1 (5′-CGGCCTCTCTGCACAGCACGTTCCAGGGG-3′). The reaction mixtures were denatured at 94 °C for 3 min followed by 38 cycles of 94 °C for 30 s and 68 °C for 3 min and then 1 cycle of 68 °C for 3 min using the Advantage-GC2 polymerase mix (Clontech). PCR from the samples were then resolved by electrophoresis in a 1% agarose gel, and the bands were gel-purified, cloned into the pCR2.1 vector of a TA cloning kit (Invitrogen), and subjected to DNA sequence analysis.
Identification of Possible Translational Start Sites for the Alternative 5′ End of Ano1
We used the NetStart 1.0 prediction server freely available on the web. The NetStart server produces neural network predictions of translation start in vertebrate and Arabidopsis thaliana nucleotide sequences (21). The newly identified Ano1 with an alternative 5′ end was used as input, and the predicted translation start sites were reported in a table showing the positions and the scores of all instances of ATG in the sequence (supplemental Fig. S2). Scores greater than 0.5 represented a probable translation start site.
RESULTS
Patients with Diabetic Gastroparesis Had Fewer Interstitial Cells of Cajal in the Smooth Muscle Layers of the Stomach
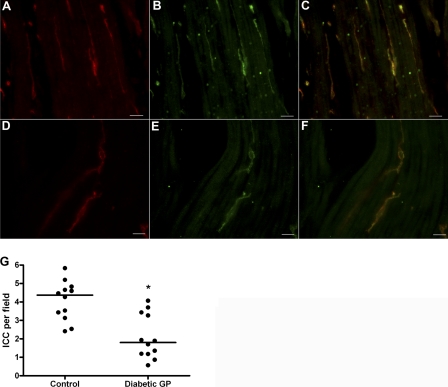
To determine whether patients with diabetic gastroparesis had altered expression of ICC in the gastric smooth muscle layers compared with controls, we labeled sections of stomach smooth muscle tissue collected from 12 patients with diabetic gastroparesis (Fig. 1A) and 12 age- and sex-matched nondiabetic controls (Fig. 1D) with antibodies against Kit, a marker for ICC. The ages and sexes of the patients whose tissue was used for this study are shown in Table 1. ICC numbers were markedly fewer in the entire thickness of the stomach smooth muscle in patients with diabetic gastroparesis (4.4, 3.2–4.7 ICC/field in controls compared with 1.8, 1.2–3.3 in patients, p < 0.05; the data are median, interquartile ranges; Fig. 1G). Double labeling of sections from control and patients with antibodies to Ano1 (Fig. 1, B and E, respectively) resulted in signals completely overlapping to Kit staining (Fig. 1, C and F, show the merged signals). These data suggest that both Kit and Ano1 are decreased in diabetic gastroparesis.
FIGURE 1.
Patients with diabetic gastroparesis had fewer interstitial cells of Cajal in the smooth muscle layers of the stomach. A, B, D, and E, representative images showing labeling for Kit (A and D) and for Ano1 (B and E) as markers for ICC on stomach section of nondiabetic control and patient with diabetic gastroparesis, respectively. C and F show the merged images. G, quantification of ICC for nondiabetic controls and diabetic gastroparesis. The circles represent individual patients, and the lines are the median values. Scale bars, 20 μm. *, p < 0.001 (Mann-Whitney test).
Expression of Ano1 Transcriptional Variants Is Altered in Diabetic Gastroparesis
To test whether expression of transcriptional variants of Ano1 was different in patients with diabetic gastroparesis compared with controls, we performed quantitative RT-PCR on total RNA extracted from the gastric smooth muscles of the two patient groups. We used primers specific for each alternatively spliced exon. Fig. 2A is a schematic of the 5′ end of Ano1 including the first three exons. Exon 1 together with the 5′ region of exon 2 constitutes segment a (in black). As shown in Fig. 2B, the expression of segment a was significantly reduced in patients with diabetic gastroparesis. Approximately 40% of the Ano1 transcripts in tissues from patients with diabetic gastroparesis contained segment a (0.41, 0.28–0.47; Table 2), compared with >80% of transcripts from control tissues (0.81, 0.64–0.92; Table 2, p < 0.004). As shown in Fig. 2C, the proportion of the transcripts that included sequence from exon 6b (segment b) was significantly higher in patients with diabetic gastroparesis (0.38, 0.29–0.68; Table 2) compared with controls (0.25, 0.16–0.29; Table 2, p < 0.004). No significant differences were found between the two groups with respect to expression of Ano1 transcriptional variants that included sequences from exon 13 (segment c; Fig. 2D) or exon 15 (segment d; Fig. 2E) (values in Table 2). The results from the gastrointestinal tract of control patients were consistent with expression data of variants in control tissues from other parts of the body (18). No correlation was observed between the number of ICC per field and the expression of each of the alternatively transcribed exons examined in samples from patients with diabetic gastroparesis (data not shown). There was also no correlation between the expression of either segment a or exon 6b and the duration of diabetes or the blood glucose level (measure as level of HbA1C), nor was there association with sex of the patient or type of diabetes for expression of exon 6b (data not shown). However, expression of segment a was higher in gastroparesis patients with type 2 diabetes compared with patients with type 1 (Fig. 2F).
FIGURE 2.
Expression of Ano1 transcriptional variants is altered in diabetic gastroparesis. A, schematic of the 5′ end of Ano1. Segment a (exon 1 and the 5′ region of exon 2) is indicated in black. B and C, the expression of transcripts that contain segment a (comprised of exons 1 and 2) was significantly reduced (B), whereas the expression of exon 6b was increased in patients with diabetic gastroparesis compared with controls (C). D and E, no difference was detected for the expression of exons 13 (D) and 15 (E). F, graph shows lower expression of mRNA for segment a in patients with type 1 diabetes versus type 2. In each graph, the circles represent individual patients, and the lines are the median values; the data are expressed as ratios to the total Ano1 in each sample. *, p < 0.004; **, p < 0.05 (Mann-Whitney test).
TABLE 2.
Quantification of expression of Ano1 alternatively spliced exons
The data are reported as median and interquartile ranges.
| Exon 1 | Exon 6b | Exon 13 | Exon 15 | |
|---|---|---|---|---|
| Control | 0.81, 0.64–0.92 | 0.25, 0.16–0.29 | 0.23, 0.18–0.28 | 0.19, 0.14–0.21 |
| Diabetic gastroparesis | 0.41, 0.28–0.47a | 0.38, 0.29–0.68a | 0.28, 0.19–0.43 | 0.2, 0.1–0.43 |
a p < 0.004 using Mann-Whitney test.
Characterization of the 5′ End of Ano1 mRNA by RACE
To investigate the predicted splicing pattern of Ano1, we analyzed the whole Ano1 gene using ASPicDB. This data base resource provides access to alternative splicing patterns of human genes based on published data on gene sequences and transcripts, including functional annotations (22). ASPicDB showed a complex predicted alternative splicing pattern for human Ano1 (supplemental Fig. S1) by comparison between the Ano1 gene (genomic location between bp 69602050 and 69713299 of chromosome 11), the transcript of the reference sequence (accession number NM_018043), and 156 published expressed sequence tags. Many of those transcripts had a shorter 5′ end with a missing exon 1 (such as in transcripts 3, 5, 6, 9, 11, 12, and 14 in supplemental Fig. S1) or both exons 1 and 2 (transcripts 7, 13, 15, and 16 in supplemental Fig. S1).
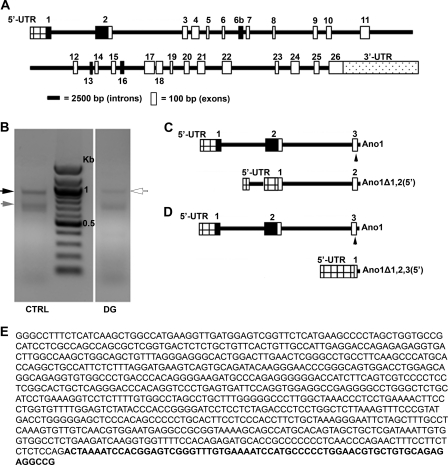
Given this information and the results obtained from quantification of segment a (the 5′ end of Ano1) in patients with diabetic gastroparesis, we performed 5′ RACE on total RNA extracted from gastric smooth muscle tissue from four patients with diabetic gastroparesis and four nondiabetic controls to obtain the complete sequence of the 5′ end of the Ano1 mRNA transcripts. Fig. 3A represents a schematic of the human Ano1 gene including all 26 known exons. The primer used for the RACE reaction was located in exon 3 of Ano1 and was designed based on the published reference sequence (GenBankTM accession number NM_018043.5). RACE amplification products separated electrophoretically as one distinct band and multiple smaller sized bands (Fig. 3B). DNA obtained from these two regions of each gel was subcloned into a plasmid vector, and the sequences were determined. In control tissues, a band of 845 bp contained the sequence corresponding to the published 5′ end of the coding region of the Ano1 gene (Fig. 3B, black arrow). No other clones were obtained from this band. From the smaller amplification products of control tissues (Fig. 3B, gray arrow), a 650-bp sequence was identified as an alternative isoform of Ano1 lacking exon 1 and containing a 217-bp sequence located in intron 1, 2081 bp upstream of the start of exon 2, and all of exon 2 (Fig. 3C). We therefore called this isoform Ano1Δ1,2(5′) and identified it as corresponding to the 5′ end of the computationally theoretical transcripts 3, 5, 9, and 11 predicted by ASPicDB (in supplemental Fig. S1) and matching a previously identified sequence (GenBankTM accession number BC033036.2).
FIGURE 3.
Identification of 5′ end of Ano1 mRNA by RACE. A, schematic of the Ano1 gene with open bars indicating exons and lines indicating introns. Filled boxes indicate alternatively spliced exons. The cross-hatched box indicates the 5′ UTR, whereas the dotted box represents the 3′-UTR. B, representative agarose gel for bands obtained after RACE for controls (CTRL) and diabetic gastroparesis patients (DG). Black arrow, full-length Ano1; gray arrow, Ano1Δ1,2(5′); white arrow, Ano1Δ1,2,3(5′) and full-length Ano1. C, schematic of the 5′ end of Ano1Δ1,2(5′), the isoform identified after RACE in control samples, compared with full-length Ano1. The arrowhead in exon 3 indicates the position of the primer used for RACE. D, schematic of the 5′ end of Ano1Δ1,2,3(5′), the isoform identified in patients with diabetic gastroparesis, compared with full-length Ano1. E, sequence of the newly identified Ano1Δ1,2,3(5′) with the beginning of exon 3 indicated in bold.
In tissues from patients with diabetic gastroparesis, we obtained two cDNA products that ran on the gel as a single band (Fig. 3B, white arrow). One product contained 845 nucleotides and corresponded to the one identified in the control samples. The other product contained 846 nucleotides and was identified as an Ano1 transcript with an alternative 5′ end completely lacking exons 1 and 2 and containing a 779-bp sequence located in intron 2 that was immediately upstream and contiguous with 67 bp in exon 3. This sequence is illustrated schematically in Fig. 3D and reported in Fig. 3E (GenBankTM accession number HQ418153). We defined this novel isoform Ano1Δ1,2,3(5′). Other products cloned from the smaller sized multiple bands from both control and patient tissues were identified as mixed fragments of unrelated genes.
Expression of mRNA of Ano1 with the Alternative 5′ End, Ano1Δ1,2,3(5′), Is Increased in Gastroparesis
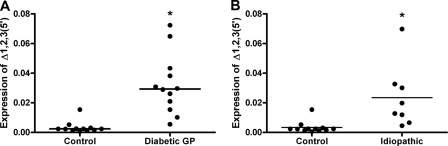
To quantify Ano1Δ1,2,3(5′) in gastric smooth muscle tissue collected from patients with diabetic gastroparesis, we designed specific primers within the newly identified sequence (supplemental Table S2). Quantitative RT-PCR established that expression of this isoform of Ano1 was extremely low in control tissue (0.002, 0.0015–0.0027), whereas it was about 10 times higher in tissues from patients with diabetic gastroparesis (0.029, 0.018–0.041, p < 0.0001; Fig. 4A). Ano1Δ1,2,3(5′) was similarly increased in patients with idiopathic gastroparesis (0.016, 0.0045–0.031, p < 0.001; Fig. 4B) compared with controls.
FIGURE 4.
Expression of Ano1Δ1,2,3(5′) is increased in gastroparesis. A, graph showing the increased expression of the mRNA for Ano1Δ1,2,3(5′) in patients with diabetic gastroparesis versus nondiabetic controls. B, graph showing the increased expression of the mRNA for Ano1Δ1,2,3(5′) in patients with idiopathic gastroparesis versus nondiabetic controls. The data are expressed as ratios to the total Ano1 in each sample. The circles indicate individual patients, and the lines are the median values. *, p < 0.001 (Mann-Whitney test).
Electrophysiological Properties of Ano1Δ1,2(5′)
The presence of multiple transcripts of the Ano1 gene that have altered expression in diseased tissue raised the possibility that the various proteins encoded by these transcripts have differing functional properties. To test this hypothesis, we constructed expression vectors bearing the isoforms of Ano1 with alternative 5′ ends identified in control and patients by RACE. For Ano1Δ1,2(5′), the isoform lacking exon 1 and part of exon 2, we used site-directed mutagenesis to disrupt the ATG translational start site in exon 1. This forced translation to start at the second available in-frame ATG at the end of exon 2. The deleted region represents segment a, and this construct corresponds to Ano1Δ1,2(5′). Whole cell patch clamp recordings were obtained from HEK293 cells transfected with this expression vector and compared with recordings from cells transfected with a plasmid containing the full-length Ano1 (Fig. 5). Transfections with both vectors resulted in expression of channels that produced Ca2+- and voltage-dependent outwardly rectifying currents. Current densities markedly increased when the intracellular concentration of Ca2+ was changed from 50 to 500 nm (at 120 mV, IANO1, 7.0 ± 1.2 to 264.6 ± 50.4 pA/pF, n = 10 each, p < 0.01; IAno1Δ1,2(5′), 2.3 ± 0.2 to 358.5 ± 128.6 pA/pF, n = 10 each, p < 0.01; Fig. 5A). At 500 nm free Ca2+, Cl− current densities were not significantly different in between the two isoforms (p > 0.05 at all voltage steps comparing IANO1 to IAno1Δ1,2(5′), n = 10 each; Fig. 5B). No Cl− currents were recorded in nontransfected HEK293 cells (INONTRANSFECTED < 2 pA/pF, n = 4; data not shown). The activation and deactivation kinetics of the Ano1 channel were fit well with a single exponential (tau). Significant differences were observed between the activation and deactivation rates of the two currents generated from the two constructs. The Ano1Δ1,2(5′) transcriptional variant was slower to activate (at 120 mV, τANO1 = 344.1 ± 31.4 ms versus τAno1Δ1,2(5′) = 669.4 ± 83.6 ms, n = 10 each, p < 0.01; Fig. 6A) and deactivate (at 120 mV, τANO1 = 70.4 ± 6.3 ms versus τAno1Δ1,2(5′) = 103.5 ± 9.7 ms, n = 10 each, p < 0.01; Fig. 6B). The activation rates for Ano1Δ1,2(5′) showed a loss of the voltage-dependent slowing seen for Ano1.
Electrophysiological Properties of Ano1Δ1,2,3(5′)
To characterize the electrophysiological properties of the newly identified isoform of Ano1 lacking exons 1 and 2 and the 5′ end of exon 3, we determined the possible translation start site of the protein. To do so we ran the newly identified sequence in the NetStart 1.0 prediction server (21) and obtained four possible translation initiation sites in position 49, 206, 313, and 866 (supplemental Fig. S2A). The first three putative start sites generated very short open reading frames (supplemental Fig. S2B, underlined); however, the translation initiation site in position 866 (supplemental Fig. S2B, boxed), located in a region of the transcript encoded by exon 3, was in-frame with the coding sequence for the full-length Ano1 protein. To test the function of this novel isoform of Ano1, a new expression vector bearing Ano1Δ1,2,3(5′) was generated, as described under “Experimental Procedures.”
Whole cell patch clamp recordings were obtained from HEK293 cells transfected with an expression vector bearing the Ano1Δ1,2,3(5′) isoform. Ano1Δ1,2,3(5′) was activated by Ca2+ (at 120 mV, IAno1Δ1,2,3(5′) from 50 to 500 nm free Ca2+, 2.2 ± 0.6 to 114.2 ± 16.2 pA/pF, n = 8, p < 0.01; Fig. 7A), but the current densities were lower when compared with full-length Ano1 at voltages higher than 80 mV at 500 nm Ca2+ (at 80 mV, IANO1, 91.1 ± 20.7 pA/pF (n = 10) versus IANO1Δ1,2,3(5′), 39.5 ± 7.5 pA/pF, (n = 8), p < 0.05; Fig. 7B). Significant differences were also observed in the kinetics of Ano1Δ1,2,3(5′). As for the Ano1Δ1,2(5′) (Fig. 6A), the activation rate for Ano1Δ1,2,3(5′) was slower when compared with full-length Ano1 (at 120 mV, τANO1 = 344.1 ± 31.4 ms versus τAno1Δ1,2,3(5′) = 550.4 ± 52.0 ms, n = 8–10, p < 0.05; Fig. 8A). Activation rates for Ano1Δ1,2,3(5′) also showed voltage-dependent slowing similar to Ano1 but lost in Ano1Δ1,2(5′) (Fig. 6A). However, unlike the differences in deactivation noted for Ano1 and Ano1Δ1,2(5′) (Fig. 6B), the deactivation rates for Ano1Δ1,2,3(5′) were similar to Ano1, differing only at very positive voltages (+120 mV) (τANO1 = 70.4 ± 6.3 ms versus τAno1Δ1,2,3(5′) = 92.3 ± 8.0 ms, n = 8–10, p < 0.05; Fig. 8B).
Single Channel Properties of Ano1 Isoforms
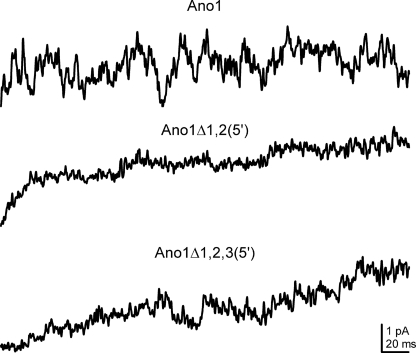
To study the kinetics of Ano1, we also carried out single channel recordings in the inside-out patch mode in cells transfected with expression vectors bearing the full-length Ano1 and both the isoforms with shorter 5′ ends (Fig. 9). Despite several measures to increase resolution, we were unable to definitely establish a single conductance suggesting a very low unitary conductance. This is consistent with a recent report (23). Multiple sweeps of single channel data were averaged for each expression vector to enable comparisons between single channel and whole cell current recordings. The averaged single channel traces showed rates of rise similar to the whole cell currents.
FIGURE 9.
Averaged traces obtained from inside-out patches from cells transfected with the different Ano1 isoforms. The traces show similar kinetics to the whole cell current but single channel openings could not be definitely resolved.
Electrophysiological Properties of Ano1Δ15
No difference in expression of the Ano1 transcript lacking exon 15 was found between tissue from patients with diabetic gastroparesis and controls. However, because the Ano1 transcript lacking exon 15 has not yet been characterized, we used whole cell patch clamp recordings from HEK293 cells transfected with this expression vector and compared them with recordings from cells transfected with a plasmid containing the full-length Ano1 (supplemental Fig. S3). Transfection of both expression vectors resulted in a calcium-dependent outwardly rectifying voltage-sensitive currents (at 120 mV, IANO1 as detailed above; IANO1Δ15, 7.6 ± 0.8 to 162.8 ± 45.0 pA/pF, n = 10 each, p < 0.01; supplemental Fig. S3A). No difference was detected between the two transcripts at 500 nm free Ca2+ (p > 0.05 comparing IANO1 to IANO1Δ15 at voltage steps to 40–120 mV, n = 10 each; supplemental Fig. S3B). Activation kinetics were significantly faster in Ano1Δ15 compared with full-length Ano1 (at 120 mV, τANO1 as detailed above versus τANO1Δ15 = 238.0 ± 13.6 ms, n = 10 each, p < 0.01; supplemental Fig. S3C). Ano1Δ15 deactivation was faster compared with full-length (at 120 mV, τANO1 as detailed above versus τANO1Δ15 = 51.7 ± 4.8 ms, n = 10 each, p < 0.01; supplemental Fig. S3D).
Identification and Quantification of 3′ End of Ano1 mRNA by Quantitative RT-PCR
Further analyses of the Ano1 gene using ASPicDB showed few transcripts with the alternative 3′ end. In particular, transcripts 8–14 in supplemental Fig. S1 are predicted to have shortened transcripts with an early stop codon after exon 19. For this reason, based on the sequence identified with this analysis, we designed primers specific for the alternative 3′ end of Ano1 and tested by quantitative RT-PCR the expression of this isoform in patients with diabetic gastroparesis compared with controls. As shown in supplemental Fig. S4, significant quantities of the transcripts truncated at the 3′ end of the sequence were detected in both sets of samples, but there was no difference between control and diabetic gastroparesis patients in the expression of mRNA for the isoform of Ano1 with shorter 3′ end.
DISCUSSION
The recent reports that Ano1 and other anoctamin proteins appear to function as classical Ca2+-activated Cl− channels have ignited significant interest in this new family of ion channels. In this study, we show that the expression of alternatively spliced exons of Ano1 is different in smooth muscle layers of gastric tissues from patients with diabetic gastroparesis when compared with matched nondiabetic controls.
An unexpected and intriguing result of this study relates to the inclusion/exclusion of segment a. By quantitative RT-PCR, we confirmed the published observation that Ano1 in nondiabetic controls nearly always includes the exons 1 and 2 constituting segment a (Fig. 2B). Surprisingly, in patients with diabetic gastroparesis, this was not the case, where many more transcripts lacking this segment were found. We also show that expression of segment a in patients with diabetic gastroparesis is associated with the type of diabetes as it was increased in patients with type 2 diabetes (Fig. 2F) compared with type 1. The exact identity and role of segment a is unclear. It has been hypothesized, without experimental data, that its expression could be restricted to a particular cell type or developmental stage or tightly regulated by use of an alternative promoter (18). Given the multiplicity of expression of Ano1 not only in terms of number of isoforms identified but also in the preferential expression of some of the isoforms in tissue from patients with diabetic gastroparesis and not controls, altered alternative splicing may contribute to pathological conditions including diabetes and complications of diabetes such as gastroparesis.
We also show that patients with diabetic gastroparesis express a previously unknown Ano1 variant that has an alternative translation start site in exon 3. Expression of this variant was 10-fold increased in the diabetic gastroparetic patients (Fig. 4). Despite the 10-fold increase, the full-length Ano1 was still the dominant variant. The electrophysiological properties of the variant were substantially different when compared with full-length Ano1. The electrical behavior of Ano1 is therefore dependent on differential expression of variants and is altered in disease. An immediate question is whether it is the diabetes or the gastroparesis that is associated with the altered expression of the previously unknown short Ano1 variant. We therefore looked for expression of this variant in a different group of patients with nondiabetic (idiopathic) gastroparesis and also found elevated expression of this variant, suggesting that it is the gastroparesis and not the diabetes that is associated with Ano1Δ1,2,3(5′) (Fig. 4). Interestingly, this novel isoform shows different electrophysiological properties not only when compared with the full-length Ano1 but even when compared with the splice variant lacking only exon 1 (Ano1Δ1,2(5′)). Ano1Δ1,2,3(5′) channels remained Ca2+- and voltage-sensitive but produced smaller current densities and activated slower than Ano1. However, the kinetics of deactivation in Ano1Δ1,2,3(5′) was preserved when compared with the full-length Ano1. Instead, Ano1Δ1,2(5′) channels retained Ca2+ and voltage sensitivity but gated (activate and deactivate) much slower than the full-length Ano1.
Our data (Figs. 2–9), taken together with published data (18) show that alternative splicing of Ano1 leads to changes in the electrophysiological properties of Ano1. In the gastrointestinal tract, this is of particular interest given that expression of Ano1 is restricted to ICC in the tissues that we examined and that Ano1 is proposed to play a critical role in the generation of the electrical slow wave (10), which is a key determinant of smooth muscle electrical and contractile activity (24–28). We found that in diabetic gastroparesis, inclusion of exon 6b was increased, suggesting, based on previous data (18), that the chloride currents will have decreased Ca2+ sensitivity. Given the critical role of Ca2+ cycling in generation and propagation of the slow wave (24), altered Ca2+ sensitivity of Ano1 variants will likely alter the kinetics, amplitude, and/or propagation of slow waves. Because Ano1 is implicated in the generation of the slow wave (10), this finding is expected to contribute to the electrophysiological dysfunction that leads to gastroparesis. Deletion of exon 13 is reported to occur in other tissues (18) and to alter voltage sensitivity of Ano1 but was not different in the two groups studied here, suggesting that it is not involved in the electrophysiological changes seen in diabetic gastroparesis. The changes in electrophysiological properties of Ano1 as a result of alternative splicing highlight the increasingly recognized role alternative splicing has in modifying the function of proteins (19). Alternative splicing appears to occur in most if not all organisms and markedly expands the number of proteins made by a cell through expression of multiple different mRNAs from the same gene. It is also a regulatory stage in the pathway of gene expression, and it is important for both the tissue specificity and regulation of protein expression levels. Use of alternative translation start sites is also a regulatory mechanism to encode functionally different protein isoforms and has been extensively studied in regulatory proteins such as MDM2 (29) and p53 (30) or receptors such as the glucocorticoid receptor (31).
Altered splicing may contribute to the temporal and spatial expression of functionally diverse isoforms of Ano1 or to the fine tuning of the ratio between the synthesized isoforms. Alteration of this mechanism could then lead to pathological conditions including diabetes and complications of diabetes such as gastroparesis. Disruption of alternative splicing patterns of a gene has been suggested to play a role in the pathogenesis of disease as a direct cause, but more often as a susceptibility factor (see Ref. 32 for a review). A well characterized example is the case of a family of neurodegenerative diseases termed tauopathies, caused by abnormalities in the microtubule associated protein tau encoded by the MAPT gene (33). The inclusion/exclusion of exon 10 of this gene determines the presence of either three or four imperfect repeats (3R or 4R) in the microtubule binding domain of tau. Alterations in the normal equimolar 3R to 4R ratio of tau, regardless of direction of the shift, affect tau function leading to neurodegeneration (34). For Ano1, the encoded channel protein has different properties based on the isoform expressed, and alternative splicing has previously been suggested as a mechanism for modulating the function of the Cl− channel and Cl− transport in different organs (4, 18). Moreover, differential expression of Ano1 isoforms in different human organs suggests that the channel may contribute differently to membrane currents in different organs (18). A shift in the splicing pattern of Ano1, such as the one observed here in patients with diabetic gastroparesis, can modify the balance between the levels of expression of its isoforms and contribute to development of disease.
In conclusion, in this study we show alternative splicing as a versatile mechanism to regulate Ano1 function. Moreover, we show that the expression of two out of four known transcriptional variants of Ano1 differs in gastroparesis patients compared with controls, and we identified a novel isoform of Ano1 with an alternative 5′ end with altered electrophysiological properties compared with the full-length isoform. Therefore, we identify Ano1 as a new potential molecular target not only in the etiology of diabetic gastroparesis but also as a molecule of interest to target to restore normal gastrointestinal motility.
Acknowledgments
We thank Kristy Zodrow for secretarial assistance and Gary Stoltz for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK57061, DK52766, DK84567, DK73983, and DK74008.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S5.
- ICC
- interstitial cells of Cajal
- RACE
- rapid amplification of cDNA ends
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Reisert J., Bauer P. J., Yau K. W., Frings S. (2003) J. Gen. Physiol. 122, 349–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Large W. A., Wang Q. (1996) Am. J. Physiol. 271, C435–C454 [DOI] [PubMed] [Google Scholar]
- 3. Kunzelmann K., Milenkovic V. M., Spitzner M., Soria R. B., Schreiber R. (2007) Pflugers Arch. 454, 879–889 [DOI] [PubMed] [Google Scholar]
- 4. Caputo A., Caci E., Ferrera L., Pedemonte N., Barsanti C., Sondo E., Pfeffer U., Ravazzolo R., Zegarra-Moran O., Galietta L. J. (2008) Science 322, 590–594 [DOI] [PubMed] [Google Scholar]
- 5. Yang Y. D., Cho H., Koo J. Y., Tak M. H., Cho Y., Shim W. S., Park S. P., Lee J., Lee B., Kim B. M., Raouf R., Shin Y. K., Oh U. (2008) Nature 455, 1210–1215 [DOI] [PubMed] [Google Scholar]
- 6. Schroeder B. C., Cheng T., Jan Y. N., Jan L. Y. (2008) Cell 134, 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Espinosa I., Lee C. H., Kim M. K., Rouse B. T., Subramanian S., Montgomery K., Varma S., Corless C. L., Heinrich M. C., Smith K. S., Wang Z., Rubin B., Nielsen T. O., Seitz R. S., Ross D. T., West R. B., Cleary M. L., van de Rijn M. (2008) Am J. Surg. Pathol. 32, 210–218 [DOI] [PubMed] [Google Scholar]
- 8. Gomez-Pinilla P. J., Gibbons S. J., Bardsley M. R., Lorincz A., Pozo M. J., Pasricha P. J., Van de Rijn M., West R. B., Sarr M. G., Kendrick M. L., Cima R. R., Dozois E. J., Larson D. W., Ordog T., Farrugia G. (2009) Am. J. Physiol. Gastrointest Liver Physiol. 296, G1370–G1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu M. H., Kim T. W., Ro S., Yan W., Ward S. M., Koh S. D., Sanders K. M. (2009) J. Physiol. 587, 4905–4918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hwang S. J., Blair P. J., Britton F. C., O'Driscoll K. E., Hennig G., Bayguinov Y. R., Rock J. R., Harfe B. D., Sanders K. M., Ward S. M. (2009) J. Physiol. 587, 4887–4904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Farrugia G. (2008) Neurogastroenterol. Motil. 20, (Suppl. 1) 54–63 [DOI] [PubMed] [Google Scholar]
- 12. He C. L., Soffer E. E., Ferris C. D., Walsh R. M., Szurszewski J. H., Farrugia G. (2001) Gastroenterology 121, 427–434 [DOI] [PubMed] [Google Scholar]
- 13. Ordög T., Takayama I., Cheung W. K., Ward S. M., Sanders K. M. (2000) Diabetes 49, 1731–1739 [DOI] [PubMed] [Google Scholar]
- 14. Choi K. M., Gibbons S. J., Roeder J. L., Lurken M. S., Zhu J., Wouters M. M., Miller S. M., Szurszewski J. H., Farrugia G. (2007) Neurogastroenterol. Motil. 19, 585–595 [DOI] [PubMed] [Google Scholar]
- 15. Iwasaki H., Kajimura M., Osawa S., Kanaoka S., Furuta T., Ikuma M., Hishida A. (2006) J. Gastroenterol. 41, 1076–1087 [DOI] [PubMed] [Google Scholar]
- 16. Pasricha P. J., Pehlivanov N. D., Gomez G., Vittal H., Lurken M. S., Farrugia G. (2008) BMC Gastroenterol. 8, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miller S. M., Narasimhan R. A., Schmalz P. F., Soffer E. E., Walsh R. M., Krishnamurthi V., Pasricha P. J., Szurszewski J. H., Farrugia G. (2008) Neurogastroenterol. Motil. 20, 349–357 [DOI] [PubMed] [Google Scholar]
- 18. Ferrera L., Caputo A., Ubby I., Bussani E., Zegarra-Moran O., Ravazzolo R., Pagani F., Galietta L. J. (2009) J. Biol. Chem. 284, 33360–33368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lareau L. F., Green R. E., Bhatnagar R. S., Brenner S. E. (2004) Curr. Opin. Struct Biol. 14, 273–282 [DOI] [PubMed] [Google Scholar]
- 20. Liu S. (2010) PLoS One 5, e9482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pedersen A. G., Nielsen H. (1997) Proc. Int. Conf. Intell. Syst. Mol. Biol. 5, 226–233 [PubMed] [Google Scholar]
- 22. Castrignanò T., D'Antonio M., Anselmo A., Carrabino D., D'Onorio De Meo A., D'Erchia A. M., Licciulli F., Mangiulli M., Mignone F., Pavesi G., Picardi E., Riva A., Rizzi R., Bonizzoni P., Pesole G. (2008) Bioinformatics 24, 1300–1304 [DOI] [PubMed] [Google Scholar]
- 23. Tian Y., Kongsuphol P., Hug M., Ousingsawat J., Witzgall R., Schreiber R., Kunzelmann K. (2011) FASEB J. 25, 1058–1068 [DOI] [PubMed] [Google Scholar]
- 24. Sanders K. M., Koh S. D., Ward S. M. (2006) Annu. Rev. Physiol. 68, 307–343 [DOI] [PubMed] [Google Scholar]
- 25. Ward S. M., Sanders K. M. (2006) J. Physiol. 576, 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farrugia G., Szurszewski J. H. (1999) Microsc. Res. Tech. 47, 321–324 [DOI] [PubMed] [Google Scholar]
- 27. Gibbons S. J., Farrugia G. (2004) J. Physiol. 556, 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kraichely R. E., Farrugia G. (2007) Neurogastroenterol. Motil. 19, 245–252 [DOI] [PubMed] [Google Scholar]
- 29. Cheng T. H., Cohen S. N. (2007) Mol. Cell. Biol. 27, 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Courtois S., Verhaegh G., North S., Luciani M. G., Lassus P., Hibner U., Oren M., Hainaut P. (2002) Oncogene 21, 6722–6728 [DOI] [PubMed] [Google Scholar]
- 31. Yudt M. R., Cidlowski J. A. (2002) Mol. Endocrinol. 16, 1719–1726 [DOI] [PubMed] [Google Scholar]
- 32. Ward A. J., Cooper T. A. (2010) J. Pathol. 220, 152–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee V. M., Goedert M., Trojanowski J. Q. (2001) Annu. Rev. Neurosci. 24, 1121–1159 [DOI] [PubMed] [Google Scholar]
- 34. Liu F., Gong C. X. (2008) Mol. Neurodegener. 3, 8–18 [DOI] [PMC free article] [PubMed] [Google Scholar]