Abstract
The methionine S-sulfoxide reductase MsrA catalyzes the reduction of methionine sulfoxide, a ubiquitous reaction depending on the thioredoxin system. To investigate interactions between MsrA and thioredoxin (Trx), we determined the crystal structures of yeast MsrA/Mxr1 in their reduced, oxidized, and Trx2-complexed forms, at 2.03, 1.90, and 2.70 Å, respectively. Comparative structure analysis revealed significant conformational changes of the three loops, which form a plastic “cushion” to harbor the electron donor Trx2. The flexible C-terminal loop enabled Mxr1 to access the methionine sulfoxide on various protein substrates. Moreover, the plasticity of the Trx binding site on Mxr1 provides structural insights into the recognition of diverse substrates by a universal catalytic motif of Trx.
Keywords: Crystal Structure, Cysteine-mediated Cross-linking, Methionine, Oxidation Reduction, Protein-protein Interactions, Reductase, Thiol, Methionine Sulfoxide Reductase
Introduction
Thioredoxins (Trxs)2 are ubiquitous small thiol-disulfide exchange proteins that are involved in many important cellular processes such as reduction of methionine sulfoxide, ribonucleotide, and peroxide (1). These proteins have a highly conserved active site of CXXC motif. During the reaction, the first Cys attacks the intramolecular disulfide bond in the substrate protein, accompanying w ith the formation of an intermolecular disulfide intermediate. This mixed disulfide is subsequently attacked by the second Cys, resulting in release of the reduced substrate protein and the oxidized Trx.
Methionine is one of the most sensitive amino acid residues subject to oxidation. It can be readily oxidized to methionine sulfoxide (Met-SO) as a mixture of two enantiomers at the sulfoxide moiety (Met-S-SO and Met-R-SO), by various reactive oxygen or nitrogen species (2). Oxidation of the methionines on protein surfaces would cause some lethal effects to the cells and accelerate the aging process. However, both in vitro (3) and in vivo (4), a group of enzymes called methionine sulfoxide reductases (Msr, EC 1.8.4.11) can use Trx as the electron donor to regenerate the oxidized proteins, by reducing Met-SO to methionine (5). Recently, the glutathione/glutaredoxin system has been discovered that it also could act as the electron donor for Met-SO reduction (6, 7). This protective mechanism has been shown to play a significant role in elongating the lifespan of yeast, insects, and mammals (8–10).
To date, three Msr families have been reported. MsrA and MsrB are classic Msr enzymes that regenerate the proteinous Met-S-SO and Met-R-SO, respectively. They also could regenerate the corresponding free Met-SO (11). A series of human disease-related proteins have been identified as substrates of MsrA and MsrB, such as calmodulin, HIV-2 protease, and α-1-proteinase inhibitor (12–14). However, fRMsr is a recently discovered Msr, which is exclusively responsible for the reduction of free Met-R-SO, but not the proteinous one (15). Despite the fact that the three families have distinct differences in origin, structure, substrate specificity, and species distribution, they basically share a similar catalytic mechanism. The mechanism involves the oxidation of the catalytic cysteine to a sulfenic acid intermediate, followed by the formation of an intramolecular disulfide bond, and the final regeneration process driven by Trx or other reductants (6, 7, 15–18).
Structures of MsrA from seven different species are currently reported. The core structures of these MsrA enzymes are very similar and could be well superimposed (19–25). This is regardless of the method of structure determination, either crystallography or NMR spectroscopy. The major differences are involved in the loops around the active site, especially the C-terminal loop, which is supported by the electrostatic analysis and distance measurements between the two catalytic cysteines. It is also suggested that these conformational changes would facilitate the formation of an intramolecular disulfide bond and the exposure of a hydrophobic patch for Trx interaction (22–24).
The yeast Saccharomyces cerevisiae encodes three Msr enzymes (Mxr1/MsrA, Mxr2/MsrB, and Ykg9/fRMsr). Although the mechanism of the Msr enzymes has been studied extensively, their interactions with diverse substrate proteins and a universal Trx remain unknown. Therefore, we have systematically characterized the yeast Msr-Trx complexes by determining the crystal structures of Mxr1 in its reduced form at 2.04 Å, in a dimeric oxidized form at 1.90 Å, and in a Trx2-complexed form at 2.70 Å. Superposition of these structures have revealed three highly flexible loops. These loops undergo drastic conformational changes and may be responsible for the enzyme's substrate diversity. Additionally, interface analysis of Mxr1-Trx2 along with data from previously reported Trx-protein complexes, we have found a new mode for Trx-involved complexes, which undergo drastic conformational changes. These findings would be helpful for the prediction of potential interfaces on the Trx substrate proteins.
EXPERIMENTAL PROCEDURES
Construction, Expression, and Purification of Mxr1 and Mutants
The MXR1 gene was amplified by PCR from genomic DNA of Saccharomyces cerevisiae S288C. The PCR product was inserted into a pET29a-derived vector (Novagen) and transformed into Escherichia coli strain BL21 (DE3) (Novagen). The Mxr1-expressing cells were grown to an A600 nm of 0.6–0.8 at 37 °C using 2× YT culture medium. Expression was started by adding isopropyl β-d-1-thiogalactopyranoside, and the cells continued growing for another 4 h at 37 °C before harvesting. After 5 min of centrifugation at 4,000 × g and resuspension in lysis buffer (20 mm Tris-HCl, 200 mm NaCl, pH 8.5), the cells were subjected to sonication. The supernatant was collected by 20 min centrifugation at 12,000 × g and loaded onto a HiTrap nickel-chelating column (GE Healthcare) equilibrated with the binding buffer (20 mm Tris-HCl, 200 mm NaCl, pH 8.5). The target proteins were eluted with 300 mm imidazole and further loaded onto a HiLoad 16/60 Superdex 75 column (GE Healthcare) equilibrated with 20 mm sodium citrate, pH 6.0, 50 mm NaCl with (for crystallization) or without (for checking the intermolecular disulfide bond) 10 mm 2-mercaptoethanol. The target fractions were collected and concentrated to 25 mg/ml for crystallization. Protein purity was assessed by SDS-PAGE and stored at −80 °C.
SDS-PAGE Analysis of Complexes between Trx2C34S and Mxr1 Mutant
The complex linked with a disulfide was prepared according to the previous report (26). Trx2C34S, Mxr1C25S/C44S/C68S (designated as Mxr1C23C176), and Mxr1C44S/C68S/C176S (designated as Mxr1C23C25) were purified and reduced by DTT separately before desalting. Then, Trx2C34S was incubated with 20-fold molar excess of 5,5′-dithiobis-(2-nitrobenzoic acid) at 25 °C to form a mixed disulfide between the active-site cysteine Cys31 of Trx2 and thionitrobenzoic acid. After desalting, five samples were prepared (A, 0.008 mm Trx2C34S; B, 0.016 mm Mxr1C23C176; C, 0.016 mm Mxr1C23C25; D, 0.008 mm Trx2C34S + 0.016 mm Mxr1C23C176; and E, 0.008 mm Trx2C34S + 0.016 mm Mxr1C23C25) and incubated at 25 °C for 30 min. Each sample was divided into two parts, with or without 5 mm DTT, and subjected to SDS-PAGE to test the quantity of the complex.
Preparation of Mxr1-Trx2 Complex
This is a widely used method for preparing Trx-substrate protein complex (27–32). Trx2C34S (with a hexahistidine tag) and Mxr1C23S/C25S/C44S/C68S (without a hexahistidine tag, designated as Mxr1C176) were purified separately. After removing surplus DTT, Trx2C34S was incubated with 20-fold molar excess of 5,5′-dithiobis-(2-nitrobenzoic acid) at 25 °C to form a mixed disulfide between the active site cysteine Cys31 of Trx2 and thionitrobenzoic acid. The Trx2-thionitrobenzoic acid intermediate was mixed with excess Mxr1C176 (∼2-fold of molarity) to form the Mxr1-Trx2 complex. The mixed sample was further purified by HiTrap nickel-chelating column (with the hexahistidine tag of Trx2C34S) and HiLoad 16/60 Superdex 75 column (GE Healthcare) to remove the excess Mxr1C176. The purity of the cross-linked protein was verified by SDS-PAGE.
Crystallization, Data Collection, and Processing
Crystals of the reduced, oxidized, and Trx2-complexed Mxr1 were grown by hanging-drop vapor diffusion at 16 °C, with the initial condition of mixing 1 μl of protein sample at 25 mg/ml with an equal volume of the reservoir solution (for the reduced form, 0.2 m ammonium acetate, 0.1 m sodium acetate, pH 4.6, 30% polyethylene glycol 4,000, 10 mm dithiothreitol; for the oxidized form, 0.5 m ammonium sulfate, 0.1 m sodium citrate, pH 5.6, 1.0 m lithium sulfate; for the Trx-complexed form, 0.2 m trimethylamine N-oxide, 0.1 m Tris-HCl, pH 8.5, 20% polyethylene glycol 2,000 monomethyl ether) The crystals of the reduced and Trx2-complexed forms appeared in 3 days and reached the maximum size in 1 week, whereas the oxidized form appeared in 4 months. These crystals were transferred to cryoprotectant, flash-frozen in liquid nitrogen, and subjected to data collection. The datasets of the oxidized and Trx2-complexed form were collected at a radiation wavelength of 0.9795 Å at the Shanghai Synchrotron Radiation Facility (Shanghai Institute of Applied Physics, Chinese Academy of Sciences) using the beamline BL17U at 100 K with a MX-225 CCD (Marresearch) and then processed with HKL2000. The data of the reduced form was collected with a Rigaku MM007 x-ray generator (1.5418 Å) with an MAR345 image plate detector (Marresearch) and processed with iMosflm.
Structure Determination and Refinement
All structures were determined by molecular replacement using the central domain (MsrA) of Neisseria meningitidis PilB (PDB code 3BQE) (23) and the S. cerevisiae cytoplasmic thioredoxin Trx2 (PDB code 2FA4) (33) as search models with MOLREP (34) in CCP4 (35). Refinement was carried out using the maximum likelihood method implemented in REFMAC (36) and the interactive rebuilding process in Coot (37). The overall assessment of model quality was performed using MolProbity (38). The crystallographic parameters of the three structures are listed in Table 1. All structure figures were prepared with PyMOL (39).
TABLE 1.
Data collection and structure refinement
| Mxr1re | Mxr1ox | Mxr1-Trx2 | |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 1.5418 | 0.9795 | 0.9795 |
| Space group | P212121 | P3121 | I23 |
| Unit cell | a = 64.31, b = 70.14, c = 74.03 Å; α = β = γ = 90° | a = b = 111.89, c = 108.12 Å; α = β = 90, γ = 120° | a = b = c = 122.27 Å; α = β = γ = 90° |
| Molecules per asymmetric unit | 2 | 3 | 1 |
| Resolution range (Å)a | 29.17–2.03 (2.14–2.03) | 50.00–1.90 (1.97–1.90) | 50.00–2.70 (2.80–2.70) |
| Unique reflections | 21,889 (2,861) | 59,060 (5,987) | 8,320 (834) |
| Completeness (%) | 98.4 (89.6) | 95.6 (97.8) | 98.0 (98.5) |
| 〈I/σ(I)〉 | 15.7 (5.2) | 11.3 (4.7) | 10.2 (2.2) |
| Rmergeb (%) | 8.3 (33.3) | 10.6 (32.6) | 7.6 (36.8) |
| Average redundancy | 5.6 | 5.1 | 2.4 |
| Refinement statistics | |||
| Resolution range (Å) | 29.17–2.04 (2.09–2.04) | 50.00–1.90 (1.95–1.90) | 50.00–2.70 (2.77–2.70) |
| R-factorc/R-freed (%) | 20.7/24.7 (26.5/36.5) | 23.6/26.4 (26.5/29.1) | 23.4/27.9 (34.2/32.3) |
| No. of protein atoms | 2,942 | 4,187 | 1,924 |
| No. of water atoms | 342 | 234 | 15 |
| r.m.s.d.e bond lengths (Å) | 0.006 | 0.011 | 0.010 |
| r.m.s.d. bond angles | 0.8° | 1.2° | 1.0° |
| Mean B factors (Å2) | 16.0 | 25.5 | 30.7 |
| Ramachandran plotf | |||
| Most favored (%) | 98.3 | 97.2 | 97.4 |
| Additionally allowed (%) | 1.7 | 2.8 | 2.6 |
| PDB code | 3PIL | 3PIM | 3PIN |
a The values in parentheses refer to statistics in the highest bin.
b Rmerge = ΣhklΣi|Ii(hkl) − 〈Ihkl〉|/ΣhklΣiIi(hkl), where Ii(hkl) is the intensity of an observation and 〈I(hkl)〉 is the mean value for its unique reflection; summations are over all reflections.
c R-factor = Σh|Fo(h) − Fc(h)|/ΣhFo(h), where Fo and Fc are the observed and calculated structure-factor amplitudes, respectively.
d R-free was calculated with 5% of the data excluded from the refinement.
e r.m.s.d. from ideal values. r.m.s.d., root mean square deviation.
f Categories were defined by MolProbity.
RESULTS
Favored Disulfide Bond between Mxr1 and Trx2
Mxr1 has five cysteines (Cys23, Cys25, Cys44, Cys68, and Cys176). The multiple-sequence alignment (supplemental Fig. S1) and the previous reports of Mxr1 homologs (21, 23) indicated that Cys25 and Cys176 are the catalytic cysteines. Although the correspondents of Cys176 in the homologs are not strictly aligned in the multiple sequences, structural superposition revealed their locations at the active site architectures are almost the same. In contrast, Cys68 is not highly conserved and is missing in some lower organisms. Moreover, previous report on bovine MsrA has shown that mutation of the cysteine corresponding to Mxr1-Cys68 did not disturb the Met-SO reduction (40). The left two cysteines, solvent-secluded Cys23 and solvent-exposed Cys44, are totally nonconserved and would not be involved in the reduction.
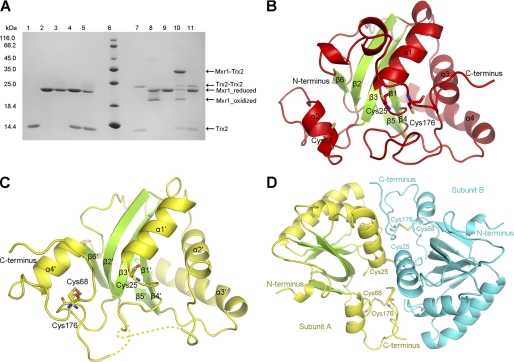
As catalytic cysteines, Cys25 or Cys176 would form an intramolecular disulfide bond after Met-SO reduction (40). Upon regeneration by Trx, this intramolecular disulfide bond would switch to a transient intermolecular one linking Mxr1 and Trx. However, which cysteine of Mxr1 (Cys25 or Cys176) is favored by Trx is unknown. To assign this cysteine, we constructed two mutants, Mxr1C23C176 and Mxr1C23C25, after mutating the other three cysteines except for Cys23 and Cys176 or Cys23 and Cys25. The reaction efficiency of these two mutants toward Trx2C34S was semiquantatively determined using SDS-PAGE. The complex formed between Trx2C34S and Mxr1C23C176 is clearly observed (lane 10 in Fig. 1A), whereas that formed with Mxr1C23C25 is not apparent (lane 11 in Fig. 1A). This indicated that Cys176 of Mxr1 is the favored cysteine in the formation of the transient disulfide intermediate with Trx2. Thus, the disulfide-linked complex between Cys176 of Mxr1 and Cys31 of Trx2 was prepared and applied to crystallization.
FIGURE 1.
A, electrophoresis of the complexes between Trx2C34S and Mxr1 mutants. The Coomassie-stained gel shows the formation of intermolecular disulfide bonds between Trx2C34S and Mxr1 mutants after incubation under non-reducing conditions (see “Experimental Procedures”). The addition of DTT could reduce the disulfide-linked complex (Mxr1-Trx2 and Trx2-Trx2) or the oxidized Mxr1. Lanes 1–5, Trx2C34S, Mxr1C23C176, Mxr1C23C25, Trx2C34S+Mxr1C23C176, and Trx2C34S+Mxr1C23C25 with 5 mm DTT; lane 6, protein marker; lanes 7–11, samples corresponding to lanes 1–5, respectively, without DTT. B, overall structure of the reduced Mxr1. Cys25, Cys68, and Cys176 are shown as sticks. Helices are colored red, and β-strands are green. C, overall structure of Mxr1 in the unusual oxidized form. Cys25, Cys68, and Cys176 are shown as sticks. Helices are colored yellow, and β-strands are green. D, disulfide-linked Mxr1 dimer in the unusual oxidized form. The two Cys25 residues form an intermolecular disulfide bond, whereas Cys68 and Cys176 form an intramolecular disulfide bond.
Structure of Reduced Mxr1
Similar to the previously reported MsrAs, the overall structure of the reduced Mxr1 (designated as Mxr1re) adopts an α/β-roll conformation (Fig. 1B and supplemental Fig. S2A). The root mean square deviation between Mxr1 and N. meningitidis PilB is only 0.81 Å over 132 Cα atoms. The core of the protein consists of a six-stranded β-sheet (β1–β6) flanked by three α-helices (α1, α2, and α4). Its C-terminal end (Ala159–Glu183) is observed as a surface-attached long loop and is mainly stabilized by hydrophobic interactions with the core domain. It runs in approximately the same direction as those of reported homologs (19–25). Several highly conserved residues (Cys25, Phe26, Trp27, Tyr64, Glu76, Tyr116, and Cys176) make up the active site, which could be superimposed to those in the homologs, except for Cys176 at the C-terminal loop. Counterparts of Mxr1-Cys176 in the homologs locate at the same area of the active site architecture, whereas the conformations of the sulfhydryl group vary mildly. In Mrx1re, the active site is occupied by an acetate molecule, of which the carboxyl oxygen is stabilized by three hydrogen bonds to Tyr64, Glu76, and Tyr116, whereas the methyl group fixed by hydrophobic interactions with Phe26 and Trp27 (supplemental Fig. S2B). The binding pattern of this acetate molecule mimics that of the sulfoxide moiety of Met-SO in previous structures (23).
An Unusual Oxidized Mxr1
According to the previous reports of oxidized MsrAs, an intramolecular disulfide bond forms between the two catalytic cysteines. This is accompanied by mild conformational changes of the loop flanking the C-terminal cysteine and the loop between α3 and β4 (α3–β4 loop) of Mxr1re (23, 24). However, here we have determined a structure of the oxidized Mxr1 (designated as Mxr1ox) with such drastic conformational changes that the C-terminal loop (Ala159–Glu183) completely leaves its place in the Mxr1re, together with movement of the α2′–β4′ loop (Thr103–Gly113, exactly corresponding to the α3–β4 loop of Mxr1re) and the β2′–β3′ segment (Asn53–Asp73).
The crystal structure of Mxr1ox contains three molecules, which are arranged as an arc in the asymmetric unit. The core of each molecules is also composed of a six-stranded β-sheet (β1′–β6′) flanked by three α-helices (α1′, α2′, and α3′) (Fig. 1C and supplemental Fig. S2C). Each molecule has an intramolecular disulfide bond between Cys176 and the function-unknown Cys68, which is conserved in higher organisms but does not exist in some of the lower organisms (supplemental Fig. S1). Formation of the intramolecular disulfide bond leads to shifting of the β2′–β3′ segment, then unwinding of the helix α2 in Mxr1re. This is followed by redirection of the C-terminal loop with formation of a new α-helix (α4′) by the C-terminal loop. Several residues in the loop regions Ser54–Val62 and Gln108–Lys112 are not visible in the final 2Fo − Fc electron density map, presumably due to their high flexibility, but not proteolytic cleavage as evidenced by SDS-PAGE (supplemental Fig. S3). Upon oxidation, the hydrophobic substrate binding pocket, which is close to the active site, is reorganized partially due to the shifts of Tyr64 and Trp27, whereas Cys25, Phe26, Glu76, and Tyr116 could be superimposed to those in Mxr1re.
Superposition shows these three molecules of Mxr1ox could be well fitted, giving a root mean square deviation of 0.541, 0.587, and 0.493 Å (over 170, 160, and 163 Cα atoms for A/B, A/C, and B/C, respectively). The molecules A and B are linked by a disulfide bond (Fig. 1D) via the two Cys25 residues and bury an interface of 2900 Å2. In contrast, molecule C does not form disulfide bonds with neighboring molecules. The 600 Å2 interface between molecules C and B is due to crystal packing and is too small to sustain dimerization in solution. Up to date, reports about MsrA are focused on its monomeric form. Here, we reported for the first time its dimeric form, despite the physiological role of this dimer was not clear. Both the intra- and intermolecular disulfides observed in Mxr1ox do not represent any intermediates formed during the catalytic cycle of the enzyme. It was probably resulted from a sort of random oxidation that occurred during crystallization. However, the total interface of 2900 Å2 indicated subunits A and B form a tight dimer. The dimeric form of Mxr1 also exists in the reduced buffer, as shown by gel filtration (supplemental Fig. S4). Moreover, the fact that crystals of Mxr1ox did not occur under the crystallization conditions of Mxr1re and supports vice versa that the two oxireductive states had significant structural differences in solution. This indirectly supports that the structural changes are not simply a consequence of crystal packing, but of oxireductive states.
Complex Structure of Mxr1 and Trx2
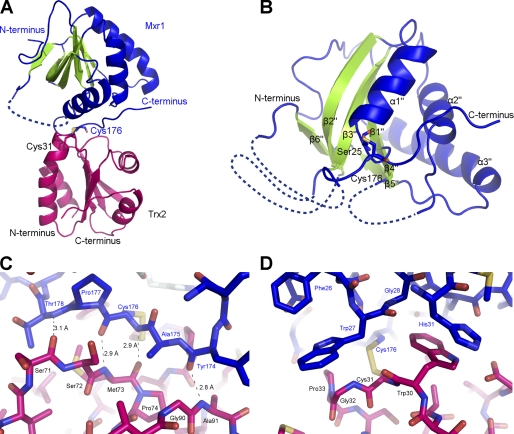
It has been demonstrated that MsrA can be regenerated by Trx via disulfide bond exchange, but the structural insights into this regeneration process remains unknown. Based on the previously reported MsrA regeneration mechanism and our experiments (Fig. 1A), we prepared a Mxr1-Trx2 complex covalently linked by a mixed disulfide bond between Cys31 of Trx2 and Cys176 of Mxr1, with mutants Trx2C34S and Mxr1C176. The crystal structure was determined at 2.7 Å (the two molecules in the complex are designated as Trx2co and Mxr1co, respectively). The 2Fo − Fc electron density map clearly displays a disulfide bond linked complex in the asymmetric unit (Fig. 2A). The structural model contains an intact Trx2 (residues 1–104) and an Mxr1 of discontinuous segments (Ser3–Glu52, Asp73–Thr104, Arg117–Glu161, and Gln172–Glu183). The missing residues of Mxr1 are located in the loop regions, which are probably too flexible to be traced in the electron density map (Fig. 2B and supplemental Fig. S2D).
FIGURE 2.
A, overall structure of the Mxr1-Trx2 complex (Mxr1, blue and green; Trx2, magenta). Cys176 of Mxr1 and Cys31 of Trx2 are labeled as sticks. Helices are colored blue, and β-strands are colored green. B, overall structure of Mxr1 in the Trx2-complexed form. Ser25 (mutation from Cys25) and Cys176 are shown as sticks. Helices are colored blue, and β-strands are green. C, four main-chain hydrogen bonds. D, hydrophobic interactions between the C-terminal loop of Mxr1 (blue) and Trx2 (magenta).
The overall structure of Trx2co in the complex is very similar to that of free Trx2 (PDB code 2FA4) (33), except for several subtle conformational changes. The N terminus of helix α2 (Cys31–Glu46) comprises the CXXC active site motif, which slightly moves toward the interface of the complex upon binding to Mxr1. This slight shift makes Cys31 approach to the complex interface, which may facilitate the formation of the intermolecular disulfide bond.
Mxr1co and Mxr1re share a very similar core structure. The major differences are involved in the C-terminal loop (Ala159–Glu183), the α2"–β4" loop (with Thr103–Gly113 exactly corresponding to the α3–β4 loop of Mxr1re), and the β2"–β3" segment (Asn53–Asp73). The interaction with Trx2 causes conformational adjustments of the C-terminal loop of Mxr1, which subsequently push the β2"–β3" segment and α2"–β4" loop to the side. The displacements of the three segments from their original positions in the free forms probably lead to the untraceable electron density. In summary, all these structural adjustments form an intermediate conformation of Mxr1co. In particular, segment Ala158–His179 on the C-terminal loop, is involved directly in the interaction with Trx2 and moves a remarkable distance to adopt an intermediate conformation compared with that in Mxr1re. This movement is the key to the complex formation, for it not only results in the exposure of a formerly buried hydrophobic surface but also considerably adjusts the conformation of the C-terminal loop, both of which are responsible for binding Trx2.
The interface between the two proteins buries a total solvent-accessible area of 1000 Å2 (480 Å2 for Trx2 and 520 Å2 for Mxr1). This is a typical interface for redox proteins due to their short-lived interactions (41). In addition to the active site disulfide bond, another two types of interactions are involved. The disulfide bond is surrounded by four intermolecular hydrogen bonds on one side, whereas several hydrophobic residues distribute on the other side. The segment Tyr174–Thr178 at the C-terminal loop of Mxr1co goes through a deep groove between β2–α3–β3 of Trx2 and helices α1"/α2" of Mxr1co. It interacts with Trx2 (α3–β3 and β4–α4 loop) via four hydrogen bonds in an anti-parallel pattern (Fig. 2C). This is similar to that of the previous Trx-complexed proteins (28–32, 42). The hydrophobic interactions are mainly mediated by a solvent-exposed hydrophobic patch (Trp30, Cys31, Gly32, and Pro33) around the active site of Trx2 (Fig. 2D). In contrast, the hydrophobic patch (Trp27, Gly28, His31, and His179) of Mxr1 is uncovered due to the shift of the C-terminal loop. In addition, the indole ring of Mxr1-Trp27, the function of which is reported to bind the methyl moiety of sulfoxide, rotates nearly 120° to accommodate Trx2. This rotation, together with the shift of the C-terminal loop, completely disintegrates the Met-SO binding cavity.
DISCUSSION
Structural Plasticity of MsrA and MsrB
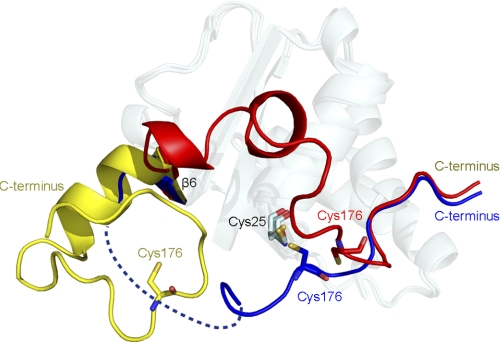
Oxidation of the methionine would lead to various deleterious effects, ranging from decline of the enzymatic activity to inactivation of the proteins. The oxidation process is so widely distributed that any surface-exposed methionine residues could be modified readily. Despite the surfaces around the modified methionines possessing various properties, such as shape, polarity, and rigidity, MsrA and MsrB are able to reverse this oxidative process and restore the primal state of the protein. For example, about six to eight methionines are distributed on the surface of calmodulin from different species, but all of these could be reduced by MsrA and MsrB (43). How could MsrA and MsrB recognize so many diverse surfaces with just the same active site? The three flexible loops around the active site in our structures could provide a possible answer. These three loops all display considerable movements compared with Mxr1re (Figs. 1, B and C, and 2B), especially for the C-terminal loop (Fig. 3). Any movement would cause a change in the property around the active site. Upon substrate protein binding, conformational changes would take place at the three loops of the reduced MsrA to better fit the complementary surface of the substrates. It may help MsrA to reduce some proteinous Met-SO groups that are not easy to access. Thus these three loops constitute a soft cushion around the active site, which could fit diverse protein substrates. However, despite changes in the cushion, part of the catalytic residues Cys25, Phe26, and Glu76 in the structures of Mxr1ox and Mxr1co are still in their original positions, where they probably maintain the activity of the enzyme at a lower level. Moreover, the cushion also plays crucial role in Trx recognition, for it changes its shape and uncovers a hydrophobic patch to fit the active- site motif of Trx2. Based on these speculations, we can deduce that the loops around the active site of MsrB probably could function in a similar way. They also form a cushion around the active site of MsrB and have been reported to display a high level of flexibility (44), which would facilitate the interactions between MsrB and various protein substrates or Trx. However, fRMsr has a covered active site and two relatively rigid loops. The stereohindrance prevents fRMsr from restoring proteinous Met-SO; thus, only free Met-SO is regenerated (15). However, further evidence is required to validate our deduction.
FIGURE 3.
Conformational changes of the C-terminal loop in the three structures (red, reduced form; yellow, oxidized form; blue, Trx2-complexed form). The core structures are treated with the semitransparent effect.
Structural Plasticity of Trx Binding Sites Enables Trx to Bind Diverse Protein Substrates
Previous studies have found that upon oxidation, the β2–β3 segment can undergo a drastic structural disorder and expose a hydrophobic area, which is ready for the interaction with Trx at the following regeneration process (23, 24). The current structure of Mxr1-Trx2 clearly demonstrates this hypothesis, for the movement of the β2"–β3" segment would make room for the incoming Trx2. Superposition of oxidized E. coli MsrA (PDB code 2IEM), reduced E. coli MsrA (PDB code 2GT3), Mxr1re and Mxr1co shows that if the β2"–β3" segment does not deviate from its original position, it would have steric clash with the α3–β3 loop of Trx2. Thus, this conformational change appears indispensable for the forthcoming interaction.
However, the hydrophobic patch presented here is not involved in the hydrophobic interaction with Trx2 as suspected previously. The actual hydrophobic interface is buried by the C-terminal loop, which would be moved away by a Trx2-aroused induced fit process. This process has not been observed in the six previously published Trx or Trx-like complexed substrate protein structures (PDB codes 1Z5Y, 2IPA, 2IWT, 2K9F, 2O8V, and 2PUK), which involve an electron transfer process. Each of the interfaces in these complex structures are composed of several classic hydrogen bonds arranged in an anti-parallel or parallel pattern, in addition to hydrophobic patches that are exposed to the solvent. Exposure of these hydrophobic patches would not need any induced fit process, for they are all naturally exposed to the solvent and are ready for Trx binding.
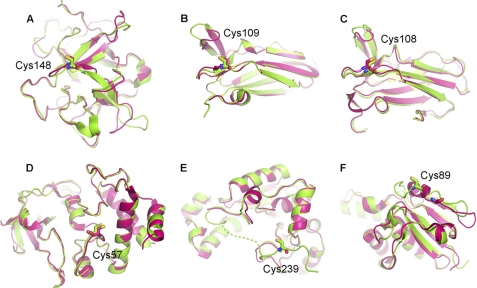
For example, in the Trx-complexed barley α-amylase/subtilisin inhibitor barley alpha-amylase/subtilisin inhibitor, thiol:disulfide interchange protein DsbD and ferredoxin-thioredoxin reductase (PDB codes 1Z5Y/1JPE, 2IWT/1AVA, 2K9F/2K0R, and 2PUK/2PVD) (27, 28, 30, 32, 42, 45, 46), no obvious conformational changes take place around the interfaces (Fig. 4, A–D). These complexes are formed simply by facing one surface against the other. The Trx-complexed 3′-phosphoadenosine-5′-phosphosulfate reductase (PDB code 2O8V/1SUR) (31, 47) also does not undergo apparent conformational changes, whereas Trx only stabilizes the highly flexible loop of 3′-phosphoadenosine-5′-phosphosulfate reductase (Fig. 4E). Although the Trx-complexed arsenate reductase (PDB code 2IPA/1Z2D) (29) displays a helix to loop conformational transition (Fig. 4F), the transition is related only to formation of classic intermolecular hydrogen bonds, not exposure of the hydrophobic patches.
FIGURE 4.
Superposition of the free (magenta) and Trx-complexed (green) substrate proteins. A, 1Z5Y/1JPE (DsbD from Escherichia coli). B, 2IWT/1AVA (barley alpha-amylase/subtilisin inhibitor from Hordeum vulgare). C, 2K9F/2K0R (DsbD from Neisseria meningitidis). D, 2PUK/2PVD (ferredoxin-thioredoxin reductase from Synechocystis sp.). E, 2O8V/1SUR (3′-Phosphoadenosine-5′-phosphosulfate reductase from Escherichia coli). F, 2IPA/1Z2D (ArsC, arsenate reductase from Bacillus subtilis).
Based on these analyses, the above six Trx-complexed structures could be grouped into three types. The apparent differences in the Mxr1-Trx2 complex suggest a new type of Trx-involved protein-protein interactions. The conformational changes of Mxr1, which uncover a buried hydrophobic patch, are significant, due to minimization of the stereohindrance between Mxr1 and Trx2 and also rearrangement at its surface to offer more polar or non-polar interactions. This new type of Trx-involved interaction explains the difficulties for predicting new substrate proteins of Trx or the precise Trx binding site with rigid-body docking. More efforts on soft docking or molecular simulation are needed to make better predictions.
Acknowledgments
The diffraction data sets were collected at the Shanghai Synchrotron Radiation Facility. We greatly appreciate the generous assistance of the staff at SSRF.
This work was supported by the National Natural Science Foundation of China (Program 30870490) and the Ministry of Science and Technology of China (Project 2006CB910202).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
The atomic coordinates and structure factors (codes 3PIL, 3PIM, and 3PIN) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Trx
- thioredoxin
- PDB
- Protein Data Bank.
REFERENCES
- 1. Arnér E. S., Holmgren A. (2000) Eur. J. Biochem. 267, 6102–6109 [DOI] [PubMed] [Google Scholar]
- 2. Weissbach H., Etienne F., Hoshi T., Heinemann S. H., Lowther W. T., Matthews B., St. John G., Nathan C., Brot N. (2002) Arch. Biochem. Biophys. 397, 172–178 [DOI] [PubMed] [Google Scholar]
- 3. Moskovitz J., Weissbach H., Brot N. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2095–2099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moskovitz J., Berlett B. S., Poston J. M., Stadtman E. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kallis G. B., Holmgren A. (1980) J. Biol. Chem. 255, 10261–10265 [PubMed] [Google Scholar]
- 6. Tarrago L., Laugier E., Zaffagnini M., Marchand C. H., Le Maréchal P., Lemaire S. D., Rey P. (2010) J. Biol. Chem. 285, 14964–14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tarrago L., Laugier E., Zaffagnini M., Marchand C., Le Maréchal P., Rouhier N., Lemaire S. D., Rey P. (2009) J. Biol. Chem. 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koc A., Gasch A. P., Rutherford J. C., Kim H. Y., Gladyshev V. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruan H., Tang X. D., Chen M. L., Joiner M. L., Sun G., Brot N., Weissbach H., Heinemann S. H., Iverson L., Wu C. F., Hoshi T. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2748–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moskovitz J., Bar-Noy S., Williams W. M., Requena J., Berlett B. S., Stadtman E. R. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang X. H., Weissbach H. (2008) Biol. Rev. Camb. Philos. Soc. 83, 249–257 [DOI] [PubMed] [Google Scholar]
- 12. Davis D. A., Newcomb F. M., Moskovitz J., Wingfield P. T., Stahl S. J., Kaufman J., Fales H. M., Levine R. L., Yarchoan R. (2000) Biochem. J. 346, 305–311 [PMC free article] [PubMed] [Google Scholar]
- 13. Sun H., Gao J., Ferrington D. A., Biesiada H., Williams T. D., Squier T. C. (1999) Biochemistry 38, 105–112 [DOI] [PubMed] [Google Scholar]
- 14. Abrams W. R., Weinbaum G., Weissbach L., Weissbach H., Brot N. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7483–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin Z., Johnson L. C., Weissbach H., Brot N., Lively M. O., Lowther W. T. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 9597–9602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brot N., Weissbach L., Werth J., Weissbach H. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2155–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sagher D., Brunell D., Hejtmancik J. F., Kantorow M., Brot N., Weissbach H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8656–8661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boschi-Muller S., Gand A., Branlant G. (2008) Arch. Biochem. Biophys 474, 266–273 [DOI] [PubMed] [Google Scholar]
- 19. Tête-Favier F., Cobessi D., Boschi-Muller S., Azza S., Branlant G., Aubry A. (2000) Structure 8, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 20. Lowther W. T., Brot N., Weissbach H., Matthews B. W. (2000) Biochemistry 39, 13307–13312 [DOI] [PubMed] [Google Scholar]
- 21. Taylor A. B., Benglis D. M., Jr., Dhandayuthapani S., Hart P. J. (2003) J. Bacteriol. 185, 4119–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rouhier N., Kauffmann B., Tete-Favier F., Palladino P., Gans P., Branlant G., Jacquot J. P., Boschi-Muller S. (2007) J. Biol. Chem. 282, 3367–3378 [DOI] [PubMed] [Google Scholar]
- 23. Ranaivoson F. M., Antoine M., Kauffmann B., Boschi-Muller S., Aubry A., Branlant G., Favier F. (2008) J. Mol. Biol. 377, 268–280 [DOI] [PubMed] [Google Scholar]
- 24. Coudevylle N., Antoine M., Bouguet-Bonnet S., Mutzenhardt P., Boschi-Muller S., Branlant G., Cung M. T. (2007) J. Mol. Biol. 366, 193–206 [DOI] [PubMed] [Google Scholar]
- 25. Kim Y. K., Shin Y. J., Lee W. H., Kim H. Y., Hwang K. Y. (2009) Mol. Microbiol. 72, 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang P. F., Veine D. M., Ahn S. H., Williams C. H., Jr. (1996) Biochemistry 35, 4812–4819 [DOI] [PubMed] [Google Scholar]
- 27. Maeda K., Hägglund P., Finnie C., Svensson B., Henriksen A. (2006) Structure 14, 1701–1710 [DOI] [PubMed] [Google Scholar]
- 28. Stirnimann C. U., Rozhkova A., Grauschopf U., Grütter M. G., Glockshuber R., Capitani G. (2005) Structure 13, 985–993 [DOI] [PubMed] [Google Scholar]
- 29. Li Y., Hu Y., Zhang X., Xu H., Lescop E., Xia B., Jin C. (2007) J. Biol. Chem. 282, 11078–11083 [DOI] [PubMed] [Google Scholar]
- 30. Quinternet M., Tsan P., Selme-Roussel L., Jacob C., Boschi-Muller S., Branlant G., Cung M. T. (2009) Structure 17, 1024–1033 [DOI] [PubMed] [Google Scholar]
- 31. Chartron J., Shiau C., Stout C. D., Carroll K. S. (2007) Biochemistry 46, 3942–3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dai S., Friemann R., Glauser D. A., Bourquin F., Manieri W., Schürmann P., Eklund H. (2007) Nature 448, 92–96 [DOI] [PubMed] [Google Scholar]
- 33. Bao R., Chen Y., Tang Y. J., Janin J., Zhou C. Z. (2007) Proteins 66, 246–249 [DOI] [PubMed] [Google Scholar]
- 34. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 35. CCP4 (1994) Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 36. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 37. Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 38. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Delano W. L. (2010) The PyMOL Molecular Graphics System, Version 1.3r1, Schrodinger, LLC, Palo Alto, CA [Google Scholar]
- 40. Lowther W. T., Brot N., Weissbach H., Honek J. F., Matthews B. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6463–6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Janin J., Bahadur R. P., Chakrabarti P. (2008) Q. Rev. Biophys. 41, 133–180 [DOI] [PubMed] [Google Scholar]
- 42. Vallée F., Kadziola A., Bourne Y., Juy M., Rodenburg K. W., Svensson B., Haser R. (1998) Structure 6, 649–659 [DOI] [PubMed] [Google Scholar]
- 43. Grimaud R., Ezraty B., Mitchell J. K., Lafitte D., Briand C., Derrick P. J., Barras F. (2001) J. Biol. Chem. 276, 48915–48920 [DOI] [PubMed] [Google Scholar]
- 44. Ranaivoson F. M., Neiers F., Kauffmann B., Boschi-Muller S., Branlant G., Favier F. (2009) J. Mol. Biol. 394, 83–93 [DOI] [PubMed] [Google Scholar]
- 45. Haebel P. W., Goldstone D., Katzen F., Beckwith J., Metcalf P. (2002) EMBO J. 21, 4774–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quinternet M., Tsan P., Selme L., Beaufils C., Jacob C., Boschi-Muller S., Averlant-Petit M. C., Branlant G., Cung M. T. (2008) Biochemistry 47, 12710–12720 [DOI] [PubMed] [Google Scholar]
- 47. Savage H., Montoya G., Svensson C., Schwenn J. D., Sinning I. (1997) Structure 5, 895–906 [DOI] [PubMed] [Google Scholar]